Abstract
Chemical cross-linking mass spectrometry identifies interacting surfaces within a protein assembly through labeling with bifunctional reagents and identifying the covalently modified peptides. These yield distance constraints that provide a powerful means to model the three-dimensional structure of the assembly. Bioinformatic analysis of cross-linked data resulting from large protein assemblies is challenging because each cross-linked product contains two covalently linked peptides, each of which must be correctly identified from a complex matrix of potential confounders.
Protein Prospector addresses these issues through a complementary mass modification strategy in which each peptide is searched and identified separately. We demonstrate this strategy with an analysis of RNA polymerase II. False discovery rates (FDRs) are assessed via comparison of cross-linking data to crystal structure, as well as by using a decoy database strategy. Parameters that are most useful for positive identification of cross-linked spectra are explored. We find that fragmentation spectra generally contain more product ions from one of the two peptides constituting the cross-link. Hence, metrics reflecting the quality of the spectral match to the less confident peptide provide the most discriminatory power between correct and incorrect matches. A support vector machine model was built to further improve classification of cross-linked peptide hits. Furthermore, the frequency with which peptides cross-linked via common acylating reagents fragment to produce diagnostic, cross-linker-specific ions is assessed.
The threshold for successful identification of the cross-linked peptide product depends upon the complexity of the sample under investigation. Protein Prospector, by focusing the reliability assessment on the least confident peptide, is better able to control the FDR for results as larger complexes and databases are analyzed. In addition, when FDR thresholds are calculated separately for intraprotein and interprotein results, a further improvement in the number of unique cross-links confidently identified is achieved. These improvements are demonstrated on two previously published cross-linking datasets.
Most proteins are organized into stable assemblies that communicate among themselves through transient protein–protein interaction networks to catalyze cellular phenomena. Chemical cross-linking mass spectrometry directly measures protein–protein interactions by using bifunctional cross-linking reagents to covalently link surfaces of interacting partners (1–3). Following proteolysis, mass spectrometry is used to identify the covalently linked peptides and modified residues. This information, taken together with the geometry of the cross-linking reagent, provides distance constraints that are reflective of the three-dimensional structure of the protein complex. Cross-linking-derived distance constraints provide a powerful means by which to integrate atomic resolution structures of individual protein subunits or subassemblies with low-resolution electron-microscopy-derived structures, as well as to clarify molecular details that are unresolved in electron density maps. For instance, this approach has recently been applied to modeling the RNA Pol II preinitiation complex (4), several chromatin remodeling complexes (5, 6), the 26S proteasome (7), and the Mediator middle module (8); solving the subunit arrangement of TCP1 ring complex (9, 10); modeling the electron density map of the Mediator head module (11); and investigating the binding sites of ribosomal protein S1 to the 30S ribosome (12) and the general transcription factor TFIIF to RNA polymerase II (13).
Successful identification of cross-linked spectra from large datasets is a challenging database search task, as every cross-linked product contains two individual peptides covalently linked. Thus, the number of cross-linked products that are consistent with a given precursor mass grows quadratically with the size of the protein complex under investigation. Furthermore, product ion spectra of cross-linked peptides contain fragment ions from both of the individual peptides. We have noted that under collisional activated dissociation regimes, it is very common for these fragment ions to be unevenly distributed between the two individual precursor peptides; that is, the majority of peptide backbone dissociations take place on one of the two peptides that constitute the cross-linked product.
One way to address this issue is through the use of MS-cleavable cross-linking reagents (14–18). These cleavable reagents incorporate a low-energy bond such as an Asp-Pro peptide bond (15), a sulfoxide (16), or the stabilized amino group of a Rink linker (14), which are more readily cleaved than typical peptide backbone bonds, to produce the major fragments in MS2 spectra. Separate MS3 experiments of the two peptides are acquired, and each peptide can be identified separately, with known cross-linker-specific modifications, by regular peptide identification search engines. A concern with this approach is that MS3 is necessarily a less sensitive technique than MS2. Furthermore, these methods depend upon the two most intense MS2 products being consistently the two component peptides of the cross-link and successful MS3 of both of these ions. Also, it is currently not possible to selectively target only potential cross-linked components for MS3 analysis, so if MS2 and two MS3 spectra are acquired for every precursor, the number of components that can be analyzed in a run is compromised.
Several bioinformatic tools have been developed for cross-linking analysis, and unsurprisingly, these are mostly based on tools for the identification of linear peptides. These can broadly be classified into two strategies. The first is to create a database enumerating all recombined peptide pairs from the sample proteins joined by the appropriate cross-linker mass. This is queried analogously to a regular, linear peptide search, in which database entries within the tolerance of the precursor mass are assessed for how well their product ions match the theoretical MS2 products from the recombined peptides (9, 19–22). A challenge with this approach is that with a linear increase in the number of proteins considered, the number of cross-linked peptide permutations increases quadratically. Thus, prefiltering steps, such as a hard requirement for matching particular fragment ions, have been incorporated to try to reduce the number of permutations that need to be considered (20).
The second strategy is to initially search only for single peptides that have been tagged as potentially constituting half of a cross-linked pair. As described earlier, cleavable reagents allow separate fragmentation of each peptide. In a spectrum where fragments from both linked peptides are present, an analogous analysis approach is achieved bioinformatically by treating a cross-linked peptide complex as a single peptide bearing a large mass modification (23–25). Here the modification represents the second peptide plus the cross-linker bridge. Based on the specificity of the cross-linking reagent, the modification needs to be considered on only a limited number of residues; the most heavily used reagents target primary amines, so lysine side-chains and protein N termini are the sites of modification normally considered. These programs search against a regular protein database, with each constituent peptide identified separately, and a second computation step recombines individual peptide hits into cross-linked hits. For instance, Protein Prospector scans the mass of the variable modifications through a range of integers (plus a mass defect derived from averagine (26)) and regards peptide matches bearing large modifications (typically >400 Da) as potential cross-linked products.
For samples that contain only a few proteins, the identification of one peptide plus the mass of the second peptide may be enough for cross-linked product identification (23). Essentially, the second peptide can be identified by knowledge of its intact mass alone. However, in complex mixtures the second peptide must be identified by a product ion series as well. If a hypothetical cross-linked dipeptide product were to produce a complete fragment ion series from both peptides, the mass modification approach would produce two high-scoring matches to separate peptides that bore complementary mass modifications. That is, the mass modification on peptide 1 would correspond to the mass of peptide 2 plus the appropriate linker mass, and vice versa. This approach, as implemented in Protein Prospector, enables querying of large datasets against complex databases for cross-linked peptides.
Measuring the reliability of the reported results is a complex issue. Most recent efforts have utilized a concatenated target-decoy database searching strategy (25, 27), derived from the well-established approach used for identification of linear peptides (28). High-scoring, incorrect cross-linked product matches frequently correctly identify one of the two peptides, and thus are not random occurrences. However, these matches contain no meaningful information and must be treated as decoy matches. Thus, the number of decoy permutations considered is larger than the number of target permutations considered, and this skews the distribution of answers. Other studies have used a decoy cross-linker bridge mass rather than decoy peptide sequences (9, 13). However, it is also likely that the choice of decoy cross-linker mass significantly affects the size of the decoy database, as certain masses are very unlikely or not even possible, especially for shorter peptides. Finally, most studies have also compared the distances defined by the cross-linked residues to available crystallographic structures. Typically, the distances between Cα or Cβ atoms are measured, as the lysine side chains themselves are not rigid. However, in solution, proteins are more dynamic and can exist in more conformations than reflected by crystal structures measured in condensed states.
Here we formally show that collisional activation of cross-linked peptide products typically favors the formation of product ions from one of the two constituent peptides. Because generally the longer peptide dissociates more efficiently, a disproportionate amount of information identifies one half of the cross-link rather than the other. The challenge in identifying a cross-linked peptide spectrum is thus frequently a matter of assessing whether there are sufficient product ions matching the less confident of the two peptide matches. The amount of information necessary to identify the second peptide depends upon the complexity of the biological sample and thus the size of the sequence database searched. In experiments on binary protein complexes, when one peptide is confidently matched there may be only one potential second peptide that could account for the uninterpreted mass. However, large protein complexes containing dozens of proteins, or even cell lysates that contain thousands of proteins, will require more product ion signals to match the second peptide.
Most current algorithms only assess the reliability of cross-link matches as a score reflecting the match of the spectrum to the entire cross-linked dipeptide product. These scores are unable to differentiate between cases where both peptides are identified with high confidence and cases where one peptide is identified with very high confidence but the other is ambiguous. Hence, assessing the reliability of the lower confidence peptide identification, independent of how confidently the other peptide is identified, becomes essential. Recognizing this, some software employs an arbitrary cut-off based on a minimum number of fragment ions matched to the less confidently identified peptide (9, 13), whereas others recommend manual verification of results where one peptide is short (27). In these approaches there is no way to assess whether an optimal threshold is being employed, so they may be too conservative, too liberal, or too subjective.
Using the 500-kDa RNA polymerase II (pol II)1 enzyme complex as a testing ground, in the present work we demonstrate that the most reliable metrics for discriminating between incorrect and correct cross-linked peptide matches are scores reflecting the confidence of the worse of the two peptide identifications. As most cross-linking search engines report a score based on the total number of product ion matches regardless of their peptide of origin, Protein Prospector can more robustly assess the reliability of cross-linked peptide matches than current software, particularly when analyzing more complex samples. This is demonstrated by a reanalysis of two previously published datasets (25). Additionally, we assess the frequency of cross-linker specific diagnostic product ions resulting from cleavage at or near the lysyl-cross-linker bond (29, 30) and discuss the utility of determining separate false discovery rate (FDR) estimates for interprotein and intraprotein cross-link hits in large database searches (27).
EXPERIMENTAL PROCEDURES
Cross-linking of Pol II
RNA pol II was purified from Saccharomyces cerevisiae as previously described (11). 60 μg of pol II (determined by Bradford assay) was reacted with 1 mm disuccinimidyl suberate (DSS) (Thermo Scientific, San Jose, CA) added from 30x dimethyl sulfoxide stock in a final volume of 60 μl. The reaction mixture was incubated at 0 °C for 2 h and quenched by the addition of Tris Base to final concentration of 50 mm (added from 20x aqueous stock). Cross-linked samples were concentrated to a final volume of 25 μl and exchanged into 8 m urea, 10 mm triscarboxyethylphosphine using three spins on a 5-kDa cut-off Ultrafree MC centrifugal device (Millipore, Billerica, MA). After being heated at 50 °C for 20 min, the samples were cooled, alkylated with 20 mm iodoacetamide, and then diluted to 100 μl with 100 mm (NH4)HCO3 before 2 μg of side chain protected trypsin (Promega, Madison, WI) was added and the samples were digested at 37 °C for 7 h. The peptide digests were diluted to 500 μl with 0.3% TFA (aq) and applied to a C18 MacroTrap cartridge (Bruker-Michrom, Auburn, CA) at 100 μl/min, using the same buffer. Elution was accomplished with 0.3% TFA, 70% acetonitrile at a flow rate of 250 μl/min. The eluate was dried and resuspended in 500 μl of buffer A (10 mm NH4HCOO, pH 10) and then loaded onto a Gemini C18 1.0 × 100 mm, 3-μm (particle size), 110-Å (pore size) column (Phenomenex, Torrance, CA) at a flow rate of 75 μl/min. A linear gradient to 65% buffer B (50% acetonitrile, 10 mm NH4HCOO, pH 10) over 5 ml was applied while collecting 15 × 200 μl fractions. Fractions were dried on a vacuum centrifuge and brought up in 20 μl of 0.1% formic acid. Chromatographic steps were performed on an Akta Purifier 10 HPLC system (GE Healthcare).
Mass Spectrometry of Pol II Cross-links
Mass spectra were obtained on an LTQ-Orbitrap Velos (Thermo Scientific) coupled to a nanoAcquity UPLC system (Waters, Millford, MA). 10-μl cross-linked peptide fractions were loaded onto a Symmetry C18 180 μm × 20 mm, 5-μm (particle size) trap column (Waters) at 5 μl/min in 3% B (A = 0.1% formic acid (aq); B = 0.1% formic acid in acetonitrile) for 5 min. Peptides were eluted over a BEH130 C18 100 μm × 100 mm, 1.7-μm (particle size) column (Waters) via a linear gradient from 3%–27% B followed by washing at 50% B and re-equilibration at 3% B at a flow rate of 600 nl/min. The total run lengths were either 60 or 90 min depending on the anticipated level of complexity of each fraction. Both precursor and product ion signals were measured in the Orbitrap at 30,000 and 7500 resolving power, respectively. The six most intense ion signals in the precursor scan that were at least triply charged (as determined by the instrument firmware) were selected for HCD activation using a 3 m/z isolation window and a normalized collision energy of 30. Precursor ions were excluded from further selection for 30 s.
Protein Prospector Search Algorithm and Scoring Results
Cross-linked peptide identification in Protein Prospector is performed by mass modification searching (31), which considers a modification within a specified mass range (for the searches in this study, this range was typically from m/z 400 to m/z 5000) where the modification can occur only on internal lysines (i.e. that are a missed tryptic cleavage site) or on the protein N terminus. For each spectrum, typically the top 1000 peptide identifications are saved, although this is a user-adjustable parameter. The software then pairs together results in which the mass modification on one peptide corresponds to the mass of the second peptide plus the mass of the cross-linker. Many metrics are reported about each potential cross-linked peptide identification, including scores (score is based on number and types of fragment ions identified and is sequence and charge dependent (32, 33)) and expectation values for each peptide identification, as well as for the entire cross-linked product, and a score difference corresponding to how much better the cross-linked peptide identification scores compared with the top-ranked assignment to a single peptide. Only results in which the score difference was greater than 0 (i.e. the cross-linked peptide match was better than a single peptide match alone) were considered. The expectation values are calculated based on matches to single peptides and so should be treated as another score, rather than a rigorous measure of probability.
Prospector considers a-, b-, and y-ions, along with water and ammonia loss ions, internal ions, and immonium ions for HCD data. It also considers the cross-linker-specific ions resulting from dissociation of the cross-linker-lysine amide bond (illustrated for DSS/ bis(sulfosuccinimidyl) suberate cross-links in Fig. 3). These consist of one fragment with the same mass as the individual peptide (which we refer to as a P ion), one peptide with the cross-linker fragment attached (PL ion), and one peptide with the cross-linker fragment modified by a tetrahydropyridine originating from the modified lysine of the other peptide (PLK ion). Protein Prospector assigns different weightings for ion types, depending on their frequency and their specificity. Weightings for ions not related to the cross-linker are derived from single peptide identifications from HCD data and were calculated similarly to those reported for electron transfer dissociation data using several thousand spectra as the reference (33). Weightings for cross-linker specific ions were calculated based on the frequency of their observation in cross-linked peptide spectra acquired in-house. These constituted only a few hundred spectra, so weightings were more stochastic in nature. Different weightings are used depending on whether the data are ion trap collision-induced dissociation, HCD, or electron transfer dissociation fragmentation data.
Fig. 3.
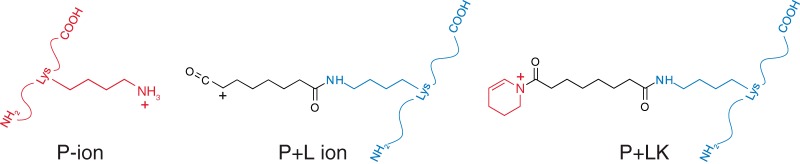
Structures of cross-link specific product ions derived from DSS and bis(sulfosuccinimidyl) suberate cross-linking reagents.
For high-resolution product ion spectra, the software determines charge states based on the spacing of isotope peaks. For each spectrum, the mass range of observed peaks is split in half (the same as for regular peptide identification using Protein Prospector (32)); then, after deisotoping, the most intense peaks in each half of the mass range are combined. The total number of peaks considered is a user-definable value within Batch-Tag. Cross-link searches typically work best with at least 70 product ion peaks considered. The detection of monoisotopic peaks and removal of isotopes in the product ion peaklist is by a “look back” approach. Starting with the highest m/z signal, the algorithm looks for isotope peaks (i.e. m/z 1.00, m/z 0.50, or m/z 0.33 lower for singly, doubly, or triply charged peaks, respectively) where the matched isotope is at least 20% the intensity of the current peak. If no peak matches the correct m/z spacing, the current signal is assumed to be the monoisotopic peak. Otherwise, the current peak is removed and the look-back step is repeated. Using this process, Protein Prospector will match fragments of any charge state up to that of the precursor ion, provided it is possible to determine the charge of the peak. All peaks without isotopes are assumed to be singly charged.
Analysis of RNA Pol II
Because highly charged precursor ions are more frequently misannotated in peak lists and cross-linked products have higher charges due to charge contributions from both peptides, and to allow for multiple precursors falling within the ion selection window, a hybrid method of peaklist generation was employed. Peaklists were initially generated using an in-house script, PAVA (34), based on the Raw_Extract script in Xcalibur v2.4 (Thermo Scientific). In parallel, Hardklor v1.35 (35) was used to deisotope precursor ion scans from the raw data. Then, for a given product ion spectrum in the PAVA peaklists, each Hardklor-determined monoistopic ion that fell within 3 m/z units of the nominal precursor was annotated as a separate spectrum in the final MGF format peaklist produced by an in-house Ruby script. Hardklor and PAVA agreed on a monoisotopic precursor ∼50% of the time. Product ions were not deisotoped at this stage, as Protein Prospector handles this task. This procedure resulted in 111,893 spectra from 16 high-pH HPLC-separated fractions.
85 peaks from each spectrum were searched using a tolerance of 10 ppm for precursor ions and 25 ppm for product ions. Enzyme specificity was tryptic, and up to four missed cleavages per peptide were allowed. Carbamidomethylation of cysteines was specified as a constant modification, and oxidation of methionine, pyro-glutamate derived from peptide N-terminal glutamine, protein N-terminal methionine removal and/or acetylation, and dead-end modification with the cross-linker (where one end reacts with a primary amine in the peptide but the other is hydrolyzed) were set as variable modifications. The database searched was a custom database containing the sequences of 52 components of the S. cerevisiae preinitiation complex, including all of the subunits of pol II, Mediator, and the general transcription factors. Additionally, each of the 52 preinitiation complex sequences was randomized 10 times, and these sequences were concatenated to the forward sequences. Thus the final database contained the 52 target sequences in addition to 520 decoy sequences.
Spectra were annotated as potential cross-linked products by Prospector based on a total score of the cross-linked product of >20 and a score difference > 0. If either of the two component peptides matched to the decoy database, then the identification was classified as a decoy. Exploratory data analysis was performed to determine which Prospector metrics had the most discriminatory power between target and decoy hits (see “Results” section). Furthermore, spectral matches to cross-linked peptides were randomly split into a test set and a training set. A support vector machine (SVM) classification model was built on several Prospector parameters and evaluated on the test set, using the e1071 package for the R platform. Models were evaluated based on their specificity (proportion of decoy hits correctly classified) and the total number of hits to the target database that were classified as such. The final model combined two parameters (“score difference” and “% TIC matched”) in a linear SVM model that was applied to the entire dataset to generate the final SVM decision value classifier.
The best cross-link match to each spectrum was kept if the SVM score was 0.3 greater than the second hit. Otherwise the spectrum was marked as ambiguous and all possibilities within this score range were annotated in the final list. This situation applied almost exclusively to ambiguous site localizations as to the exact lysine that was modified. Cross-links were next sorted by the position and identity of the two adducted lysine residues (without regard to peptide sequence), and only the best-scoring match was kept. The final list was manually inspected for correct precursor annotation.
Analysis of UTP-B Complex and E. coli Soluble Proteome
Raw data were kindly supplied by the authors of the pLink study (25). Raw data were converted to peaklists using an in-house script, PAVA (34). Data were searched with an instrument setting of ESI-Q-hi-res. UTP-B data were searched against a concatenated database of seven proteins (the six yeast proteins in the complex and pig trypsin) plus sequence-randomized versions of these entries (i.e. the database contained 14 entries). E. coli data were searched against two different databases. The first was a concatenated database of all E. coli proteins in the March 2012 release of Swiss-Prot plus sequence-randomized versions of these (a total of 11,934 protein entries were searched). A second set of searches were performed against a list of protein accession numbers identified in the sample on the basis of unmodified peptides, plus sequence-randomized versions of these entries, for a total of 1512 entries. Searches of 72,721 spectra against all E. coli entries took roughly 1 day on a 2.66-GHz eight-core processor, whereas the restricted accession number searches took just over five hours.
Peaklists were searched with a ±20-ppm mass tolerance on precursor ions and ±25 ppm for fragment ions. The top 35 peaks from each half of the m/z range (70 peaks total) of each spectrum were used for searching. Modifications were specified as above. Additionally incorrect monoisotopic peak assignments (where the mass in the peak list corresponded to the second or third isotope of the peptide) were considered as variable modifications. When incorrect monoisotopic peak identifications are reported, Prospector requires this modification in both of the individual peptide identifications of the cross-linked product. Results were filtered to remove any assignments where one of the cross-linked peptides was less than four amino acids long.
The classification score used in this analysis was S.D. − pep2.pExp, where S.D. is the difference in score between the cross-linked result and the best match to a single (non-cross-linked) peptide, and pep2.pExp is the log of the expectation value of the least confident peptide identification. Thus, if the cross-link match scored 10 more than the best single peptide match and the less confident peptide was identified with an expectation value of 0.01, then the score would be: 10 − (−2) = 12.
If either peptide matched to the decoy database, the cross-link hit was classified as decoy. FDR values for intraprotein and interprotein cross-link hits were calculated separately and compared with global FDR estimates. The decoy database contained sequence-shuffled versions of each protein in the target database. If a match to target and shuffled versions of the same protein was reported, this was interpreted as a decoy intraprotein identification. See the supplementary material for further discussion of FDR calculation and assessment for cross-linking analysis.
The two datasets analyzed here both employed isotope-labeled bis(sulfosuccinimidyl) suberate as the crosslinker, with a light version and a heavy version in which four hydrogens were replaced with deuterium atoms. The data were searched twice, once assuming the light cross-linker and once assuming the heavy cross-linker. Results were then combined using Prospector's Search Compare program, followed by removal of the lower confidence match for a particular spectrum when the two searches both produced an assignment.
RESULTS
Cross-link Backbone and Diagnostic Product Ions Are Asymmetrically Distributed between the Component Peptides
Protein Prospector's complementary mass modification algorithm was used to search over 111,000 HCD spectra from DSS cross-linked RNA pol II against a database consisting of 52 sequences of the yeast preinitiation complex plus 520 decoy sequences (10 randomized versions of each target sequence). This resulted in 9204 spectral matches to potential cross-linked peptides with an overall Prospector score > 20. Of these, 3885 matched both peptides to the target database, and the remaining 5319 matched at least one peptide to the decoy database and were therefore regarded as decoy matches. Approximately one-third of the cross-linked spectral matches were unique identifications, and the rest matched to two to four cross-linked sequences (these are typically highly related). Linear peptides are discovered in the same search but reported separately. For the analyses presented here, only cross-linked spectral matches were considered.
Protein Prospector calculates a number of metrics describing the quality of the spectral match to the cross-linked sequences. Among these are scores that reflect the number of observed product ion signals matching the database sequence. Prospector reports scores for each of the component peptides of the cross-link, as well as an overall score that describes how many total product ions are attributable to the entire cross-link. These scores are weighted by the overall frequency of a particular product ion type using the same fragmentation method (33).
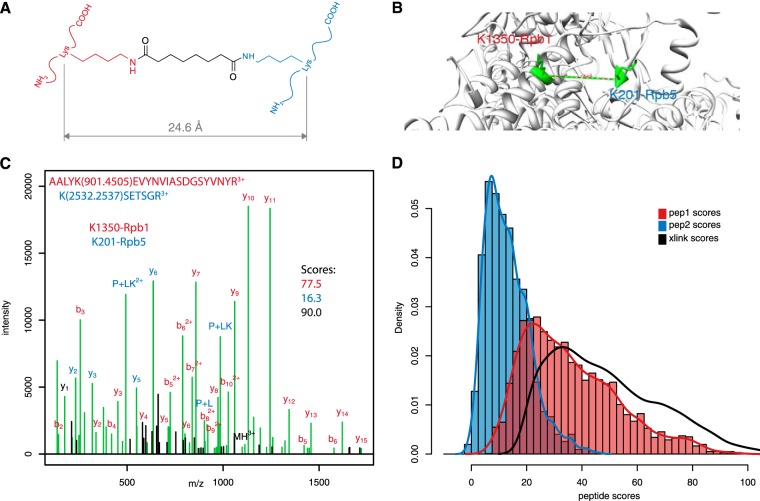
Fig. 1C shows a high-quality product ion spectrum matched to a DSS cross-linked pair. In this case, the cross-link spanned K1350 of the Rpb1 subunit to K201 of the Rpb5 subunit of Pol II, which is consistent with both the geometry of the cross-linking reagent (Fig. 1A) and the crystal structure of the enzyme complex (Fig. 1B). As is frequently observed, one peptide of the pair scored much higher (77.5) than the other (16.3). Prospector assigned an overall score to the cross-link of 90.0. Thus, one peptide accounts for much more of the observed fragment ions than the other. This situation was found to apply to the entire dataset. Fig. 1D shows the distribution of scores for the more weakly fragmented peptide of a pair (peptide 2) compared with the more strongly covered peptide (peptide 1), as well as the overall cross-link score for the 3885 matches to the target database. It is clear that these distributions are different (p ≪ 0.001) and that the distribution of overall cross-link scores (black line) more closely reflects the distribution of the higher scoring peptide, with only minor contributions from the lower scoring peptide.
Fig. 1.
HCD product ions from DSS cross-links are unevenly distributed between the two component peptides. 3885 cross-link spectral matches from RNA pol II analysis matched by Protein Prospector. A, structure of DSS cross-link illustrating maximal Cα distance. B, Cα distances measured against PDB:1WCM. The cross-link between K1350.Rpb1 and K201.Rpb5 spans 14.0 Å. C, example spectrum identifying the cross-link in B and illustrating the origin of the product ions. The more highly covered peptide (in red) scores 77.5, whereas the weaker peptide match (blue) scores 16.3. The overall score assigned to the entire cross-link is 90.0. The numbers in parentheses are the mass modification matched by Prospector. Unmatched ion signals are in black. For this spectrum, 87.5% of the TIC was matched. D, distributions of individual peptide scores for the more confident (red) and less confident (blue) peptides. The distribution of overall cross-link scores is shown in black.
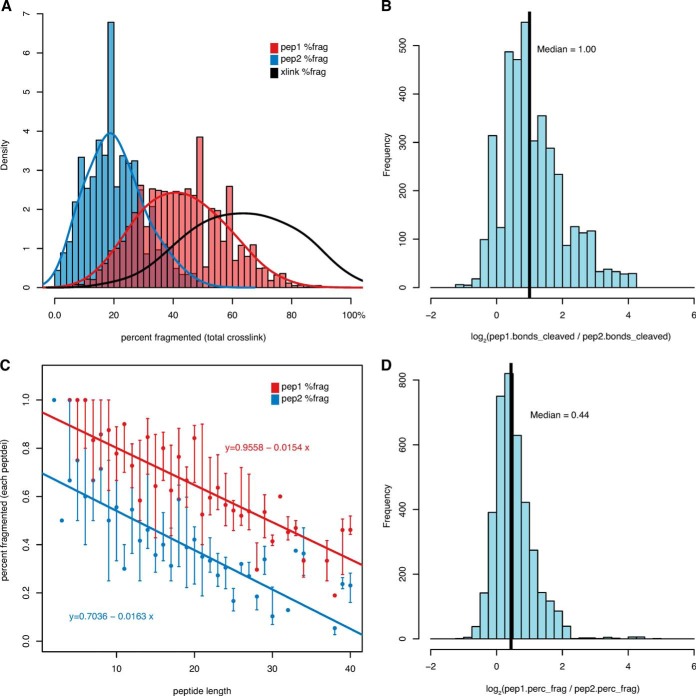
We determined the number of peptide bonds cleaved for each cross-link match by counting whether at least one b- or y-ion corresponding to a given bond position was observed. This revealed that on average, peptide 1 accounted for twice as many bond cleavages relative to the total number of peptide bonds in each cross-link (Figs. 2A and 2B). We next examined whether this trend could be explained solely by differences in the lengths of the component peptides or whether the two halves of the cross-link fragment had different efficiency. The more confident peptide (peptide 1) was typically 37% longer than its counterpart (supplemental Fig. S1). Therefore, the 2-fold increase in observed fragment ions was not fully explained by the 37% increase in peptide length.
Fig. 2.
The number of bonds cleaved from DSS cross-links are unevenly distributed between the two component peptides. 3885 cross-link spectral matches from RNA pol II analysis matched by Protein Prospector. A, the higher scoring peptide (red) contributes twice as many cleaved bonds as the lower scoring peptide (blue) relative to the number of bonds in the crosslink. B, for each cross-link, peptide 1 contributes twice as many cleaved bonds as peptide 2 (median log2 ratio = 1.0). C, the percentage of bonds cleaved within each peptide has an inverse relationship to peptide length, with peptide 1 fragmenting 36% more efficiently at a given length. D, within each cross-link, peptide 1 fragments 35% more efficiently than peptide 2 (median log2 ratio = 0.44). Percent fragmented figures in D were corrected for peptide length using the slope of the regression line in C.
Percent fragmentation, defined as the number of observed bond cleavages divided by the number of bonds in each component peptide, was calculated for each cross-link match. An inverse relationship between fragmentation and peptide length was observed for both peptide 1 and peptide 2 (Fig. 2C). Linear regression of these data showed essentially parallel trends, but with peptide 1 fragmenting consistently more efficiently for a given length than peptide 2. Peptide 1 was estimated to fragment 36% more effectively based on the trend lines. Adding a correction to compensate for the effect of length on fragmentation and then comparing the ratios of corrected fragmentation efficiency for the two peptides within each crosslink also gave a median ratio corresponding to 35% increased efficiency in the fragmentation of peptide 1 (Fig. 2D). Thus the 2-fold difference in observable fragments is mostly accounted for by a combination of increased peptide length and increased fragmentation efficiency of the higher scoring peptide (137% * 135% = 185%).
In addition to cleavages along the peptide backbone, Protein Prospector scores cross-linker specific product ions from the modified lysine residues (Fig. 3). These dissociations take place either at the amide bond joining the lysine ε-amine to the cross-linking reagent (resulting in ions that we refer to as P ions and PL ions) or at the peptide bonds joining a modified lysine to adjacent residues (29, 30). This second process, which is similar to the formation of lysine immonium ions, results in tetrahydropyridine cross-linked to the other peptide (termed PLK). These dissociations are common among DSS and bis(sulfosuccinimidyl) suberate cross-linked peptides. Of the 3885 spectral matches to the target database, 71.9% matched at least one of these diagnostic ions (Table I).
Table I. Frequency of diagnostic P, PL, and PLK ion signals in cross-link spectral matches.
| 0 diagnostics | At least 1 P, PL, PLKa | Pa | PLa | PLKa | |
|---|---|---|---|---|---|
| Peptide 1b | 2937 | 948 | 244 | 637 | 599 |
| Peptide 2b | 1388 | 2497 | 279 | 1671 | 2240 |
| Per cross-link | 1093 | 2792 | 464 | 1954 | 2532 |
a Number of peptides matching at least one of these ion types.
b Peptide 1 is defined as the higher scoring of the two peptides comprising a cross-link, and peptide 2 is the lower scoring of the pair.
As with regular y- and b-ions resulting from peptide backbone dissociations, these cross-link specific product ions were unevenly distributed between the two peptides of a cross-linked pair, but in a manner opposite that of backbone ions: diagnostic ions were more common for the lower scoring peptide than for the higher scoring peptide. 64.3% of the cross-linked matches had at least one diagnostic ion matching the lower scoring peptide, whereas only 24.4% had at least one of these ions originating from the other peptide. PLK type ions were the most common type (65.2% of cross-linked matches contained at least one), followed by PL ions (50.3%), and then P ions (11.9%).
The Lower Scoring Peptide Is the Best Classifier of Cross-linked Spectral Match Reliability
The metrics reported by Protein Prospector describing the quality of the spectral match were assessed for their ability to discriminate between target and decoy hits. As mentioned, of the 9204 spectra annotated as cross-link matches, 3885 had both peptides matched to the target database, and the remainder were classified as decoy hits. Hits to the target database consist of both correct and incorrect matches, whereas essentially all hits to the decoy database can be considered incorrect. So as to better model the distribution of incorrect cross-linked matches, the decoy database searched was 10 times larger than the target database, consisting of 520 randomized versions of preinitiation complex protein sequences (versus 52 in the target database).
The distributions of 18 Prospector parameters were compared between hits to the target and decoy databases (Table II, supplemental Fig. S2). These parameters included Prospector score for each peptide and for the entire cross-linked product, expectation values for each of these, length of the peptides, rank of the individual peptide hits in the mass modification search, and percentage of the product ion intensity explained by the cross-linked assignment. The Prospector parameter “score difference,” representing the difference in score between the top cross-linked match and the top linear peptide match, was the most strongly correlated with a hit to the target database, having a point biserial correlation coefficient of 0.544. This was followed by metrics describing the quality of the lower scoring peptide (peptide 2)—“pExp” (−log10 of the expectation value for this peptide), “score” (calculated after the first mass modification search and before the elemental composition of the other peptide is determined), and “low score” (calculated after putative cross-linked products have been assigned and elemental composition is known)—which had correlation coefficients above 0.430. Additionally, the expectation value for the total cross-match (“XL pExp”) had a strong association with matches to the target database.
Table II. Effectiveness of Protein Prospector metrics as classifiers of target versus decoy cross-link matches.
| Parameter | r coefa | # Targetb | Specificity | Threshold |
|---|---|---|---|---|
| SVM dvalc | 0.581 | 2349 | 0.927 | 0.0 |
| scorediffd | 0.544 | 2258 | 0.923 | 8.5 |
| pep2.pExpe | 0.441 | 1405 | 0.926 | −0.2 |
| XL pExpf | 0.439 | 1045 | 0.926 | 9.9 |
| pep2.score | 0.439 | 1621 | 0.925 | 13.6 |
| Low score | 0.430 | 1785 | 0.924 | 15.9 |
| XL score | 0.408 | 1122 | 0.926 | 58.3 |
| % TIC matchedg | 0.390 | 1092 | 0.926 | 79.6% |
| pep2.norm_scoreh | 0.297 | 733 | 0.926 | 2.4 |
| pep1.pExp | 0.280 | 425 | 0.924 | 9.2 |
| pep1.score | 0.275 | 532 | 0.925 | 55.3 |
| pep1.norm_score | 0.143 | 342 | 0.924 | 4.7 |
| pep1.lengthi | 0.104 | 375 | 0.926 | 24 |
| ppm | 0.086 | NA | NA | NA |
| z | 0.067 | NA | NA | NA |
| mz | 0.004 | NA | NA | NA |
| pep2.length | −0.024 | NA | NA | NA |
| pep1.rankj | −0.204 | NA | NA | NA |
| pep2.rank | −0.308 | NA | NA | NA |
a Point biserial correlation coefficient between Prospector metric and matches to the target database.
b Number of spectral matches classified as positive cross-link hits at the given score threshold, chosen to achieve equal specificity (see text).
c Score of final SVM classifier. Model was trained as described in the text; reported here is the result of the final classification.
d Difference in score between the top cross-linked match and the top linear match.
e −log10(Exppep2), where Exppep2 + expectation value of weaker peptide.
f −log10(ExpXL), where XL refers to the complete cross-link.
g Percentage of total ion current intensity in the peaklist that can be explained by the cross-link.
h Score normalized by length of peptide.
i Length of peptide in amino acids.
j Rank of individual peptide match (e.g. rank 1 indicates top match to spectrum).
Prospector searches for cross-linked peptides by examining the top 1000 linear peptide matches in a mass modification search for complementary modification masses. For cross-linked assignments, very frequently, one of the component peptides is the top-ranked hit in the linear search, and it is nearly always ranked within the top 10 (median rank of better scoring peptide is 1 and 75% of all cross-link matches have the peptide ranked ≤ 2). Thus, in practice, the “score difference” parameter is also a measure of how much peptide score is attributable to the less confident peptide assignment. Score difference and the score of peptide 2 are in fact highly correlated parameters (r = 0.766) (supplemental Fig. S3).
The target-decoy strategy allows modeling of the distribution of incorrect hits and provides a means to estimate the specificity of a given scoring regime (e.g. the number of decoy hits that are correctly classified as decoy). However, because hits to the target database consist of both incorrect and correct matches, it is more problematic to estimate the sensitivity. To evaluate the performance of different scoring models, thresholds for each parameter were chosen such that the specificity of the analysis was as near 0.925 as possible. The total numbers of hits to the target database were compared at these levels. Table II again demonstrates that “score difference” was the most effective single measure of cross-linked assignment quality. A score difference threshold of 8.5 classifies 92.5% of the decoy database hits as incorrect and classifies 2258 target database matches as correct. All of the metrics reflecting the score of the worse peptide identification (peptide 2) were better classifiers than metrics reflecting the better peptide match (peptide 1) or the entire cross-linked product, giving greater numbers of target database matches at equal specificity thresholds. For instance, XL score, the best classifier based on the whole cross-link, annotated 1122 hits as correct at the 92.5% sensitivity level. Therefore, score difference predicted over twice as many cross-links as XL score at the same specificity level.
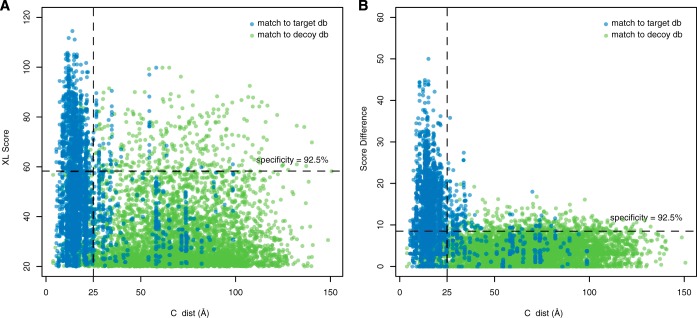
To assess whether this increase in positive outcomes was due to incorrectly classifying matches from the target database, we examined the Cα distances of the cross-linked lysine residues against the crystal structure of the 12 subunit pol II complex (pdb:1wcm (36)). Fig. 4 plots the Lys-Lys distances for all target database matches that were resolved in the crystal structure, as well as all the decoy database hits, which were assigned random distance values sampled from the set of all possible Lys-Lys distances in pol II. 92.3% of the hits with score difference ≥ 8.5 (the 0.925 sensitivity level) had interlysine distances less than the 25-Å span of DSS. The crystal structure contains some positional uncertainty and the actual protein assembly is a dynamic entity in solution. Thus, even more of these hits were likely correct (98.3% were within 35 Å). Most of the additional positive hits classified by the score difference metric were thus probably true positives. Furthermore, score difference is much better at discriminating between positive and negative outcomes than XL score or other measurements matching the fit to the entire cross-linked product, which are the types of scores used by other cross-linking analysis software.
Fig. 4.
The Protein Prospector parameter “score difference” is the best single classifier of cross-link spectral matches. Cross-linking of RNA pol II by DSS resulted in 2535 spectral matches to the target database, which also corresponded to measurable Cα distances on the crystal structure (blue). 5319 hits to the decoy database (green) were assigned random distance measurements sampled from the set of all possible Lys-Lys distances on the PDB:1WCM structure. The vertical line indicates 25 Å, the nominal span of DSS. The horizontal line indicates a score threshold that classifies 92.5% of decoy matches correctly. A, classification based on XLScore misclassifies many seemingly correct cross-link identifications (those with distance < 25 Å). B, classification based on score difference results in better discrimination between target and decoy matches and leaves fewer apparently true positives misclassified.
Analysis of RNA pol II Cross-link Sites
The 9204 cross-link spectral matches to RNA pol II were split into equally sized training sets and test sets. Linear SVM classification models were built using multiple combinations of Protein Prospector metrics and evaluated similarly to the methods described for single variable classifiers above. The major difference was that the SVM models were trained on half of the data, and the number of positive classifications was evaluated on the other half of the dataset at a specificity of 0.925. SVM models were evaluated in this way for different combinations of two and three Prospector parameters at different cost and tolerance values. Few models offered much better performance than simply using score difference as a stand-alone classifier. The final classification score was built on Prospector parameters “Score Difference” and “% TIC matched” (supplemental Fig. S4). This model classified 2349 cross-linked spectra from the target database at a sensitivity of 0.927 versus 2258 cross-links classified using score difference as a stand-alone classifier (Table II). Three-parameter models offered no improvement over two-parameter models. Normalizing the number of decoy hits by a factor of 10 to account for the increased relative size of the decoy database led to an estimated FDR of 1.6%.
The utility of the SVM model is demonstrated in Fig. 5. This shows two cross-linked spectra with high and low (but both positive) SVM decision values. In one case, the cross-link is matched by comprehensive backbone fragmentation of both peptides but has a large unmatched peak. This results in a score difference of 42.1, but only 58.3% of the ion signal intensity is matched. However, the low percentage match to the TIC is not sufficient to classify the spectrum as negative in the SVM model. In the second case, despite matching most of the y-ions from the weaker peptide, the score difference for this match is quite small (5.8), as it is a short peptide and the y1 ion is in common between both peptides. Using score difference as a stand-alone classifier would result in a negative classification for this spectrum. However, because most of the ion intensity is matched (85%), the final SVM score classifies this hit as correct, thereby rescuing it. This spectrum is assigned to a crosslink between K201 on Rpb5 and K941 on Rpb1 corresponding to a distance measurement of 13.2 Å, and is therefore likely correct.
Fig. 5.
Examples of cross-linked spectra from RNA pol II with (A) high SVM decision value (6.8) and (B) low SVM decision value (0.1). The number in parentheses represents the mass modification identified by Protein Prospector, which corresponds to the complementary peptide plus the cross-linker bridge. The “score difference” parameter is the difference between the overall score for the cross-link (in black) and the best scoring linear peptide linear hit, which in both of these cases is equal to the score in red of the strong peptide. The spectrum in A has a high score difference of 42.1 but matches only 58.3% of the ion intensity due to a large unmatched signal. The spectrum in B has a low score difference (5.8) but matches 85.0% of the ion intensity. The SVM classifier integrates both of these parameters to rescue hits that would otherwise be misclassified.
The 2349 cross-linked spectral matches were then filtered for redundancy. Keeping only spectral matches with an SVM decision value at least 0.3 points greater than the next highest hit and accounting for redundant cross-links (defined by the positional numbers of the adducted lysine residue pair) led to 157 positionally unique sites of cross-linking within the pol II complex (supplemental Table S1, sheet 1). An additional 53 cross-links could not be unambiguously localized to a single lysine (supplemental Table S1, sheet 2), although many of these sites were redundant with sites from the unambiguous list.
Of the unique cross-linked position combinations, 112 corresponded to distances that could be measured on the pol II crystal structure (36). The vast majority of these were consistent with both the geometry of the DSS cross-linking reagent and the expected distances from the crystal structure. 99 of 112 corresponded to measured Cα distances of less than 25 Å, and 107 of 112 measured less than 35 Å. Furthermore, the overall distribution of measured Cα distances was different from the distribution of all Lys-Lys distances (supplemental Fig. S5).
Comparison of Protein Prospector to Other Software
The performance of Protein Prospector cross-link searching was compared with two previously published analyses: cross-linked UTP-B complex and cross-linked E. coli whole cell lysate (25). The UTP-B complex contains six proteins (37). Searching cross-linking data from this complex with Protein Prospector and employing a global 5% FDR threshold led to the reporting of 77 unique cross-links. Calculating separate thresholds for intraprotein and interprotein results led to 58 intraprotein matches and 26 interprotein. Thus, using these results, 84 unique cross-links were discovered by Protein Prospector with an estimated FDR of 5% (supplemental Table S2). Calculating separate intra- and interprotein crosslink FDRs resulted in six extra intraprotein results and one extra interprotein result.
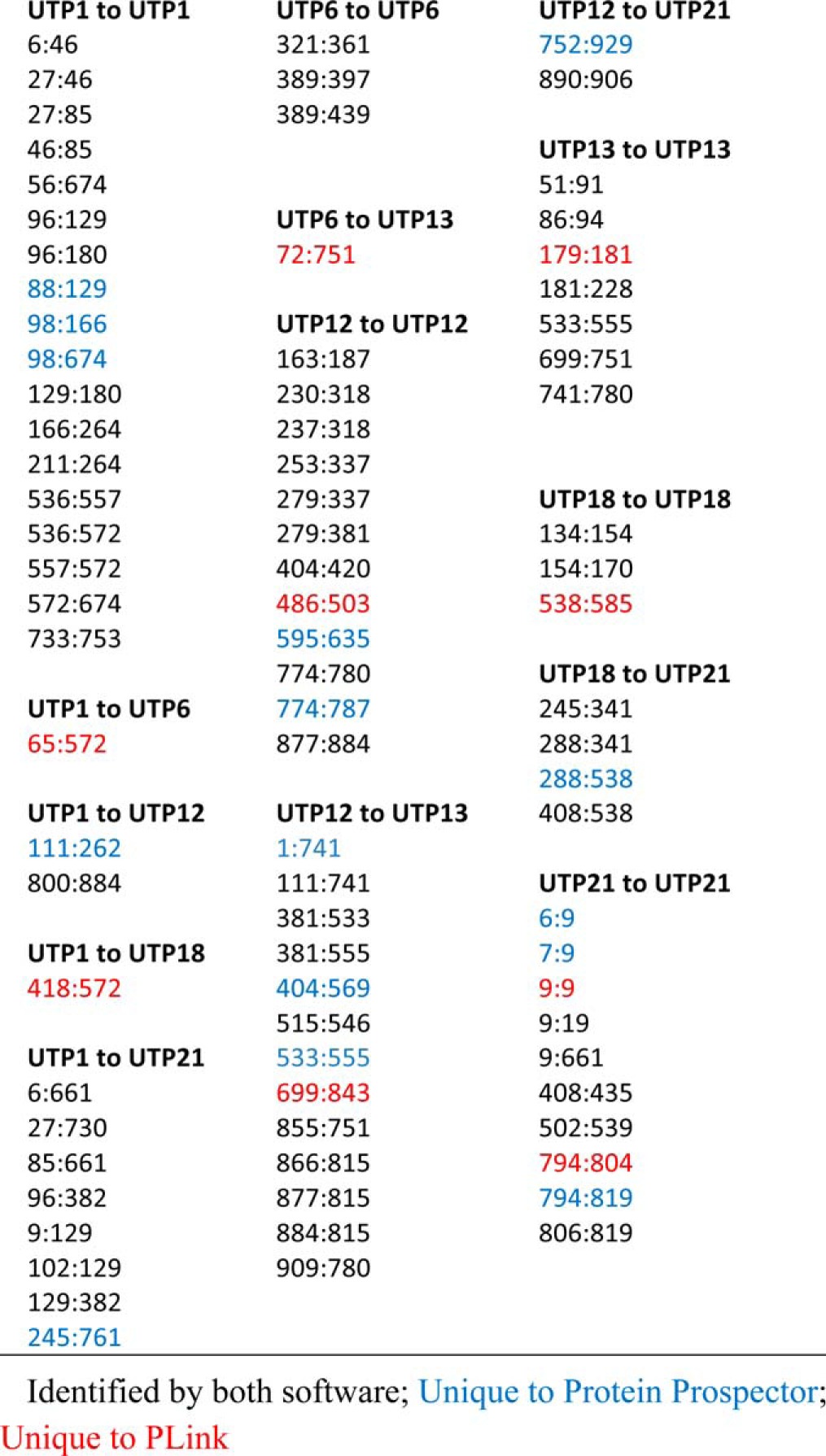
In a previous publication analyzing this data, pLink reported 71 high-quality and a total of 78 cross-links from the same data (25). Table III presents a comparison of the cross-linked residue combinations identified by Protein Prospector and pLink. The overlap in identifications was 67, showing good agreement between the two types of software. pLink reported single cross-link identifications between five pairs of subunits (UTP1:UTP6, UTP1:UTP12, UTP1:UTP18, UTP6:UTP13, and UTP12:UTP21). Protein Prospector found an additional cross-link for two of these (UTP1:UTP12 and UTP12:UTP21), whereas for the other three, Protein Prospector did not report any significant assignments. Thus, the Protein Prospector results increased the confidence in two of the direct protein interactions while flagging the other three as likely to be incorrect. Indeed, in the manuscript accompanying these results, two of the three cross-links in question were described as being of low confidence, and the other as mid-confidence, when the authors examined the spectra manually.
Table III. Cross-linked residues identified between members of the UTP-B complex.
The first major publication to describe large-scale cross-linking software utilized a dataset consisting of cross-linked E. coli whole cell lysate (20), and this same study design has been used since in other software-development efforts (22, 25). An E. coli whole cell lysate cross-linking dataset was created to assess pLink performance (25), and the present study used the same raw data to benchmark Protein Prospector performance. Assessing interprotein and intraprotein results together, Protein Prospector reported 195 unique cross-link identifications at an estimated 5% FDR, the value used in the pLink study. However, examining only intraprotein results, Protein Prospector reported a total of 406 intraprotein matches, with one decoy intraprotein identification. Thus, the estimated FDR for the intraprotein results was 0.2%. Among the interprotein matches, only eight results for the target portion of the database scored higher than the first decoy match. Thus, with an FDR as close to 5% as possible, only 8 interprotein results and overall 414 unique cross-linked peptide identifications were reported by Protein Prospector (supplemental Table S2). In comparison, pLink reported 390 unique cross-links.
Fig. 6 shows Venn diagrams of the overlap in results for intra- and interprotein cross-links reported by the two programs. Reassuringly, the majority of cross-links reported by the two software programs are intraprotein, and for these results there is a respectable overlap in identifications, although Protein Prospector reported significantly more intraprotein assignments. Of the 44 intraprotein matches unique to pLink, 9 were peptides reported linked to themselves through the same residue in each peptide. In this situation, it is unlikely that any fragment ions were specific for identifying the second peptide (as all single bond cleavages could be explained as a fragment from the first peptide). Therefore, these matches were almost certainly based on precursor mass alone, and are of questionable reliability.
Fig. 6.
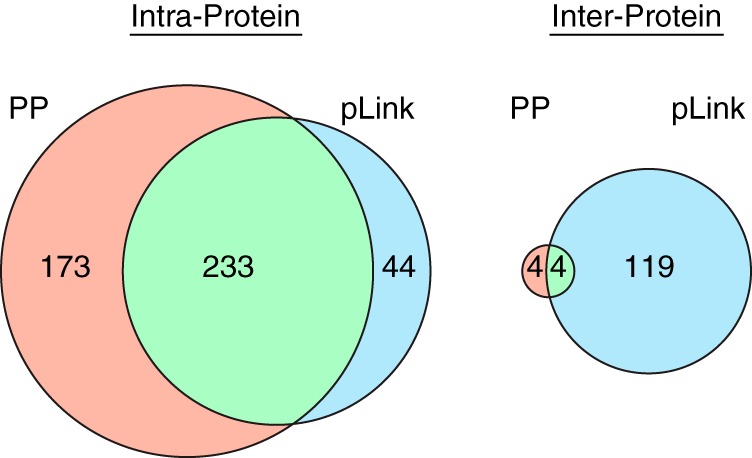
Overlap in cross-linked peptide identifications between Protein Prospector (PP) (pink) and pLink (blue). There is high overlap in identifications for intraprotein matches, although Protein Prospector reports many more. There is some overlap in identifications of interprotein cross-links, but pLink reports dramatically more hits than Protein Prospector.
However, for the interprotein matches there was a dramatic difference in the number of matches reported, with pLink reporting roughly 15 times more identifications than Protein Prospector. This raises the question of whether pLink is too liberal or Prospector is too conservative in its assessment of FDR. Half of the eight Protein Prospector results were also reported by pLink. Table IV lists the eight interprotein identifications reported by Protein Prospector. The pLink study reported that 21 out of the 123 interprotein cross-links corresponded to a protein complex that had a structure in the PDB. Furthermore, 10 of these 21 mapped to Cα distances within the range of the cross-linker. Interestingly, the four interprotein cross-links that were agreed upon by both software programs included 3 of these 10 cross-links that were validated by PDB measurement. Thus, the small overlap in interprotein cross-links reported is heavily biased toward results for which there is independent, corroborating evidence. The four interprotein matches unique to Protein Prospector included a second cross-link between aspartate carbamoyltransferase and aspartate carbamoyltransferase regulatory chain (in addition to the one reported by both programs), a cross-link between ribosomal RNA small subunit methyltransferase D and elongation factor Tu, which are known to interact (38), and two other cross-links between ribosomal subunits. Thus, there is independent evidence to suggest all of these results could be reliable. In contrast, of the 123 pLink interprotein results, 31 were supported by corroborating evidence from yeast two-hybrid, affinity purification mass spectrometry, or the existence of a structure in the PDB (25).
Table IV. Intraprotein identifications by Protein Prospector from E. coli dataset. Shaded identifications were also reported by pLink (14).
| Peptide 1 | Peptide 2 | Xlink Res 1 | Xlink Res 2 | Protein 1 | Protein 2 |
|---|---|---|---|---|---|
| WLGGMLTNWK*TVR | Acetyl-AHIEK*QAGELQEK | 105 | 6 | 30S ribosomal protein S2 | 30S ribosomal protein S5 |
| K*ILPDPK | K*AGFVTR | 11 | 100 | 30S ribosomal protein S7 | 30S ribosomal protein S9 |
| LLSLFK*LTETDQR | VDEIATDVDK*TPHAWYFQQAGNGIFAR | 34 | 280 | Aspartate carbamoyltransferase regulatory chain | Aspartate carbamoyltransferase |
| SSAQK*VR | K*PELDAK | 16 | 108 | 50S ribosomal protein L22 | 30S ribosomal protein S3 |
| ANDIALK*CK | TVHSLTQALAK*FDGNR | 137 | 179 | Aspartate carbamoyltransferase regulatory chain | Aspartate carbamoyltransferase |
| FWVESEK*R | K*NIEFFEAR | 44 | 42 | 50S ribosomal protein L28 | 50S ribosomal protein L9 |
| SSAQK*VR | K*HNASR | 16 | 19 | 50S ribosomal protein L22 | 30S ribosomal protein S20 |
| EK*VAGQVAYR | K*LLDEGR | 177 | 264 | Ribosomal RNA small subunit methyltransferase D | Elongation factor Tu 1 |
This same dataset was also searched against a restricted database constructed from a list of the protein accession numbers that were confidently identified by Protein Prospector based on the identification of unmodified peptides in these samples (756 proteins total). The results of this search identified 464 unique intraprotein matches, including 3 decoy intraprotein matches (estimated FDR 0.6%), and 10 interprotein matches that were more significant than the first interprotein decoy match (see supplemental Table S2). Thus, from this search, 474 unique cross-links could be reported at an FDR less than 5%, a 14% increase over the Protein Prospector full database search and a 22% increase over the pLink results.
DISCUSSION
The identification of a cross-linked peptide complex is based on the identification of two peptides. It is often the case that one of the peptides in the complex fragmented more extensively than the other in the tandem mass spectrum. The most typical result is an extensive y-ion series from one peptide and several cross-linker specific PL and PLK ions identifying the weaker peptide mass, along with a more limited y-ion series (Figs. 1 and 2, Table I). Note that although cross-linker ions help identify the less confident peptide, they result from dissociation of backbone amides in the more confidently identified peptide, further demonstrating the asymmetry of fragmentation. Although the longer peptide is generally the more extensively fragmented peptide, length alone is not enough to explain the difference in collision-driven bond dissociation.
The confidence with which one can identify the better fragmenting peptide should not contribute to the assessment of the reliability of identifying its partner peptide. Many current cross-linking studies are analyzing complexes consisting of only a few proteins. In these instances, if one peptide is identified confidently, then knowing the mass of the second peptide alone may be sufficient to identify it; this approach has previously been employed using Batch-Tag and MS-Bridge (23). In data searches with a 20-ppm precursor mass accuracy and allowing for up to two missed trypsin cleavage sites and the most common modifications, on average there will be a match to a given peptide mass in about 1 in 80 proteins, with low mass fragments more likely to be matched (e.g. about 1 in 70 proteins match a given peak around mass 800, whereas ∼1 in 100 will match a given peak around mass 3000). Thus, when searching the UTP-B dataset considering seven proteins (and decoy versions of these sequences), if one peptide was confidently identified, the mass of the second peptide alone was most of the time sufficient to match it. However, for the E. coli dataset, when considering 5967 proteins and their decoy sequences, there will have been on average 150 possible matches to the second peptide based on mass alone. Thus, if the evidence for matching the second peptide is not independently assessed, then many results will be reported where one peptide is correctly determined but the other identification is ambiguous and often wrong.
Given that in collisional dissociation, one peptide produces twice as many bond cleavages as the other, and that the more strongly fragmented peptide is nearly always correctly identified, it follows that most incorrectly identified cross-links come from misidentification of the other peptide. Thus, parameters reflecting only the score of the less confident peptide assignment should correlate well with correct cross-link identifications. Indeed, this was found to be the case in the present analyses, in which all metrics reflecting the quality of the match to the less confident peptide outperformed other metrics in their ability to discriminate correct cross-linked matches from incorrect matches based on both a decoy database strategy and comparison to the pol II crystal structure (Table II, Fig. 4).
The Prospector parameter “Score Difference” was found to be a particularly effective classifier. Although this is not explicitly a measure of peptide 2 score (the less confident peptide), given that more often than not peptide 1 is the highest scoring linear peptide hit, it ends up reflecting the score of peptide 2 closely, as shown in supplemental Fig. S3. Score difference can differ from peptide 2 score in several ways. Firstly, they differ when peptide 1 does not have the highest linear peptide match score. Secondly, if product ion peaks can be attributed to both peptides, then the score for that peak is counted only once, so score difference is a measure of additional peak matches rather than simply matches to peptide 2. Nevertheless, the improved classification performance of score difference relative to explicit measures of peptide 2 score is probably mostly due to the cases (<50%) in which peptide 1 is not the highest scoring linear hit. In these cases, score difference reflects the increased confidence in the cross-link assignment relative to other interpretations of the spectra. Furthermore, XL score is determined independently and is not simply the sum of the peptide 1 and peptide 2 scores. Thus there are technical differences that make score difference subtly different from peptide 1 score and “low score,” another measure of peptide 2 reliability.
Modest improvement in classification efficiency could be obtained by using the SVM supervised learning model to allow linear combinations of multiple metrics (Table II). The most successful SVM models combine score difference with one other Prospector parameter (typically either log10(ExpXL) or % TIC matched). However, score difference alone is an effective classifier, as shown by the striking difference in distributions of incorrect decoy hits when classified by total XL score versus score difference (Fig. 4).
The use of separate inter- and intraprotein FDR thresholds is also an efficient means of improving reliability. When Protein Prospector grouped inter- and intraprotein results together for calculating a 5% FDR from the E. coli results, it reported 10 target–decoy matches and no decoy–decoy matches. This suggests that possibly all of the target-decoy matches are not truly random—that is, that one of the peptide identifications is correctly matched. The estimated number of completely incorrect matches among the target–target results should equal the number of decoy–decoy matches; that is, there are probably no results where one of the peptide identifications is not correct, whereas the number of identifications where one peptide is incorrect should equal about 10. Using a global FDR threshold, there were 21 interprotein matches reported, and if 10 of these were probably wrong, that means there was a 48% FDR among these results. This highlights the danger of using a global FDR threshold and then assuming that specific subsets of data have the same level of reliability (39). As highlighted earlier, the top eight interprotein matches may have been correct based on other supporting information. If they were, then of the 13 additional interprotein matches reported when a global FDR was used relative to separate FDR thresholds, approximately 3 were true positives. Thus, when a global FDR is used very few extra interprotein matches are being discovered at the expense of losing 232 intraprotein cross-link matches. Although from a biological perspective intraprotein matches are generally less interesting than interprotein ones, this does represent a large loss of information, and it also allows a very high error rate in the data that is of most interest to researchers.
Protein Prospector, to the best of our knowledge, is unique among software in identifying both peptides from a single cross-linked product spectrum but independently assessing the reliability of the least confident peptide identification for setting an acceptance threshold. This is the most accurate measure of whether a reported cross-linked complex is reliable that we have found. Thus, one would expect it to produce more reliable results than alternative software, particularly for analyses of more complex mixtures, where more information about the second peptide is required for identification. The effect of this was stark in comparisons to pLink for analysis of the E. coli data. Prospector reported dramatically fewer interprotein cross-links than pLink, but many more intraprotein cross-links. We suspect that many of these extra interprotein cross-links reported by pLink were instances where one of the peptides was correctly identified, but not the other. The lack of experimental data to support most of these identifications, despite the existence of a high-quality database of E. coli interactions (40), adds weight to this argument.
The use of different thresholds for subsets of data within a single search has previously been employed. For example, this approach was used for estimating phosphopeptide and unmodified peptide FDR identification rates from within a single search (33). In this instance the reasoning was that when searching allowing for phosphorylation, the search engine considers many more phosphorylated peptides than unmodified, so the majority of the false identifications are to phosphopeptides. The justification employed here for separate assessment of inter- and intraprotein identifications was identical, and this approach has previously been used for cross-linked data analysis (27).
The combination of the reported Protein Prospector E. coli results and the above discussion about reliability portrays a disturbing prognosis for large-scale identification of interprotein cross-links in complex samples. One could argue that E. coli is not the best choice of organism for identifying large, multimeric protein complexes: it produces a lot of homomultimeric complexes in instances when, for example, human cells will produce a complex with different subunits to form more interprotein interactions. Changes to cross-linking protocols could improve the situation. It should be noted that isotope-labeled cross-linkers were employed in the two studies employed here to compare software. This was done by the data creators to allow comparison to the xQuest software, which only works with labeled cross-linkers (20). However, for both Protein Prospector and pLink there is no need for isotopic labeled linkers. Indeed, they actually make identification more difficult, as their use requires considering two types of cross-linkers for every spectrum, doubling the search space. It also creates two peaks in the MS spectrum for every cross-linked product, splitting the signal intensity for the cross-linked products in half and increasing the number of redundant MS/MS spectra of the same cross-linked product that are acquired, rather than selecting new products.
Producing tandem mass spectra in which both peptides are extensively fragmented will remove some of the ambiguity in results. In this respect, the use of electron transfer dissociation fragmentation instead of collision-induced dissociation could be important, and the development of new cross-linkers that benefit electron transfer dissociation performance may have an impact (41). The use of MS-cleavable cross-linkers should also better guarantee fragments from both peptides in MS3 spectra (14–18). Nevertheless, more targeted cross-linking analysis, in which a level of protein purification prior to cross-linking is employed, is likely to be the most effective approach: if a sample can be defined as containing only tens or even hundreds of proteins, then the lower confidence peptide identification is greatly simplified.
In summary, Protein Prospector is very robust software for analyzing cross-linking data of varying complexity and outperforms equivalent tools, especially when the number of proteins that need to be considered expands. However, because of the low amounts of cross-linking that are generally achieved in a complex heterogeneous mixture, questions are raised as to whether this is a sensible experimental approach; more focused studies are likely to provide significantly more useful biological insight.
Protein Prospector is freely available online. Access to annotated spectra for all spectral assignments in this manuscript is described in the supplementary material.
Supplementary Material
Acknowledgments
We thank Drs. Meng-Qiu Dong and Si-Min He for access to the raw data used for software comparison in this manuscript.
Footnotes
Author contributions: M.J.T., A.L.B., and R.J.C. designed research; M.J.T. and R.J.C. performed research; M.J.T., P.R.B., P.J.R., and R.J.C. contributed new reagents or analytic tools; M.J.T. and R.J.C. analyzed data; M.J.T., P.R.B., and R.J.C. wrote the paper.
* This work was supported by the Biomedical Technology Research Centers program of the NIGMS, National Institutes of Health (8P41GM103481).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- pol II
- RNA polymerase II
- DSS
- disuccinimidyl suberate
- FDR
- false discovery rate
- HCD
- high-energy collisional dissociation
- MS
- mass spectrometry
- PDB
- Protein Databank
- SVM
- support vector machine
- TIC
- total ion current
- UTP
- U-three protein.
REFERENCES
- 1. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rappsilber J. (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stengel F., Aebersold R., Robinson C. V. (2012) Joining forces: integrating proteomics and cross-linking with the mass spectrometry of intact complexes. Mol. Cell. Proteomics 11, R111.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murakami K., Elmlund H., Kalisman N., Bushnell D. A., Adams C. M., Azubel M., Elmlund D., Levi-Kalisman Y., Liu X., Gibbons B. J., Levitt M., Kornberg R. D. (2013) Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342, 1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen V. Q., Ranjan A., Stengel F., Wei D., Aebersold R., Wu C., Leschziner A. E. (2013) Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell 154, 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tosi A., Haas C., Herzog F., Gilmozzi A., Berninghausen O., Ungewickell C., Gerhold C. B., Lakomek K., Aebersold R., Beckmann R., Hopfner K.-P. (2013) Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154, 1207–1219 [DOI] [PubMed] [Google Scholar]
- 7. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larivière L., Plaschka C., Seizl M., Petrotchenko E. V., Wenzeck L., Borchers C. H., Cramer P. (2013) Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res. 41, 9266–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kalisman N., Adams C. M., Levitt M. (2012) Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl. Acad. Sci. U.S.A. 109, 2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leitner A., Joachimiak L. A., Bracher A., Mönkemeyer L., Walzthoeni T., Chen B., Pechmann S., Holmes S., Cong Y., Ma B., Ludtke S., Chiu W., Hartl F. U., Aebersold R., Frydman J. (2012) The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson P. J. J., Bushnell D. A., Trnka M. J., Burlingame A. L., Kornberg R. D. (2012) Structure of the Mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 109, 17931–17935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lauber M. A., Rappsilber J., Reilly J. P. (2012) Dynamics of ribosomal protein S1 on a bacterial ribosome with cross-linking and mass spectrometry. Mol. Cell. Proteomics 11, 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J.-C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II–TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang X., Munske G. R., Siems W. F., Bruce J. E. (2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 [DOI] [PubMed] [Google Scholar]
- 15. Soderblom E. J., Goshe M. B. (2006) Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78, 8059–8068 [DOI] [PubMed] [Google Scholar]
- 16. Kao A., Chiu C., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., Huang L. (2011) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu F., Wu C., Sweedler J. V., Goshe M. B. (2012) An enhanced protein crosslink identification strategy using CID-cleavable chemical crosslinkers and LC/MSn analysis. Proteomics 12, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo J., Fishburn J., Hahn S., Ranish J. (2012) An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol. Cell. Proteomics 11, M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6, 2200–2211 [DOI] [PubMed] [Google Scholar]
- 20. Rinner O., Seebacher J., Walzthoeni T., Mueller L., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Panchaud A., Singh P., Shaffer S. A., Goodlett D. R. (2010) xComb: a cross-linked peptide database approach to protein-protein interaction analysis. J. Proteome Res. 9, 2508–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu H., Hsu P.-H., Zhang L., Tsai M.-D., Freitas M. A. (2010) Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J. Proteome Res. 9, 3384–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chu F., Baker P. R., Burlingame A. L., Chalkley R. J. (2010) Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol. Cell. Proteomics 9, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. (2008) Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal. Chem. 80, 8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang B., Wu Y.-J., Zhu M., Fan S.-B., Lin J., Zhang K., Li S., Chi H., Li Y.-X., Chen H.-F., Luo S.-K., Ding Y.-H., Wang L.-H., Hao Z., Xiu L.-Y., Chen S., Ye K., He S.-M., Dong M.-Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 [DOI] [PubMed] [Google Scholar]
- 26. Senko M. W., Beu S. C., McLafferty F. W. (1995) Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 6, 229–233 [DOI] [PubMed] [Google Scholar]
- 27. Walzthoeni T., Claassen M., Leitner A., Herzog F., Bohn S., Förster F., Beck M., Aebersold R. (2012) False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 9, 901–903 [DOI] [PubMed] [Google Scholar]
- 28. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 29. Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 [DOI] [PubMed] [Google Scholar]
- 30. Iglesias A. H., Santos L. F. A., Gozzo F. C. (2009) Collision-induced dissociation of lys-lys intramolecular crosslinked peptides. J. Am. Soc. Mass Spectrom. 20, 557–566 [DOI] [PubMed] [Google Scholar]
- 31. Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chalkley R. J., Baker P. R., Huang L., Hansen K. C., Allen N. P., Rexach M., Burlingame A. L. (2005) Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer II. New developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol. Cell. Proteomics 4, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 33. Baker P. R., Medzihradszky K. F., Chalkley R. J. (2010) Improving software performance for peptide electron transfer dissociation data analysis by implementation of charge state- and sequence-dependent scoring. Mol. Cell. Proteomics 9, 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guan S., Price J. C., Prusiner S. B., Ghaemmaghami S., Burlingame A. L. (2011) A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell. Proteomics 10, M111.010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoopmann M. R., Finney G. L., MacCoss M. J. (2007) High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal. Chem. 79, 5620–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Armache K.-J., Mitterweger S., Meinhart A., Cramer P. (2005) Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280, 7131–7134 [DOI] [PubMed] [Google Scholar]
- 37. Krogan N. J., Peng W.-T., Cagney G., Robinson M. D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D. P., Beattie B. K., Lalev A., Zhang W., Davierwala A. P., Mnaimneh S., Starostine A., Tikuisis A. P., Grigull J., Datta N., Bray J. E., Hughes T. R., Emili A., Greenblatt J. F. (2004) High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13, 225–239 [DOI] [PubMed] [Google Scholar]
- 38. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 39. Chalkley R. J. (2013) When target–decoy false discovery rate estimations are inaccurate and how to spot instances. J. Proteome Res. 12, 1062–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Su C., Peregrin-Alvarez J. M., Butland G., Phanse S., Fong V., Emili A., Parkinson J. (2008) Bacteriome.org—an integrated protein interaction database for E. coli. Nucleic Acids Res. 36, D632–D636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trnka M. J., Burlingame A. L. (2010) Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene. Mol. Cell. Proteomics 9, 2306–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.