Abstract
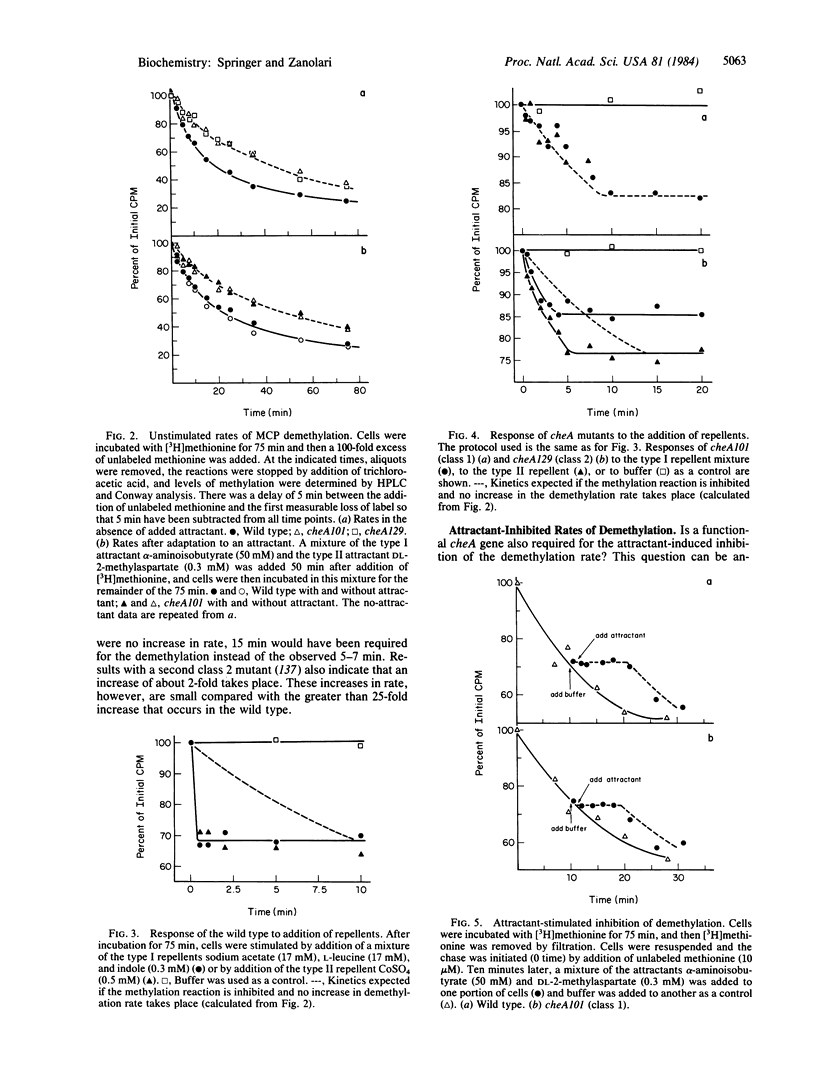
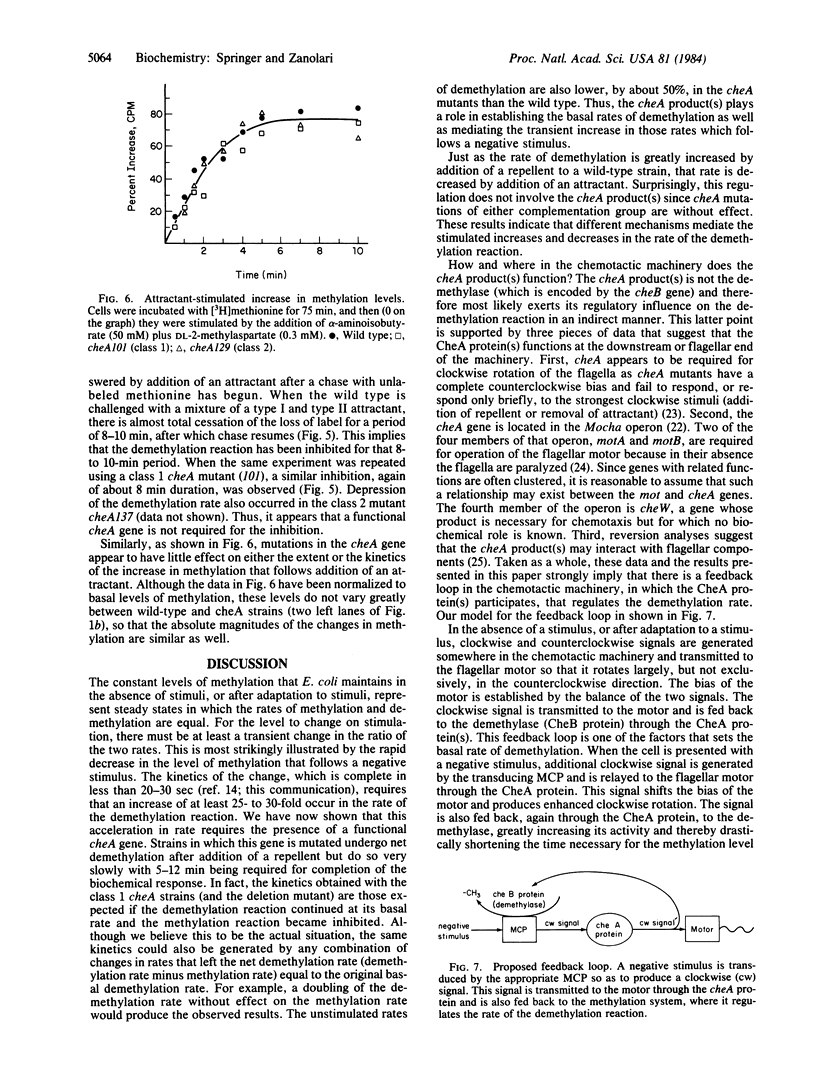
The reversible methylation of three membrane proteins plays an essential role in bacterial chemotaxis. Chemotactic stimuli bring about changes in the levels of methylation of these proteins, at least in part, by regulation of the demethylation reaction. Addition of attractants causes an increase in the methylation level and a transient, but essentially complete, inhibition in the rate of the demethylation reaction, while addition of repellents results in a decrease in level and a transient increase (of at least 25- to 30-fold) in rate. We have now found that the increase, but not the decrease, in rate requires the presence of the cheA gene product, a protein that is distinct from the demethylase. The demethylation reaction is therefore regulated by two distinct mechanisms--one, which involves the CheA protein, that mediates the increase in rate and a second, which does not involve the CheA protein, that mediates the decrease in rate. Several pieces of evidence already in the literature imply that the CheA protein functions downstream of the methylation system at the flagellar end of the chemotactic machinery. These data, in conjunction with the newer results, suggest that the CheA protein helps to regulate the demethylation reaction through a feedback mechanism.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg H. C., Anderson R. A. Bacteria swim by rotating their flagellar filaments. Nature. 1973 Oct 19;245(5425):380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Tedesco P. M. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci U S A. 1980 May;77(5):2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci U S A. 1980 May;77(5):2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Koiwai O., Kozuka M. Studies on bacterial chemotaxis. II. Effect of cheB and cheZ mutations on the methylation of methyl-accepting chemotaxis protein of Escherichia coli. J Biochem. 1979 May;85(5):1213–1223. [PubMed] [Google Scholar]
- Kondoh H., Ball C. B., Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S. H., Reader R. W., Kort E. N., Tso W. W., Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974 May 3;249(452):74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., Koshland D. E., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Behavioral genetics in bacteria. Annu Rev Genet. 1977;11:397–414. doi: 10.1146/annurev.ge.11.120177.002145. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974 May 3;249(452):73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. Identification of polypeptides necessary for chemotaxis in Escherichia coli. J Bacteriol. 1977 Jun;130(3):1317–1325. doi: 10.1128/jb.130.3.1317-1325.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Operon controlling motility and chemotoxis in E. coli. Nature. 1976 Dec 9;264(5586):577–580. doi: 10.1038/264577a0. [DOI] [PubMed] [Google Scholar]
- Smith R. A., Parkinson J. S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Zanolari B., Pierzchala P. A. Ordered methylation of the methyl-accepting chemotaxis proteins of Escherichia coli. J Biol Chem. 1982 Jun 25;257(12):6861–6866. [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews M. L., Goy M. F., Springer M. S., Adler J. Attractants and repellents control demethylation of methylated chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5544–5548. doi: 10.1073/pnas.76.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]



