Abstract
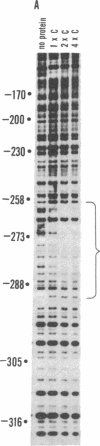
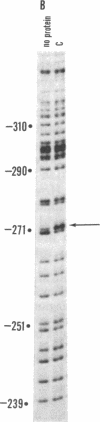
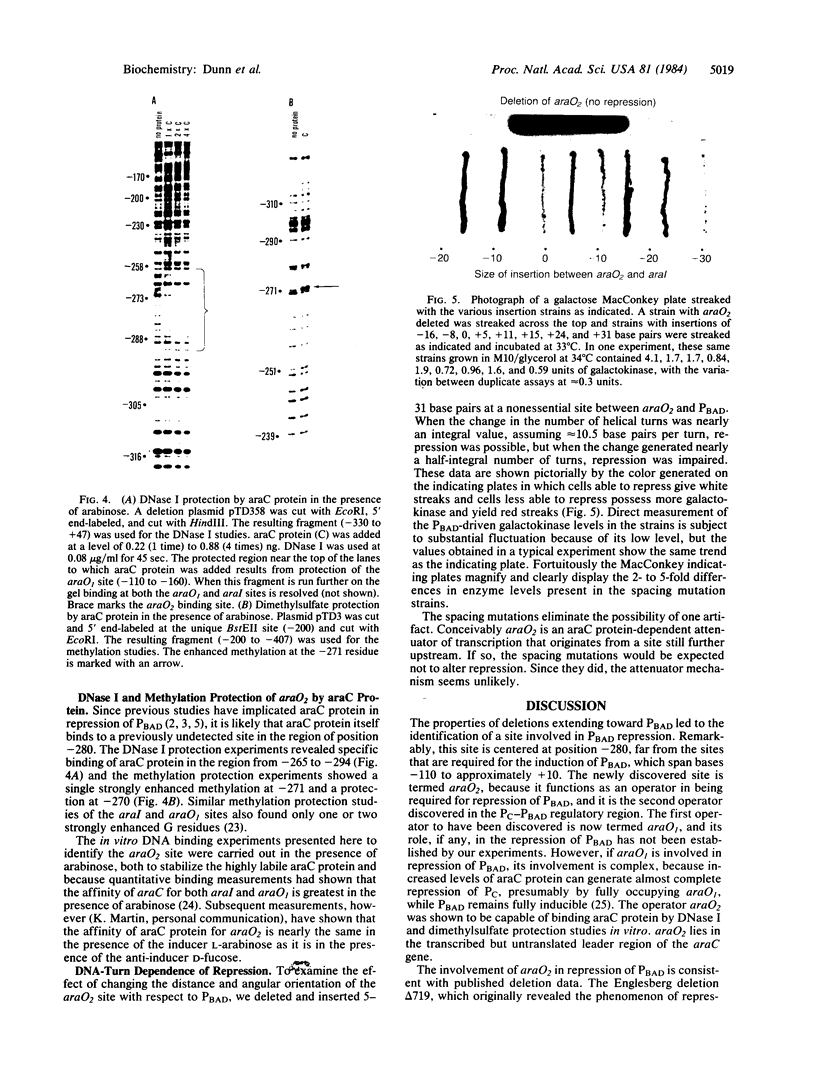
A site has been found that is required for repression of the Escherichia coli araBAD operon. This site was detected by the in vivo properties of deletion mutants. In vitro protection studies with DNase I and dimethylsulfate showed that araC protein can specifically bind in this area to nucleotides lying at position -265 to -294 with respect to the araBAD operon promoter (PBAD) transcription start point. The previously known sites of protein binding in the ara operon lie between +20 and -160. Since the properties of deletion strains show that all the sites required for araBAD induction lie between +20 and -110, the new site at -280 exerts its repressive action over an unusually large distance along the DNA. Insertions of -16, -8, 0, 5, 11, 15, 24, and 31 base pairs of DNA between the new site and PBAD were constructed. Repression was impaired in those cases in which half-integral turns of the DNA helix were introduced, but repression was nearly normal for the insertions of 0, +11, and +31 base pairs.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Englesberg E., Squires C., Meronk F., Jr The L-arabinose operon in Escherichia coli B-r: a genetic demonstration of two functional states of the product of a regulator gene. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1100–1107. doi: 10.1073/pnas.62.4.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Schleif R. Arabinose C protein: regulation of the arabinose operon in vitro. Nat New Biol. 1971 Oct 6;233(40):166–170. doi: 10.1038/newbio233166a0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Schleif R. In vivo regulation of the Escherichia coli araC promoter. J Bacteriol. 1983 Aug;155(2):593–600. doi: 10.1128/jb.155.2.593-600.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Irani M. H., Orosz L., Adhya S. A control element within a structural gene: the gal operon of Escherichia coli. Cell. 1983 Mar;32(3):783–788. doi: 10.1016/0092-8674(83)90064-8. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Kosiba B. E., Schleif R. Arabinose-inducible promoter from Escherichia coli. Its cloning from chromosomal DNA, identification as the araFG promoter and sequence. J Mol Biol. 1982 Mar 25;156(1):53–66. doi: 10.1016/0022-2836(82)90458-2. [DOI] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. L., West R. W., Heyneker H. L., Bolivar F., Boyer H. W. Characterizing wild-type and mutant promoters of the tetracycline resistance gene in pBR313. Nucleic Acids Res. 1979 Jul 25;6(10):3267–3287. doi: 10.1093/nar/6.10.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R., Lis J. T. The regulatory region of the L-arabinose operon: a physical, genetic and physiological study. J Mol Biol. 1975 Jul 5;95(3):417–431. doi: 10.1016/0022-2836(75)90200-4. [DOI] [PubMed] [Google Scholar]
- Sheppard D. E., Englesberg E. Further evidence for positive control of the L-arabinose system by gene araC. J Mol Biol. 1967 May 14;25(3):443–454. doi: 10.1016/0022-2836(67)90197-0. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Smith B. R., Schleif R. Nucleotide sequence of the L-arabinose regulatory region of Escherichia coli K12. J Biol Chem. 1978 Oct 10;253(19):6931–6933. [PubMed] [Google Scholar]
- Stoner C., Schleif R. The araE low affinity L-arabinose transport promoter. Cloning, sequence, transcription start site and DNA binding sites of regulatory proteins. J Mol Biol. 1983 Dec 25;171(4):369–381. doi: 10.1016/0022-2836(83)90035-9. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widera G., Gautier F., Lindenmaier W., Collins J. The expression of tetracycline resistance after insertion of foreign DNA fragments between the EcoRI and HindIII sites of the plasmid cloning vector pBR 322. Mol Gen Genet. 1978 Jul 25;163(3):301–305. doi: 10.1007/BF00271959. [DOI] [PubMed] [Google Scholar]
- Wilcox G., Meuris P., Bass R., Englesberg E. Regulation of the L-arabinose operon BAD in vitro. J Biol Chem. 1974 May 10;249(9):2946–2952. [PubMed] [Google Scholar]