Abstract
Bacteremia develops when bacteria manage to escape the host immune mechanisms or when the otherwise well-orchestrated immune response fails to control bacterial spread due to inherent or acquired immune defects that are associated with susceptibility to infection. The pathogenesis of bacteremia has some characteristic features that are influenced by the genetic signature of the host. In this review, the host defense mechanisms that help prevent bacteremia will be described and the populations who are at risk because of congenital or acquired deficiencies in such mechanisms will be defined. A special mention will be made to novel insights regarding host immune defense against the most commonly isolated organisms from patients with community-acquired bloodstream infections.
Keywords: bacteremia, bloodstream infection, host defense, single nucleotide polymorphism
Introduction
Bacteremia is defined as the presence of viable bacteria in the bloodstream and can occur in daily activities like toothbrushing and some minor medical procedures like dental work but also during infection.1 In the first case, it is a transient and clinically benign condition where the host immune mechanisms eliminate the bacteria from the blood. However, when those mechanisms fail or in the presence of anatomic lesions, turbulent cardiac blood flow and foreign material, bacteremia can lead to infection and sepsis. The incidence of bloodstream infections (BSI) either of community-acquired origin or of hospital-acquired origin has dramatically increased.2,3 The incidence rate of community-acquired bacteremia (CAB) varies according to the geographic location and it is reported to be 31.1 episodes per 100 000/year in northeast Thailand,4 92 episodes per 100 000/year in northern Denmark,5 153 episodes per 100 000/year in Olmsted County in the United States6 and 101.2 episodes per 100 000/year in Victoria, Canada.7 The etiology varies according to age, geographic location, ecologic environment, and co-morbid illnesses.4,8,9 The incidence is greater in males (especially older males), very young, and elderly patients.6,7 Infections of the respiratory tract, of the urinary tract, and intraabdominal infections are the commonest sites of origin of bacteremia.3,10 However, 10% of cases are classified as primary bacteremia of undefined origin.3 Escherichia coli, Streptococcus pneumoniae, and Staphylococcus aureus are the most frequently isolated pathogens.3,10,11 Community-acquired bloodstream infections are often associated with severe sepsis and septic shock, occurring at a rate of approximately 10.2 episodes per 1000 intensive care unit (ICU) admissions.12 In a study of 112 patients with sepsis and septic shock due to community-acquired BSI, APACHE II, and hypoalbuminemia were independent risk factors for mortality.13 Mortality from BSI ranges between 4% and 41.5% in different studies, depending on age, severity of illness, and presence of severe sepsis or septic shock.3-5,7,9-12 Despite an increase in the prevalence of BSI in ICU patients from 9 to 24.4 episodes per 1000 ICU admissions between 1993 and 2007, associated mortality decreased by almost 20% in the same time period.10 Similar trends have also been noted in other studies.4,5,14
In this review, we will describe the host defense mechanisms that help prevent bacteremia and the populations who are at risk because of congenital or acquired deficiencies in such mechanisms. A special mention will be made to novel insights regarding host immune defense against the most commonly isolated organisms from patients with CAB.
Host Immune Defense Mechanisms to Prevent Bacteremia
Innate immune mechanisms
Pathogen recognition and host response
In order for bacteria to cause bacteremia, they must evade the host immune mechanisms either in the site of infection or in the bloodstream. Innate immune cells recognize microorganisms through sensing of common microbial structures known as pathogen-associated molecular patterns (PAMPs), like lipotechoic acid, lipopeptides, lipopolysaccharide (LPS), peptidoglycan, flagellin, and nucleic acids.15 Receptors on the surface of immune and non-immune cells, the so-called pattern recognition receptors (PRRs), recognize and attach PAMPs.16 Toll-like receptors (TLRs) are an important family of PRRs and have a major role in host defense against bacteria. The transmembrane TLR2 and TLR4 are of crucial importance, since they bind the most common bacterial surface molecules like peptidoglycan, lipotechoic acid, lipopetides, and LPS.17 TLR2-deficient mice are more susceptible to S. aureus infection than wild-type mice.18 PRRs are not only found on the cell surface but also in the cytoplasm, like the nucleotide-oligomerization domain leucine-rich repeat proteins (NOD-LRRs). Activation of Nod-like receptors (NLRs) like NLRP3 through PAMPs leads to oligomerization and recruitment of adaptor proteins leading to the formation of a multiprotein complex called the “inflammasome” which contributes to the production of pro-inflammatory cytokines like IL-1β and antimicrobial peptides. NOD2-deficient mice showed impaired bacterial clearance and larger skin lesions after cutaneous S. aureus infection compared with wild-type controls.19 Other important PRRs for the initiation of the innate immune response are also the C-type lectin-receptors (CLRs) and recruiting domain helicases like the retinoic-acid-inducible-gene I (RIG-I)-helicases.20
Attachment of PRRs to their ligands activates downstream signaling pathways via intracellular adaptor proteins, like myeloid differentiation factor 88 (MyD88), that lead to the activation of transcription factors that modulate gene expression and pro-inflammatory cytokine production.21 Moreover, a transmembrane receptor of blood neutrophils and monocytes, triggering receptor expressed on myeloid cells (TREM), magnifies the TLR- and NLR-mediated inflammatory response to microbial products.22 A major pathway of inflammatory response is driven by the cellular transcription factor nuclear factor kappa B (NFκB), which migrates to the cell nucleus and forms a complex with DNA, resulting in the expression of pro-inflammatory cytokines.23 TNFα is rapidly produced by activated blood cells and has direct proinflammatory and procoagulant properties, which are further enhanced by other cytokines like IL-1, IL-2, IL-6, IL-8, and IFN-γ.24 In addition, novel molecular pathways are being identified as elementary in the antibacterial host defense. Nuclear factor-erythroid 2 p45-related factor 2 (Nrf2) is a transcription factor that induces antioxidant responses and other cytoprotective defenses, like glutathione and heme-oxygenase-1 biosynthesis, in response to inflammatory and oxidative stress. There is evidence that activation of the Nrf2 pathway in innate immune cells preserves antibacterial defenses and leads to decreased systemic bacterial burden after experimental cecal ligation and puncture (CLP).25
Host–pathogen interface
The first barrier to pathogen invasion is the skin and mucosal surfaces. Microbes most commonly enter the body through the skin, the gastrointestinal tract, and the respiratory tract.26 Antigen-presenting cells residing in the epithelium (mainly dendritic cells) capture bacterial antigens and present them to T lymphocytes. Specifically for the skin, Langerhans cells (dendritic cells that reside in the skin epithelium) bind and endocytose bacterial antigens, then they migrate to the lymph nodes were they present part of the antigens to naïve T lymphocytes which then differentiate to effector cells.27 This mechanism is lost in the event of trauma, burn, or the use of medical devices and renders the host susceptible to infection. Loss of the skin and mucosal surface barrier to bacterial invasion, with the use of medical devices like urinary catheters and intravenous catheters, contributed to the morbidity and mortality associated with E. coli bacteremia.28,29 In one study, host factors outweighed bacterial virulence factors in predicting the outcome in adults and neonates with E. coli bacteremia.
Most infections caused by S. aureus involve the skin and soft tissue but often this organism can cause bacteremia, pneumonia, endocarditis, osteomyelitis, and sepsis.30-32 Moreover methicillin-resistant S. aureus (MRSA) poses a major threat to public health given its worldwide spread, virulence, and difficult to treat invasive and life-threatening infections related to this organism.31,33,34 About one-third of the population is colonized with S. aureus as shown in one US study35 and it is well established that colonization with S. aureus is a risk factor for subsequent infection.36,37 Host immune defense against S. aureus originates in the skin where there are cutaneous host innate immune mechanisms that detect and combat microbial pathogens.27,38,39 Keratinocytes express TLR 1, 2, and 6 and NOD2 receptors, which recognize and bind S. aureus-derived lipoptides, lipotechoic acid, and peptidoglycan-derived muramyl dipeptide, leading to cytokine production and neutrophil recruitment.40 These cells also produce antimicrobial peptides with bacteriostatic or bactericidal activity, like β-defensin 3 (hBD3), RNase 7, or human cathelicidin (LL-37), all of which have potent activity against S. aureus.41-43 Such antimicrobial peptides are also produced after activation of keratinocytes by Staphylococcus epidermidis via a TLR-2-dependent mechanism, a finding that suggests that commensal organisms also contribute in S. aureus killing in the skin.44,45
Cellular innate immune response
The first and most important cellular host defense against invading pathogens is neutrophils. Neutrophils migrate rapidly from the blood to the site of infection and this recruitment is mediated by chemoattractants, like IL-8, granulocyte chemotactic protein 2 (GCP2), leukotriene B4 (LTB4), which are secreted by monocytes, macrophages, keratinocytes, mast cells, endothelial, and other host immune cells. Recognition and subsequent phagocytosis of invading microorganisms by neutrophils is mediated through PRRs and facilitated by antibody-Fc and complement receptors that bind to complement and antibody-coated microbes.46 After phagocytosis, microorganisms contained in phagosomes are killed by NADPH oxidase-dependent and myeloperoxidase-dependent reactive oxygen species or by antimicrobial peptides of cytoplasmic granules.46,47 In addition, the role of neutrophil extracellular traps (NETs), a novel mechanism of neutrophil antimicrobial defense has been described and is currently under investigation.48-50 NETs comprise of histones, chromatin, azurophilic granule, and cytosolic proteins and were shown to bind and destroy pathogens like S. aureus.49,50 Of the human neutrophil peptides (HNPs), the one with the greatest activity against S. aureus is HNP2.51 Also, calprotectin (S100A8/S100A9), a protein complex found in neutrophilic cytoplasm, inhibits. S. aureus growth via Mn2+ and Zn2+ chelation.52 Phagocytosis also enhances neutrophil apoptosis which is necessary for the resolution of the inflammatory response.53 Neutrophils have a major role in the control and clearance of extracellular bacteria like S. aureus.54,55 Congenital (leukocyte adhesion deficiency disorders, severe congenital neutropenia, myeloperoxidase deficiency, chronic granulomatous disease) or acquired (after chemotherapy) deficiency in the number or function of neutrophils predisposes to invasive infections, not only by S. aureus but also from gram-negative bacteria and fungi.47
Other cells with phagocytic potential include tissue macrophages, dendritic cells, and natural killer (NK) cells. Apart from phagocytosis, after their activation, macrophages secrete a number of chemotactic, inflammatory, and immunoregulatory molecules that direct the migration of other immune cells to the site of infection or act systemically regulating their response, hence playing a major role in the crosstalk between the innate and adaptive immune systems.26 The role of NK and NKT cells in bacteremia and sepsis is also of considerable importance as evidenced by an increasing number of relevant experimental and human studies.56,57 The pathogenesis of sepsis in patient populations is characterized by marked heterogeneity. It seems that both the innate and adaptive immune responses differ between patients in relation to the underlying type of infection. A prospective study including 505 patients was conducted by the Hellenic Sepsis Study Group. Flow cytometry analysis of white blood cell count subsets was conducted in blood sampled within the first 24 h from diagnosis. The primary study endpoint was the differences in immune responses between sepsis and severe sepsis/shock as these are related with the underlying type of infection leading to sepsis. From the total of 505 enrolled patients, 183 had acute pyelonephritis, 97 had community-acquired pneumonia (CAP), 100 had intrabdominal infections, 61 had BSIs, and 64 had hospital-acquired pneumonia. Increased apoptosis of natural killer T cells was the main change of the immune response upon transition from sepsis to severe sepsis/shock in patients with BSI contrary to the other types of infections.58 No profound explanation exists for this finding. Indirect evidence comes from an animal model of lethal ehlrichiosis. Although depletion of NKT cells did not affect final outcome, it prevented the advent of signs of toxic shock and prevented the development of excess levels of circulating TNFα.59
The role of complement
One of the first and most potent host immune barriers which all human pathogens, including bacteria, fungi, viruses, and parasites, encounter is the complement system. After bacterial infection, complement is activated through the classical, alternative, and mannose-binding lectin (MBL) pathways. Antigen-antibody complexes and C-reactive protein (CRP) bind to phosphorylcholine on the bacterial cell surface; amyloid P and bacterial cell wall components bind the C1q complex, which in turns activates the classic complement pathway, which includes C1, C2, C3, and C4. The alternative pathway is activated by microbial elements (bacterial cell surface) and involves factor B, factor D, properdin, and C3. Cleavage of C3 results in the production of opsonins that prepare pathogens for phagocytosis, anaphylatoxins, and the creation of a membrane attack complex that lyses target cells. One major example is the role of complement in the innate immune defense against S. pneumoniae.60-62 CAP caused by this species is associated with bacteremia in 10–30% of cases.63 One of the main mechanisms of S. pneumoniae clearance from the systemic circulation is opsonophagocytosis mediated by complement components.62,64 There is evidence that that classic complement pathway has a more critical role in host immune defense against S. pneumoniae compared with the alternative pathway.65 In more detail, Brown et al. studied mice genetically deficient of different complement components and demonstrated that C1q−/− mice (deficient in the classical complement pathway) were more susceptible to pneumococcal infection than Bf−/− mice (deficient in the alternative complement pathway).65 A defect in the classical complement pathway may limit primary adaptive immune responses which are known to be paramount in invasive pneumococcal infection.66 However, the alternative pathway has been found to amplify the activation of the classical/MBL complement pathway, thus playing an important role in the natural antibody-mediated opsonization of S. pneumoniae.67 In a pneumococcal otitis media model, where three types of complement-deficient and wild-type mice were used, Bf/C2−/− mice had higher bacterial burden in the blood 24 h after transtympanic infection than C1qa−/− and Bf−/− mice, whereas wild-type mice exhibited no bacteremia. In addition, complement-deficient mice exhibited decreased capacity for C3-mediated opsonization and complement-mediated opsonophagocytosis, which could be related to dissemination of S. pneumoniae to the bloodstream in these animals, indicating a critical role for both the classical and alternative pathways in host immune defense against pneumococcal otitis.68
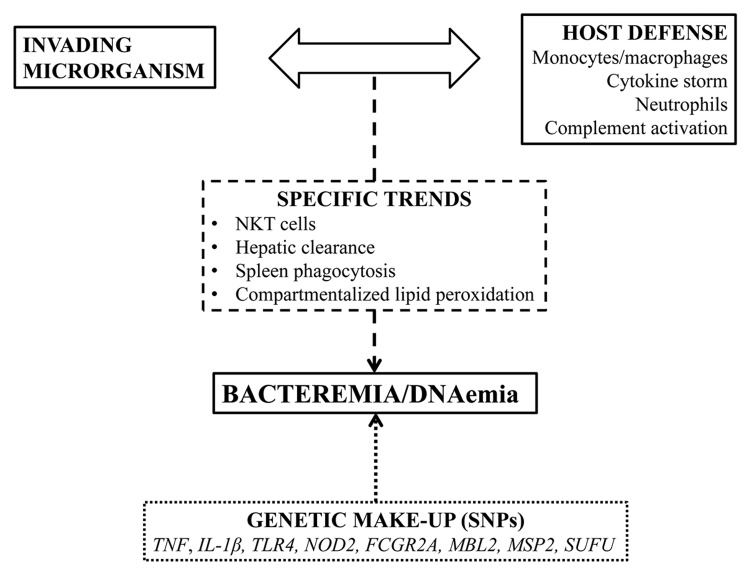
To avoid potentially injurious excessive activation of the complement system, the host uses regulatory proteins, like C4b-binding protein (C4bp), which inhibits the classic and lectin pathways. Many bacteria use these host regulatory proteins in order to escape complement-mediated killing. Despite their diversity, pathogens share common mechanisms of complement escape. Examples of such strategies are the expression of host complement regulator binding proteins, the secretion of proteases, and the release of complement inhibitors. Moreover, each microbial pathogen often uses multiple strategies to evade immune recognition and complement attack. Using such complement escape proteins, bacteria like Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, Pseudomonas aeruginosa, Borrelia burgdoferi, and fungi like Candida species, can evade host immune response and disseminate to the blood.69 Many of the individual proteins used by pathogens to prevent and inhibit complement activation and attack have been molecularly and functionally characterized in the last decade. For example, it was recently shown that an E. coli outer membrane protein, NIpI, contributes in high-level bacteremia by facilitating blockade of classical complement-mediated killing, via C4bp deposition on the bacterial surface.70 In addition, Prc, another E. coli bacterial periplasmic protease was also found to have a critical role in the evasion of complement-mediated killing.71 Hence, when immune mechanisms fail to control bacterial spread either due to bacterial evasion of host immune strategies or due to a breach or defect in the otherwise well-orchestrated immune response, bacteremia can develop (Fig. 1).
Figure 1. The complex interactions between the invading microorganism and the host defense mechanism ending in bacteremia. These interactions present some unique features in pathogenesis and they are under the influence of the genetic make-up of the host.
Adaptive immune response
As opposed to innate immune defense, the adaptive immune response is stimulated later during the infection process and encompasses B cell- and T cell-specific responses. Central to cellular adaptive immune responses is the presentation of exogenous antigens or microbes to lymphocytes by antigen presenting cells, most commonly dendritic cells and monocyte/macrophages via an MHC-I or II dependent manner. T-helper cells that express the CD4 molecule recognize antigens in the context of MHC class II molecules. Upon antigenic stimulation, naïve CD4+ cells expand and differentiate into Th1, Th2, and Th17 cells.72 Th1 response promotes cell-mediated immune responses via the production of pro-inflammatory cytokines like IFN-γ, TNF-α, IL-2, and IL-1β. Th2 cells produce IL-4, IL-10, IL-13, and support antibody mediated immune responses. Th17 cells produce IL-17, Il-21, IL-22, IL-26, mediate neutrophil recruitment and activation, contribute to abscess formation, and are critical for the clearance of extracellular bacteria.39,72 In addition, Th17 responses induced by bacteria and fungi result in the secretion of antimicrobial peptides that contribute to mucosal host defense.73 CD8+ T cells destroy target cells with cytolysis, produce potent inflammatory cytokines like IFN-γ and TNF-α, and they are the major effector cells against intracellular bacteria. When activated by antigens expressed on infected cells and co-stimulators, they differentiate into cytolytic cells (CTLs) and secrete proteins that form pores in the infected cell membrane and mediate apoptosis of these cells.74
B-cell responses lead to the production of antibodies directed against specific antigenic components of a certain pathogen.26 Immunoglobulin molecules consist of the Fc constant region that binds to Fc receptors on the surface of immune cells and is responsible for most of their effector functions and the Fab variable antigen-binding region with which the antibody binds with great specificity to the optimal target molecule and which is characterized by massive diversity. Antibodies interact with the binding of pathogens or toxins to the host cell receptors, hence neutralizing their effect and limiting microbial infectivity. Also, they opsonize bacteria and thus facilitate neutrophil or macrophage phagocytosis, mediate antibody-dependent killing of pathogens by NK cells or granulocytes and also activate complement. After the second encounter with the same antigen, B cells produce larger amounts of antibodies that often show increased heavy chain isotype switching. Moreover, the antigen–antibody binding affinity increases with repeated stimulation (affinity maturation), thus increasing the yield of the antibody response during a secondary infection.75
The importance of liver and spleen
The liver and the spleen function as filters of bacteria from the bloodstream and the spleen is a major site of antibody production. As a consequence, patients with anatomic or functional asplenia have increased risk of disseminated infections with encapsulated bacteria.76 Moreover, the significance of liver function to contain bacteremia has been increasingly recognized recently due to both its metabolic function and its ability to clear bacteria. Within the first six hours after induction of experimental pneumonia by S. pneumoniae in mice, the transcriptomic profile of the liver is modulated so that the liver metabolism is shifted toward increased production of high-density lipoproteins (HDLs). These are conceived as soluble receptors for pneumolysin.77 Moreover, a series of animal studies have shown that HDLs are a mechanism of protection in endotoxemia since they bind LPS whereas low circulating HDL is related with an increased infection risk among critically ill patients.78 This role of HDL may also have direct therapeutic implications. When reconstituted HDL was administered in the setting of experimental endotoxemia in healthy volunteers, LPS-induced activation of coagulation was attenuated.79 However, the therapeutic role of reconstituted HDL has not yet been studied in any randomized trial in sepsis. Hepatic clearance is also of major importance to contain bacteremia by Pseudomonas aeruginosa. Early after induction of bacteremia, the liver bacterial load increases dramatically to an extent depending on the number of inoculated bacteria. The inability of the liver to contain bacteria is linked with the induction of hepatic apoptosis in the event of severe large-scale bacteremia and it affects not only the liver but also the renal and myocardial function. Hepatic clearance in this situation is related to the function of Kupffer cells. Loss of Kupffer cells after pre-treatment with gadolinium paves the way for earlier animal death.80
The importance of this early modulation of liver function in response to bacteremia has also being confirmed in a recent study where experimental findings were confirmed in the clinical setting. Early after challenge of rats with P. aeruginosa, lipid peroxidation in the liver was increased compared with sham-treated animals. On the contrary, lipid peroxidation in the kidney was decreased. Lipid peroxidation was assessed by measurement of malondiadehyde (MDA). Changes of MDA were not statistically related with tissue bacterial load showing that they represent some compartmentalization of the lipid peroxidation process, which is a unique characteristic in pathogenesis. When MDA was measured in serum from patients with sepsis, this compartmentalization was further confirmed: serum MDA was greater in patients with acute liver dysfunction and lower in patients with acute renal dysfunction.81
Bacteremia and DNAemia
Bacteremia is traditionally conceived as the presence of pathogenic bacteria in the bloodstream. To this end, the traditional commercialized systems to culture blood mandate the inoculation of large quantities of blood, as much as 10 to 20 ml, in pre-prepared media. With current techniques, detection of bacteremia in severe infections fails to a great extent. Prospective observational studies report an incidence of less than 20% of positive blood cultures among patients with severe sepsis and/or septic shock.82,83 These findings broad up the question how of is it possible for a host to present with severe infection with a negative blood culture? The reply to this question is related to the extent of bacteremia and to the duration of bacteremia, that greatly differ between patients. The presence of bacteria in the bloodstream is not always constant. Transient episodes of bacteremia seem to be adequate to disseminate the infection within remote tissues. Furthermore, current research conducted with animal models implies that systemic spread of microorganisms takes place from the primary source of infection. In all these models, animals are considered to suffer from bacteremia, yet blood culture results are not always reported; instead, the presence of bacteremia is indirectly documented by the isolation of microorganisms from tissues far from the primary infection site.84,85
The extent of bacteremia is another important factor linked with final outcome. Current blood culture systems are not able to quantify circulating bacteria in the bloodstream. Few animal studies are also available to this same end.86 Molecular identification of circulating bacteria is a proposed alternative allowing to identify not only the presence of pathogens but also to quantify the amount of circulating bacteria. This is done indirectly by the measurement of the number of copies of specific bacterial genes. One well-studied example is the autolysin gene (lytA) of S. pneumoniae. lytA is a single-copy gene and its measurement is an indirect estimation of the number of circulating bacteria. Analysis of samples coming from 93 patients with CAP allowed to discriminate two patterns; patients with low-level gene copies i.e., less than 103 copies of lytA and patients with high-level gene copies i.e., ≥103 copies/ml of lytA in the bloodstream. The incidence of bacteremia was 24.2% among patients with low-level gene copies and 66.6% among patients with high-level gene copies (P < 0.0001). Similarly patients with high number of copies had a greater incidence of acute renal dysfunction (55.5% vs. 15.1%), septic shock (44.4% vs. 9.1%), and greater mortality (25.9% vs. 6.1%).87
These findings regarding the significance of the number of copies of lytA are confirmed in two more studies; the first enrolled 45 patients88 and the second enrolled 304 patients.89 The first study showed that the number of gene copies is an indicator of the presence of S. pneumoniae as the causative pathogen for CAP but also of the degree of the inflammation of the host. Using receiver operator characteristics (ROC) curve analysis, the authors concluded that the use of gene copies is a more reliable marker than the white blood cell count and serum CRP for the systemic inflammatory response syndrome stimulated by CAP.88 In the second study, it was confirmed that patients with more than 103 gene copies/ml in the bloodstream carried a greater risk for severe disease (odds ratio = 5.22).89
Similar findings to those reported for the detection of bacteremia using the copy number of specific genes of S. pneumoniae have also been reported for patients with bacteremia by S. aureus and by Acinetobacter baumannii. In a prospective study of 20 patients with secondary methicillin-resistant S. aureus (MRSA) bacteremia of various origins, the number of copies of mecA conferring resistance to methicillin on patient follow-up was reversibly related with the response to anti-MRSA antimicrobial coverage.90 In a similar fashion, the copies of Oxa-51 were measured in the blood of 34 survivors and 17 non-survivors with A. baumannii bacteremia. The authors have prospectively measured the number of copies during the disease course and found that any increase of the absolute copy numbers of Oxa-51 in the bloodstream was an independent predictor of unfavorable outcome.91 However, the clinical relevance of DNAemia is still not yet clear and further research is warranted in order to determine its exact role in the diagnosis and prognosis of BSIs.
Genetic susceptibility to bacteremia
Genes encoding for immune receptors and cytokines
Most genes of the human genome carry single nucleotide polymorphisms (SNPs) at specific exonic or intronic regions. The frequency of these SNPs is usually below 1% of the general population. Some of these SNPs are carried at greater frequencies even exceeding 20% mostly at a heterozygotes state bringing up the question whether this may impose on a certain disease phenotype. Regarding infectious diseases, the question generated is whether SNPs of genes encoding for all the above described molecules i.e., receptors and cytokines may induce susceptibility to severe infections.
The most broadly studied gene is TNF. SNPs of this gene involve the −308 position at the promoter region where a substitution of guanine (G) for adenosine (A) takes place (rs1800629). A meta-analysis of 25 studies provided contradictory results regarding the role of this SNP for the physical course of sepsis.92 We tried to decipher the role of this SNP for susceptibility to infection in 213 intubated patients all of whom developed ventilator-associated pneumonia. The rationale of the study was that if this SNP is important, it should have a clear impact on a patient population with a major risk factor (i.e., intubation) for the development of infection. The study did not focus only on the rs1800629 SNP but on the TNF haplotype as defined by all three SNPs at the −376, −308, and −238 promoter positions of the TNF gene. In all these SNPs G to A substitutions are reported. Results revealed that carriers of any A allele of the three SNPs developed VAP earlier after intubation compared with carriers of only wild-type G alleles and that this was related with lower production of TNFα and of IL-6 by circulating monocytes.93 Unpublished data of our group report on the significance of these haplotypes for the natural course of 83 patients with infective endocarditis and secondary gram-positive bacteremia. Carriage of the GGG wild-type haplotype was related with a significantly greater risk for unfavorable outcome (odds ratio = 3.29, P = 0.041). A large study on 774 medical patients in ICUs, studied the associations of several gene SNPs with the risk for development of bacteremia. Two gene SNPs were associated with a greater risk for BSI. These SNPs were at the position 299 of TLR4 encoding for TLR4 and at the position 702 of NOD2 encoding for the CARD15 (caspase activation recruitment domain 15) of the NLRP3 inflammasome PRR. Carriers of these gene SNPs had a greater risk for the acquisition of BSI (13.5% vs. 7.6% of wild-type and of TLR4 and 12.6% vs. 7.1% of wild-type and of NOD2). This risk was even greater for patients carrying both SNPs (25.0% vs. 7.6%). Carriage of these SNPs was also linked with susceptibility for earlier acquisition of BSI after ICU admission.94 Although this study does not report for an effect of carriage of the Asp299Gly SNP of TLR4 in cytokine production, it has been shown that monocytes of healthy controls who carry only this SNP allele are able for greater production of TNFα but not of IL-10 compared with monocytes of patients who carry only WT alleles after stimulation with LPS.95
Signaling of TLR2 and TLR4 stimulation is down-stream linked with the adaptor protein TIRAP. One SNP has been described for TIRAP at position 180 where serine is substituted by leucine. Heterozygozity for this SNP is linked with protection against the development of pneumococcal bacteremia as defined in a cohort of 901 patients from Kenya. Odds ratio for the development of bacteremia was 0.34 (P = 0.003) among heterozygotes for this SNP.96
Another important SNP linked with susceptibility for bacteremia has been reported from a study enrolling recipients of kidney transplants. The SNP is found at position −511 of IL-1β where either a C allele or a T allele exists. Carriage of the C allele is associated with increased risk for bacteremia (66.7% vs. 45.% for non-infected patients, P = 0.015).97
Genes encoding for other components of the immune response
Genetic association studies have tried to investigate the existence of a link between host factors related with protection against encapsulated microorganisms and the development of bacteremia. These factors primarily refer to the receptor of IgG and the MBL pathway.
One SNP exists after one point mutation of FCGR2A encoding for the FcγRIIA receptor of IgG2. This SNP leads to a substitution of arginine with histidine at position 131 of FcγRIIA ending in poor IgG2 binding capacity. This SNP was studied in 1262 patients with CAP and compared with 1224 healthy controls. The overall SNP frequency did not differ between patients and controls. However, among patients with CAP due to S. pneumoniae the frequency of SNP carriage was significantly greater among those who developed bacteremia compared with those who did not develop bacteremia. More precisely, among those who developed pneumococcal bacteremia 35.3% were homozygotes and 43.5% heterozygotes for the SNP. This translates to an odds ratio of 2.9 for bacteremia for patients homozygous for the SNP (P = 0.00016) and 2.83 for patients heterozygous for the SNP (P = 0.0012.).98
As described above, the mannose complement pathway is of major importance for the host response against bacteria particularly for encapsulated bacteria. Two major structural SNPs of MBL2 and MASP2 have been found to be associated with the physical course of bacteremia. MBL2 encodes for MBL2. Structural SNPs exist at codons 52 (CGT or TGT haplotypes), 54 (GGG or GAC haplotypes), and 57 (GGA or GAA haplotypes) of exon 1 of MBL. These SNPs were studied in a cohort of 145 patients with bacteremia and they were compared with 400 healthy controls. Although the overall frequency of SNPs did not differ between groups, carriers of the GAA haplotype at codon 57 had a 4.2-fold greater risk for the acquisition of gram-positive BSI.99
MASP2 encodes for the serine protease of MBL2. rs2273346 of MASP2 was analyzed in a large cohort of ICU patients. Although carriage of the SNP was not associated with the risk for the development of bacteremia, after adjustment for co-morbidities, it was independently associated with in-hospital mortality (odds ratio = 2.35, P = 0.02).94
SNPs of SUFU have very recently been reported to impact on the physical course of bacteremia. This gene encodes for a negative regulator of the Sonic hedgehog signaling pathway (SHH). This pathway is of major importance for the maturation of CD4 lymphocytes. A total of 69 SNPs of SUFU were studied in a cohort of 250 patients with bacteremia by Enterobacteriaceae, mainly by E. coli and K. pneumoniae. The primary study endpoint was the significance of these SNPs for the development of organ failure. It was found that four of the studied SNPs (rs10786691, rs12414407, rs10748825, and rs7078511) were protective against the development of renal dysfunction and two of the studied SNPs (rs12414407 and rs10748827) against the development of ARDS.100
Conclusions
BSI is a complex and life-threatening entity. Pathogenesis is multifactorial. Based on the presented analysis, bacteremia develops as a result of imbalances in the complex interplay between the invading microorganism and the host defense mechanisms. This interplay encompasses some characteristics that are unique for BSI and it seems to be directly influenced by the genetic background of the host.
Disclosure of Potential Conflicts of Interest
The authors have no conflict of interest to declare.
References
- 1.Bone RC. Sepsis, the sepsis syndrome, multi-organ failure: a plea for comparable definitions. Ann Intern Med. 1991;114:332–3. doi: 10.7326/0003-4819-114-4-332. [DOI] [PubMed] [Google Scholar]
- 2.Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, et al. Health care--associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002;137:791–7. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 3.Siegman-Igra Y, Fourer B, Orni-Wasserlauf R, Golan Y, Noy A, Schwartz D, Giladi M. Reappraisal of community-acquired bacteremia: a proposal of a new classification for the spectrum of acquisition of bacteremia. Clin Infect Dis. 2002;34:1431–9. doi: 10.1086/339809. [DOI] [PubMed] [Google Scholar]
- 4.Kanoksil M, Jatapai A, Peacock SJ, Limmathurotsakul D. Epidemiology, microbiology and mortality associated with community-acquired bacteremia in northeast Thailand: a multicenter surveillance study. PLoS One. 2013;8:e54714. doi: 10.1371/journal.pone.0054714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Søgaard M, Nørgaard M, Dethlefsen C, Schønheyder HC. Temporal changes in the incidence and 30-day mortality associated with bacteremia in hospitalized patients from 1992 through 2006: a population-based cohort study. Clin Infect Dis. 2011;52:61–9. doi: 10.1093/cid/ciq069. [DOI] [PubMed] [Google Scholar]
- 6.Uslan DZ, Crane SJ, Steckelberg JM, Cockerill FR, 3rd, St Sauver JL, Wilson WR, Baddour LM. Age- and sex-associated trends in bloodstream infection: a population-based study in Olmsted County, Minnesota. Arch Intern Med. 2007;167:834–9. doi: 10.1001/archinte.167.8.834. [DOI] [PubMed] [Google Scholar]
- 7.Laupland KB, Kibsey PC, Gregson DB, Galbraith JC. Population-based laboratory assessment of the burden of community-onset bloodstream infection in Victoria, Canada. Epidemiol Infect. 2013;141:174–80. doi: 10.1017/S0950268812000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, et al. EPIC II Group of Investigators International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–9. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 9.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10:417–32. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallés J, Palomar M, Alvárez-Lerma F, Rello J, Blanco A, Garnacho-Montero J, Martín-Loeches I, GTEI/SEMICYUC Working Group on Bacteremia Evolution over a 15-year period of clinical characteristics and outcomes of critically ill patients with community-acquired bacteremia. Crit Care Med. 2013;41:76–83. doi: 10.1097/CCM.0b013e3182676698. [DOI] [PubMed] [Google Scholar]
- 11.Kollef MH, Zilberberg MD, Shorr AF, Vo L, Schein J, Micek ST, Kim M. Epidemiology, microbiology and outcomes of healthcare-associated and community-acquired bacteremia: a multicenter cohort study. J Infect. 2011;62:130–5. doi: 10.1016/j.jinf.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Vallés J, Rello J, Ochagavía A, Garnacho J, Alcalá MA. Community-acquired bloodstream infection in critically ill adult patients: impact of shock and inappropriate antibiotic therapy on survival. Chest. 2003;123:1615–24. doi: 10.1378/chest.123.5.1615. [DOI] [PubMed] [Google Scholar]
- 13.Artero A, Zaragoza R, Camarena JJ, Sancho S, González R, Nogueira JM. Prognostic factors of mortality in patients with community-acquired bloodstream infection with severe sepsis and septic shock. J Crit Care. 2010;25:276–81. doi: 10.1016/j.jcrc.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Skogberg K, Lyytikäinen O, Ruutu P, Ollgren J, Nuorti JP. Increase in bloodstream infections in Finland, 1995-2002. Epidemiol Infect. 2008;136:108–14. doi: 10.1017/S0950268807008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Opal SM. The host response to endotoxin, antilipopolysaccharide strategies, and the management of severe sepsis. Int J Med Microbiol. 2007;297:365–77. doi: 10.1016/j.ijmm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.van der Poll T, Opal SM. Host-pathogen interactions in sepsis. Lancet Infect Dis. 2008;8:32–43. doi: 10.1016/S1473-3099(07)70265-7. [DOI] [PubMed] [Google Scholar]
- 17.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–6. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 19.Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot JP, Karin M, Nizet V, Eckmann L. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci U S A. 2009;106:12873–8. doi: 10.1073/pnas.0904958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cinel I, Opal SM. Molecular biology of inflammation and sepsis: a primer. Crit Care Med. 2009;37:291–304. doi: 10.1097/CCM.0b013e31819267fb. [DOI] [PubMed] [Google Scholar]
- 21.Huttunen R, Aittoniemi J. New concepts in the pathogenesis, diagnosis and treatment of bacteremia and sepsis. J Infect. 2011;63:407–19. doi: 10.1016/j.jinf.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Gibot S. Clinical review: role of triggering receptor expressed on myeloid cells-1 during sepsis. Crit Care. 2005;9:485–9. doi: 10.1186/cc3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senftleben U, Karin M. The IKK/NF-kappa B pathway. Crit Care Med. 2002;30(Suppl):S18–26. doi: 10.1097/00003246-200201001-00003. [DOI] [PubMed] [Google Scholar]
- 24.Casey LC. Immunologic response to infection and its role in septic shock. Crit Care Clin. 2000;16:193–213. doi: 10.1016/S0749-0704(05)70107-X. [DOI] [PubMed] [Google Scholar]
- 25.Kong X, Thimmulappa R, Craciun F, Harvey C, Singh A, Kombairaju P, Reddy SP, Remick D, Biswal S. Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am J Respir Crit Care Med. 2011;184:928–38. doi: 10.1164/rccm.201102-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schooley R, Wislon C. Host responses to infection. In: Cohen J, Powderly W, Opal S, eds. Infectious Diseases: MOSBY, 2010:30-44. [Google Scholar]
- 27.Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nat Rev Immunol. 2004;4:211–22. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hekker TA, Groeneveld AB, Simoons-Smit AM, de Man P, Connell H, MacLaren DM. Role of bacterial virulence factors and host factors in the outcome of Escherichia coli bacteraemia. Eur J Clin Microbiol Infect Dis. 2000;19:312–6. doi: 10.1007/s100960050483. [DOI] [PubMed] [Google Scholar]
- 29.Tullus K, Brauner A, Fryklund B, Munkhammar T, Rabsch W, Reissbrodt R, Burman LG. Host factors versus virulence-associated bacterial characteristics in neonatal and infantile bacteraemia and meningitis caused by Escherichia coli. J Med Microbiol. 1992;36:203–8. doi: 10.1099/00222615-36-3-203. [DOI] [PubMed] [Google Scholar]
- 30.McCaig LF, McDonald LC, Mandal S, Jernigan DB. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg Infect Dis. 2006;12:1715–23. doi: 10.3201/eid1211.060190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 32.Diekema DJ, Pfaller MA, Jones RN, Doern GV, Winokur PL, Gales AC, Sader HS, Kugler K, Beach M. Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin Infect Dis. 1999;29:595–607. doi: 10.1086/598640. [DOI] [PubMed] [Google Scholar]
- 33.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) Curr Opin Microbiol. 2012;15:588–95. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 34.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–68. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis. 2008;197:1226–34. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 36.Safdar N, Bradley EA. The risk of infection after nasal colonization with Staphylococcus aureus. Am J Med. 2008;121:310–5. doi: 10.1016/j.amjmed.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 37.Honda H, Krauss MJ, Coopersmith CM, Kollef MH, Richmond AM, Fraser VJ, Warren DK. Staphylococcus aureus nasal colonization and subsequent infection in intensive care unit patients: does methicillin resistance matter? Infect Control Hosp Epidemiol. 2010;31:584–91. doi: 10.1086/652530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–91. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishna S, Miller LS. Innate and adaptive immune responses against Staphylococcus aureus skin infections. Semin Immunopathol. 2012;34:261–80. doi: 10.1007/s00281-011-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller LS. Toll-like receptors in skin. Adv Dermatol. 2008;24:71–87. doi: 10.1016/j.yadr.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kisich KO, Howell MD, Boguniewicz M, Heizer HR, Watson NU, Leung DY. The constitutive capacity of human keratinocytes to kill Staphylococcus aureus is dependent on beta-defensin 3. J Invest Dermatol. 2007;127:2368–80. doi: 10.1038/sj.jid.5700861. [DOI] [PubMed] [Google Scholar]
- 42.Cho JS, Xuan C, Miller LS. Lucky number seven: RNase 7 can prevent Staphylococcus aureus skin colonization. J Invest Dermatol. 2010;130:2703–6. doi: 10.1038/jid.2010.294. [DOI] [PubMed] [Google Scholar]
- 43.Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun. 2005;73:6771–81. doi: 10.1128/IAI.73.10.6771-6781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanke I, Steffen H, Christ C, Krismer B, Götz F, Peschel A, Schaller M, Schittek B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol. 2011;131:382–90. doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]
- 45.Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, Ryan AF, Di Nardo A, Gallo RL. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–21. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. 2012;34:237–59. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Köckritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med (Berl) 2009;87:775–83. doi: 10.1007/s00109-009-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 50.Papayannopoulos V, Zychlinsky A. NETs: a new strategy for using old weapons. Trends Immunol. 2009;30:513–21. doi: 10.1016/j.it.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human alpha-defensins. Antimicrob Agents Chemother. 2005;49:269–75. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–5. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 53.Kennedy AD, DeLeo FR. Neutrophil apoptosis and the resolution of infection. Immunol Res. 2009;43:25–61. doi: 10.1007/s12026-008-8049-6. [DOI] [PubMed] [Google Scholar]
- 54.Mölne L, Verdrengh M, Tarkowski A. Role of neutrophil leukocytes in cutaneous infection caused by Staphylococcus aureus. Infect Immun. 2000;68:6162–7. doi: 10.1128/IAI.68.11.6162-6167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verdrengh M, Tarkowski A. Role of neutrophils in experimental septicemia and septic arthritis induced by Staphylococcus aureus. Infect Immun. 1997;65:2517–21. doi: 10.1128/iai.65.7.2517-2521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiche L, Forel JM, Thomas G, Farnarier C, Vely F, Bléry M, Papazian L, Vivier E. The role of natural killer cells in sepsis. J Biomed Biotechnol. 2011;2011:986491. doi: 10.1155/2011/986491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leung B, Harris HW. NKT cells in sepsis. Clin Dev Immunol. 2010;2010:pii: 414650. doi: 10.1155/2010/414650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gogos C, Kotsaki A, Pelekanou A, Giannikopoulos G, Vaki I, Maravitsa P, Adamis S, Alexiou Z, Andrianopoulos G, Antonopoulou A, et al. Early alterations of the innate and adaptive immune statuses in sepsis according to the type of underlying infection. Crit Care. 2010;14:R96. doi: 10.1186/cc9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson HL, Crossley EC, Thirumalapura N, Walker DH, Ismail N. Regulatory roles of CD1d-restricted NKT cells in the induction of toxic shock-like syndrome in an animal model of fatal ehrlichiosis. Infect Immun. 2008;76:1434–44. doi: 10.1128/IAI.01242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mold C. Role of complement in host defense against bacterial infection. Microbes Infect. 1999;1:633–8. doi: 10.1016/S1286-4579(99)80063-X. [DOI] [PubMed] [Google Scholar]
- 61.Sjöholm AG, Jönsson G, Braconier JH, Sturfelt G, Truedsson L. Complement deficiency and disease: an update. Mol Immunol. 2006;43:78–85. doi: 10.1016/j.molimm.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 62.Winkelstein JA. The role of complement in the host’s defense against Streptococcus pneumoniae. Rev Infect Dis. 1981;3:289–98. doi: 10.1093/clinids/3.2.289. [DOI] [PubMed] [Google Scholar]
- 63.Blasi F, Mantero M, Santus P, Tarsia P. Understanding the burden of pneumococcal disease in adults. Clin Microbiol Infect. 2012;18(Suppl 5):7–14. doi: 10.1111/j.1469-0691.2012.03937.x. [DOI] [PubMed] [Google Scholar]
- 64.Brown EJ, Hosea SW, Frank MM. The role of complement in the localization of pneumococci in the splanchnic reticuloendothelial system during experimental bacteremia. J Immunol. 1981;126:2230–5. [PubMed] [Google Scholar]
- 65.Brown JS, Hussell T, Gilliland SM, Holden DW, Paton JC, Ehrenstein MR, Walport MJ, Botto M. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc Natl Acad Sci U S A. 2002;99:16969–74. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Austrian R, Gold J. Pneumococcal Bacteremia with Especial Reference to Bacteremic Pneumococcal Pneumonia. Ann Intern Med. 1964;60:759–76. doi: 10.7326/0003-4819-60-5-759. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, Ma M, Ippolito GC, Schroeder HW, Jr., Carroll MC, Volanakis JE. Complement activation in factor D-deficient mice. Proc Natl Acad Sci U S A. 2001;98:14577–82. doi: 10.1073/pnas.261428398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tong HH, Li YX, Stahl GL, Thurman JM. Enhanced susceptibility to acute pneumococcal otitis media in mice deficient in complement C1qa, factor B, and factor B/C2. Infect Immun. 2010;78:976–83. doi: 10.1128/IAI.01012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zipfel PF, Würzner R, Skerka C. Complement evasion of pathogens: common strategies are shared by diverse organisms. Mol Immunol. 2007;44:3850–7. doi: 10.1016/j.molimm.2007.06.149. [DOI] [PubMed] [Google Scholar]
- 70.Tseng YT, Wang SW, Kim KS, Wang YH, Yao Y, Chen CC, Chiang CW, Hsieh PC, Teng CH. NlpI facilitates deposition of C4bp on Escherichia coli by blocking classical complement-mediated killing, which results in high-level bacteremia. Infect Immun. 2012;80:3669–78. doi: 10.1128/IAI.00320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang CY, Wang SW, Huang WC, Kim KS, Chang NS, Wang YH, Wu MH, Teng CH. Prc contributes to Escherichia coli evasion of classical complement-mediated serum killing. Infect Immun. 2012;80:3399–409. doi: 10.1128/IAI.00321-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van de Veerdonk FL, Gresnigt MS, Kullberg BJ, van der Meer JW, Joosten LA, Netea MG. Th17 responses and host defense against microorganisms: an overview. BMB Rep. 2009;42:776–87. doi: 10.5483/BMBRep.2009.42.12.776. [DOI] [PubMed] [Google Scholar]
- 74.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–8. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abbas AK, Lichman AH. Antigen recognition in the adaptive immune system. Basic Immunology. Philadelphia: Saunders Elsevier 2009:67-87. [Google Scholar]
- 76.Bohnsack JF, Brown EJ. The role of the spleen in resistance to infection. Annu Rev Med. 1986;37:49–59. doi: 10.1146/annurev.me.37.020186.000405. [DOI] [PubMed] [Google Scholar]
- 77.Weber M, Lambeck S, Ding N, Henken S, Kohl M, Deigner HP, Enot DP, Igwe EI, Frappart L, Kiehntopf M, et al. Hepatic induction of cholesterol biosynthesis reflects a remote adaptive response to pneumococcal pneumonia. FASEB J. 2012;26:2424–36. doi: 10.1096/fj.11-191957. [DOI] [PubMed] [Google Scholar]
- 78.Gordon BR, Parker TS, Levine DM, Saal SD, Wang JC, Sloan BJ, Barie PS, Rubin AL. Low lipid concentrations in critical illness: implications for preventing and treating endotoxemia. Crit Care Med. 1996;24:584–9. doi: 10.1097/00003246-199604000-00006. [DOI] [PubMed] [Google Scholar]
- 79.Pajkrt D, Lerch PG, van der Poll T, Levi M, Illi M, Doran JE, Arnet B, van den Ende A, ten Cate JW, van Deventer SJ. Differential effects of reconstituted high-density lipoprotein on coagulation, fibrinolysis and platelet activation during human endotoxemia. Thromb Haemost. 1997;77:303–7. [PubMed] [Google Scholar]
- 80.Ashare A, Monick MM, Powers LS, Yarovinsky T, Hunninghake GW. Severe bacteremia results in a loss of hepatic bacterial clearance. Am J Respir Crit Care Med. 2006;173:644–52. doi: 10.1164/rccm.200509-1470OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toufekoula C, Papadakis V, Tsaganos T, Routsi C, Orfanos SE, Kotanidou A, Carrer DP, Raftogiannis M, Baziaka F, Giamarellos-Bourboulis EJ. Compartmentalization of lipid peroxidation in sepsis by multidrug-resistant gram-negative bacteria: experimental and clinical evidence. Crit Care. 2013;17:R6. doi: 10.1186/cc11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Naber CK, Baddour LM, Giamarellos-Bourboulis EJ, Gould IM, Herrmann M, Hoen B, Karchmer AW, Kobayashi Y, Kozlov RS, Lew D, et al. Clinical consensus conference: survey on Gram-positive bloodstream infections with a focus on Staphylococcus aureus. Clin Infect Dis. 2009;48(Suppl 4):S260–70. doi: 10.1086/598185. [DOI] [PubMed] [Google Scholar]
- 83.Kotsaki A, Giamarellos-Bourboulis EJ. Molecular diagnosis of sepsis. Expert Opin Med Diagn. 2012;6:209–19. doi: 10.1517/17530059.2012.667799. [DOI] [PubMed] [Google Scholar]
- 84.Krasnodembskaya A, Samarani G, Song Y, Zhuo H, Su X, Lee JW, Gupta N, Petrini M, Matthay MA. Human mesenchymal stem cells reduce mortality and bacteremia in gram-negative sepsis in mice in part by enhancing the phagocytic activity of blood monocytes. Am J Physiol Lung Cell Mol Physiol. 2012;302:L1003–13. doi: 10.1152/ajplung.00180.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giamarellos-Bourboulis EJ, Adamis T, Laoutaris G, Sabracos L, Koussoulas V, Mouktaroudi M, Perrea D, Karayannacos PE, Giamarellou H. Immunomodulatory clarithromycin treatment of experimental sepsis and acute pyelonephritis caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2004;48:93–9. doi: 10.1128/AAC.48.1.93-99.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giamarellos-Bourboulis EJ, Mouktaroudi M, Adamis T, Koussoulas V, Baziaka F, Perrea D, Karayannacos PE, Giamarellou H. n-6 polyunsaturated fatty acids enhance the activities of ceftazidime and amikacin in experimental sepsis caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2004;48:4713–7. doi: 10.1128/AAC.48.12.4713-4717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rello J, Lisboa T, Lujan M, Gallego M, Kee C, Kay I, Lopez D, Waterer GW, DNA-Neumococo Study Group Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest. 2009;136:832–40. doi: 10.1378/chest.09-0258. [DOI] [PubMed] [Google Scholar]
- 88.Peters RP, de Boer RF, Schuurman T, Gierveld S, Kooistra-Smid M, van Agtmael MA, Vandenbroucke-Grauls CM, Persoons MC, Savelkoul PH. Streptococcus pneumoniae DNA load in blood as a marker of infection in patients with community-acquired pneumonia. J Clin Microbiol. 2009;47:3308–12. doi: 10.1128/JCM.01071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Werno AM, Anderson TP, Murdoch DR. Association between pneumococcal load and disease severity in adults with pneumonia. J Med Microbiol. 2012;61:1129–35. doi: 10.1099/jmm.0.044107-0. [DOI] [PubMed] [Google Scholar]
- 90.Ho YC, Chang SC, Lin SR, Wang WK. High levels of mecA DNA detected by a quantitative real-time PCR assay are associated with mortality in patients with methicillin-resistant Staphylococcus aureus bacteremia. J Clin Microbiol. 2009;47:1443–51. doi: 10.1128/JCM.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chuang YC, Chang SC, Wang WK. High and increasing Oxa-51 DNA load predict mortality in Acinetobacter baumannii bacteremia: implication for pathogenesis and evaluation of therapy. PLoS One. 2010;5:e14133. doi: 10.1371/journal.pone.0014133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Teuffel O, Ethier MC, Beyene J, Sung L. Association between tumor necrosis factor-alpha promoter -308 A/G polymorphism and susceptibility to sepsis and sepsis mortality: a systematic review and meta-analysis. Crit Care Med. 2010;38:276–82. doi: 10.1097/CCM.0b013e3181b42af0. [DOI] [PubMed] [Google Scholar]
- 93.Kotsaki A, Raftogiannis M, Routsi C, Baziaka F, Kotanidou A, Antonopoulou A, Orfanos SE, Katsenos C, Koutoukas P, Plachouras D, et al. Genetic polymorphisms within tumor necrosis factor gene promoter region: a role for susceptibility to ventilator-associated pneumonia. Cytokine. 2012;59:358–63. doi: 10.1016/j.cyto.2012.04.040. [DOI] [PubMed] [Google Scholar]
- 94.Henckaerts L, Nielsen KR, Steffensen R, Van Steen K, Mathieu C, Giulietti A, Wouters PJ, Milants I, Vanhorebeek I, Langouche L, et al. Polymorphisms in innate immunity genes predispose to bacteremia and death in the medical intensive care unit. Crit Care Med. 2009;37:192–201, e1-3. doi: 10.1097/CCM.0b013e31819263d8. [DOI] [PubMed] [Google Scholar]
- 95.Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D, Kibiki G, Cristea T, Hijmans A, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci U S A. 2007;104:16645–50. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khor CC, Chapman SJ, Vannberg FO, Dunne A, Murphy C, Ling EY, Frodsham AJ, Walley AJ, Kyrieleis O, Khan A, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–8. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wan QQ, Ye QF, Ma Y, Zhou JD. Genetic association of interleukin-1β (-511C/T) and its receptor antagonist (86-bpVNTR) gene polymorphism with susceptibility to bacteremia in kidney transplant recipients. Transplant Proc. 2012;44:3026–8. doi: 10.1016/j.transproceed.2012.05.081. [DOI] [PubMed] [Google Scholar]
- 98.Solé-Violán J, García-Laorden MI, Marcos-Ramos JA, de Castro FR, Rajas O, Borderías L, Briones ML, Herrera-Ramos E, Blanquer J, Aspa J, et al. The Fcγ receptor IIA-H/H131 genotype is associated with bacteremia in pneumococcal community-acquired pneumonia. Crit Care Med. 2011;39:1388–93. doi: 10.1097/CCM.0b013e31820eda74. [DOI] [PubMed] [Google Scholar]
- 99.Huttunen R, Aittoniemi J, Laine J, Vuento R, Karjalainen J, Rovio AT, Eklund C, Hurme M, Huhtala H, Syrjänen J. Gene-environment interaction between MBL2 genotype and smoking, and the risk of gram-positive bacteraemia. Scand J Immunol. 2008;68:438–44. doi: 10.1111/j.1365-3083.2008.02149.x. [DOI] [PubMed] [Google Scholar]
- 100.Henao-Martínez AF, Agler AH, LaFlamme D, Schwartz DA, Yang IV. Polymorphisms in the SUFU gene are associated with organ injury protection and sepsis severity in patients with Enterobacteriacea bacteremia. Infect Genet Evol. 2013;16:386–91. doi: 10.1016/j.meegid.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]