Abstract
Filamentous fungi including mushrooms frequently and spontaneously degenerate during subsequent culture maintenance on artificial media, which shows the loss or reduction abilities of asexual sporulation, sexuality, fruiting, and production of secondary metabolites, thus leading to economic losses during mass production. To better understand the underlying mechanisms of fungal degeneration, the model fungus Aspergillus nidulans was employed in this study for comprehensive analyses. First, linkage of oxidative stress to culture degeneration was evident in A. nidulans. Taken together with the verifications of cell biology and biochemical data, a comparative mitochondrial proteome analysis revealed that, unlike the healthy wild type, a spontaneous fluffy sector culture of A. nidulans demonstrated the characteristics of mitochondrial dysfunctions. Relative to the wild type, the features of cytochrome c release, calcium overload and up-regulation of apoptosis inducing factors evident in sector mitochondria suggested a linkage of fungal degeneration to cell apoptosis. However, the sector culture could still be maintained for generations without the signs of growth arrest. Up-regulation of the heat shock protein chaperones, anti-apoptotic factors and DNA repair proteins in the sector could account for the compromise in cell death. The results of this study not only shed new lights on the mechanisms of spontaneous degeneration of fungal cultures but will also provide alternative biomarkers to monitor fungal culture degeneration.
Culture degeneration, also called colony deterioration, of filamentous fungi can frequently occur during subsequent maintenance of fungal culture on artificial media by showing morphological changes, such as colony sectorization, loss or impaired ability of sporulation, fruiting, and sexuality (1, 2). It was first called as “woolly degeneration” in the model fungus Neurospora crassa (3). These morphological variations are also accompanied with the loss or reduction in secondary metabolites production, thus resulting in great commercial losses (4–7). Different from the mutation of genes involved in conidiation producing fluffy phenotypes (8–10), fungal culture degeneration spontaneously occurs and is usually irreversible. The rate of colony deterioration varies considerably among fungal species/strains and correlates with the growth environments, especially the composition of nutrient medium (11, 12). In addition, unlike the phenotype of senescence observed in the model fungus Podospora anserina (13), morphological variants of other filamentous fungi could be subcultured for many generations without growth arrest (14, 15). The mechanism(s) underlying fungal culture degeneration is poorly understood. Genetic mutations were not evident in the degenerated fungal isolates of Penicillium chrysogenum (6). Otherwise, methylation of genomic DNA was reported in a sector of the plant pathogenic fungus Fusarium oxysporum after successive subculturing (16). In some fungi, degeneration was linked to chromosome instability (4, 17).
Our previous studies on the insect pathogenic fungus Metarhizium anisopliae revealed that fungal culture degeneration showed the signs of aging such as cellular accumulation of reactive oxygen species (ROS)1, mitochondrial (mt) dysfunctions, and mtDNA glycation (14, 15). According to the vicious cycle theory of aging, mitochondria are the primary source of intracellular ROS production and also one of the important targets of ROS damage, which leads to generation of additional ROS (18, 19). Mitochondrial proteomics analyses have been frequently performed for studies on human diseases, and the physiologies of plants and yeasts (20, 21). Proteins localized in the mitochondria control mt dynamics, morphology, and function and their dysregulation or damage may induce abnormality in mitochondrial function (22). However, it is not known whether these changes occur in degenerated fungal cultures.
In this study, cell biology, biochemical and comparative mitochondrial proteomic analyses were performed by using the model fungus Aspergillus nidulans to better understand the features and mechanisms of fungal culture degeneration. We found a significant difference in mitochondrial protein profiles between the wild type (WT) and nonsporulation sector culture. Many of the altered proteins fall into the functional categories of energy metabolism, stress responses and cell death. Functional consequences of these changes are supported by our experimental data. The observed features such as cellular oxidative stress, mitochondrial dysfunctions, accelerated autophagy and releases of apoptotic factors in degenerated A. nidulans resemble the apoptotic process observed in mammalian cells.
EXPERIMENTAL PROCEDURES
Fungal Cultures and Maintenance
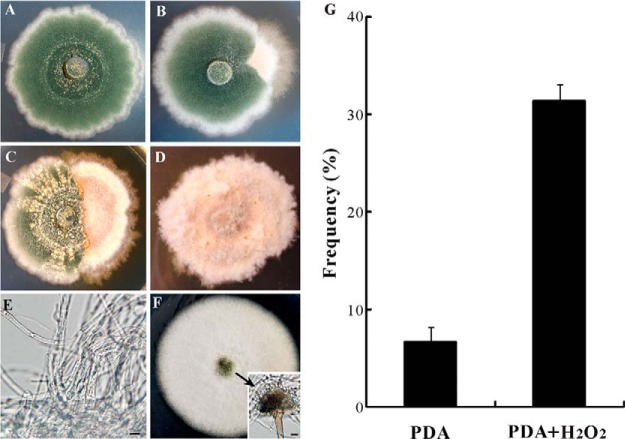
The WT strain of A. nidulans FGSC A4 was maintained on potato dextrose agar (PDA, BD Difco, Sparks, MD) for 2 weeks at 28 °C (Fig. 1A). A spontaneous sector (Sec, Fig. 1B) of A4 strain that showed a fluffy mycelium-type growth without sporulation was randomly selected and transferred to a fresh PDA plate (Fig. 1D). For oxidative stress challenge, H2O2 was supplied in the PDA plates to a final concentration of 20 mm (15). The A. nidulans strain TN02A7 (pyrG89; pyroA4; nkuA::argB2; riboB2; veA1) was used for fungal transformation and protein localization analysis (23). To determine the growth of Sec the antioxidant agent N-acetyl-l-cysteine (NAC) was added to PDA plates to a final concentration of 5 mm (24). Sabouraud dextrose broth (SDB, BD Difco, Sparks, MD) was used to grow the fungi in liquid medium when required. The following media were used for fungal transformations: YAG (2% glucose, 0.5% yeast extract, trace elements of solution), YUU (YAG supplemented with 5 mm uridine and 10 mm uracil), YAGK (YAG with 0.6 m KCl) and YUUK (YUU with 0.6 m KCl). Trace elements were prepared by adding the compounds of ZnSO4●7H2O, 2.2 g; H3BO3, 1.1 g; MnCl2●4H2O, 0.5 g; FeSO4●7H2O, 0.5 g; CoCl2●5H2O, 0.16 g; CuSO4●5H2O, 0.16 g; (NH4)6Mo7O24●4H2O, 0.11 g; Na4EDTA, 5.0 g to 80 ml of sterile water. The pH was adjusted to 6.5–6.8 with KOH pellets and the volume of solution was finally adjusted to 100 ml (25). To prepare solid media, agar was added (2%) individually in the liquid media.
Fig. 1.
Phenotypic characterization. The wild-type strain of A. nidulans grown on PDA for 2 weeks displayed the characteristic phenotype of dark-green sporulation (A) with about 8% of the colonies producing fluffy mycelium-type sectors (B, G). The addition of H2O2 increased the frequency of colony sectorization (C, G). After transfer of the sector to new plates, the fluffy mycelium-type colonies remained stable without sporulation (D, E). However, addition of the anti-oxidant NAC to the plates partially restored the sector's ability to sporulate (F).
Microscopic Observations
To determine cellular oxidative stress, a nitroblue tetrazolium (NBT) reduction assay was performed using a 0.3 mm NBT aqueous solution with 0.3 mm NADPH (14). Mycelial discs were mounted in 30% glycerol (v/v) and examined under a light microscope to examine the formation of blue/purple formazan precipitates that resulted from the reduction of NBT by ROS. For mt imaging, the WT and Sec cells grown in SDB for 36 h were stained with a fluorescent dye, MitoTracker Green (M7154, Invitrogen, Carlsbad, CA) and a membrane potential-dependent dye, MitoTracker Red CMXRos (M7512, Invitrogen). Confocal images were obtained using a LSM510 confocal microscopy (Carl Zesis Microscopy Ltd, Cambridge, UK). Apoptosis of the WT and Sec cells was assayed by counterstaining with Hoechst 33342 and propidium iodide (PI). In addition, the vital dye, neutral red was used to assess the WT and Sec cell viabilities (26).
Transmission Electron Microscopy
Morphologies of organelles and cell wall structures of A. nidulans WT and Sec were examined under a transmission electron microscopy (TEM, H-7650, Hitachi, Japan). The mycelia were grown in SDB media for 48 h at 28 °C, harvested by filtration and washed with sterile distilled water. The mycelial specimens were pre-fixed in a solution of 2.5% glutaraldehyde (w/v) phosphate buffer (0.1 m, pH 7.4) and washed three times with phosphate buffer for 10 min each at room temperature. The samples were postfixed in 2% (w/v) osmium tetroxide solution in phosphate buffer for 2 h and then dehydrated and embedded in a SPI-812 epoxy resin (27). Ultra-thin sections (50–60 nm in thickness) were prepared using a diamond knife, mounted on slotted and Formvar film-coated grids for TEM analysis.
Mitochondrion Isolation
To isolate mitochondria for proteomics analysis, the WT and Sec were grown in SDB for 48 h at 28 °C. Mycelia were harvested, and washed twice with ice-cold sterile water and homogenized in an extraction buffer (10 mm Tris-HCl, pH 7.5, 0.2 mm EDTA, 15% sucrose) (15). The homogenates were centrifuged at 5000 × g for 5 min at 4 °C and the supernatant was collected and centrifuged for another 30 min at 15,000 g to pellet crude mitochondria. The crude mitochondrial fraction was then re-suspended in TE buffer (10 mm Tris-HCl, pH 7.5, 0.2 mm EDTA) containing 20% sucrose, loaded on the top of a 20%/35%/60% sucrose gradient and centrifuged at 64,000 × g for 90 min at 4 °C. Purified mitochondria were collected from the interface between 35 and 60% sucrose layers. The samples were lysed in a denaturing lysis buffer (150 mm Tris-HCl, pH 8.5, 8 m urea, 10 mm DTT) and centrifuged at 15,000 × g for 30 min. The supernatant was collected and precipitated with trichloroacetic acid/acetone. The precipitated proteins were reconstituted in a lysis buffer (150 mm Tris-HCl, pH 8.5, 8 m urea, 10 mm DTT), quantified, aliquoted, and stored at −80 °C for further experiments. SDS-PAGE analysis was performed to profile and qualify the protein samples prepared from different experiments.
Label-free Quantitative Proteomics
Label-free quantitative proteomic analysis of the WT and Sec mt proteins was performed as described previously (28). Briefly, purified mitochondrial proteins (100 μg) were diluted in the lysis buffer to a concentration of 5 g/μl and carboxyamidomethylated in 50 mm iodoacetamide for 40 min at room temperature in the dark. The proteins were digested with an endoprotease Lys-C (Roche Applied Science, Indianapolis, IN) at a final substrate/enzyme ratio of 100:1 (w/w) at 37 °C for 3 h. The Lys-C digests were further treated with modified sequencing grade trypsin (Roche Applied Science) at a final substrate/enzyme ratio of 50:1 (w/w) at 37 °C for 20 h. After digestion, the peptide mixture was passed through an ultrafilter unit (Millipore, Billerica, MA) with a molecular weight cut-off of 10 kDa and acidified by formic acid (0.1% final concentration) for mass spectrum analysis. A linear ion trap-orbitrap liquid chromatography-tandem mass spectrometry system (LTQ-Orbitrap, Thermo Fisher Scientific, San Josse, CA) equipped with a nanospray ion source was used for full MS scan analysis followed by five MS/MS scans in the LTQ on the five most intense ions from the MS spectrum. Three parallel runs were performed consecutively for each sample. The software DeCyder MS (ver 2.0) (GE Healthcare, Pittsburgh, PA) was used to generate the peak lists from all the runs and the data were then automatically searched using the SEQUEST (ver. 2.7) (Thermo Fisher Scientific, San Josse, CA) against the A. nidulans genome archive (ver. S03-M05-R01, containing 10,644 protein entries) at AspGD database (www.aspgd.org) with the following parameters (28): peptide mass tolerance set to 10 ppm; fragment tolerance set to 0.8 Da, and two missed trypsin cleavages. Carbamidomethylation of cysteine was searched as a fixed modification, whereas N-acetyl protein and oxidation of methionine were searched as variable modifications. For protein identification, all peptide matches were filtered by a maximum false discovery rate (FDR) index less than 0.01, ΔCn larger than 0.1 and Xcorr scores of greater than 1.7, 2.0, and 3.0 for +1, +2, and +3 charged ions, respectively. The peptides were discarded for further analysis if mapped to more than two different proteins. A protein was considered identifiable with at least one unique peptide detected for more than twice in the three replicates of each sample (29). Full MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (30) with the data set identifier PXD000422 (PRIDE accessions: 32763–32768). Quantification of proteins was performed by estimating the normalized spectral index (SIN) for each protein, which combines the features of peptide count, spectral count, fragment-ion intensity and protein length as described by Griffin et al. (31). Reliability of technical repeats was analyzed by calculating the multivariate Pearson correlation coefficients using the software SigmaPlot (ver. 8.0). FDR estimation of differentially expressed proteins was conducted using a mixture model-based method (32). Heat mapping analysis was conducted using the program Matlab (R2009a). The significance of differentially expressed proteins between the samples was tested with the cutoff values of p ≤ 0.05 and FDR ≤ 0.05 (33).
Protein Distribution and Function Analysis
The identified proteins were subjected to in silico analysis with the programs ProtCom (ver. 9.0, http://linux1.softberry.com/) and WoLF PSORT (http://wolfpsort.org/) to predict subcellular localizations and InterProScan analysis (www.ebi.ac.uk/Tools/pfa/iprscan/) to classify individual proteins to protein families. Functional annotation of significantly differentially expressed proteins in the WT and Sec cells was performed using the program FunCat (34). Additional function analysis was conducted using as reference, the well-annotated yeast mitochondrial proteome database (http://www.mitop.de:8080/mitop2/).
Antibody Preparation and Western Blot Analysis
To verify LC-MS/MS data, the expressions of a few proteins in WT and Sec were verified and compared by Western blot analysis. To generate cytochrome c (CycA, AN6246) polyclonal antibody, the gene was cloned into the pGEX-4t1 vector (GE Healthcare). GST-CycA fusion protein was expressed in E. coli and purified by Sepharose Glutathione-S-Transferase column (GE Healthcare) and used to raise polyclonal antibodies in New Zealand White rabbits. The polyclonal antibodies against the lipid droplet specific perilipin-like protein (AN10518) (27) and anti-oxidant enzyme glutathione peroxidase Gpx (AN2846) (7) were obtained in previous studies. The antibody against the septum-specific protein septin B (AspB) was kindly provided by Prof. Momany (35). Antigenic peptides were predicted for the mitochondrial inner membrane protein LetM1 (AN2939, PKDTENIDEDHKYEC), which plays a role in calcium and hydrogen ion exchange (36), and the heat shock protein Hsp70 (AN1047, EPVERVLRDAKIDKSC), which is important for stress response. The peptides were synthesized for raising polyclonal antibodies. For Western blot analysis, equal amounts of either whole cell lysates or mitochondrial proteins (30 μg) were resolved by SDS-PAGE, transferred to PVDF membranes and probed with the antibodies following the protocols described previously (27).
Fungal Transformation and Confocal Microscopy
To verify the translocation of Hsp70 protein to mitochondria during fungal growth under oxidative stress, the strain TN02A7 expressing GFP-tagged Hsp70 was generated by fusing the GFP open reading frame (ORF) to the C terminus of a Hsp70 protein (AN5129) by homologous recombination (37). In brief, the 2 kb 5′- and 3′- flanking regions of the gene were amplified using genomic DNA as a template with the primer pairs l-flank5′ (GCTTCTCCTATACCTCACTTTTAACGA)/l-flank3′ (GTCGAGCTCCTCGTTCTCCTC), and R-flank5′ (TGCTTTAACGCACTTGGTTCTG)/R-flank3′ (CCTCCACCTCCTCTCAAAATCC), respectively. The cassette containing Gfp and the selective marker AfpyrG (pyrG gene from A. fumigatus) was amplified using the primers Cassette-5′ (GGTGTCGTTGAGGAGAACGAGGAGCTCGACGGAGCTGGTGCAGGCGCT) and Cassette-3′ (GAAGAAAACAGAACCAAGTGCGTTAAAGCACTGTCTGAGAGGAGGCACTGATG)from the plasmid pFNO3 (Fungal Genetics Stock Center, Kansas City, MO). The purified fragments and the selectable marker cassette were then mixed together to perform a fusion PCR with the primers Nested-F (CTTTGACCCCTCTTCACGAACC) and Nested-R (TCGTCTCTGATGCTCTCGCCT) as described before (38). After purification, the fused fragment was used to directly transform A. nidulans TN02A7 strain to generate Hsp70-GFP-expressing transformant under the control of the endogenous promoter. The protoplast transformation method was performed as described previously (35). Lytic enzyme mix for protoplast preparation was composed of 0.4 mg/ml b-d-glucanase (Interspex Products, 0439–1), 5 ml/ml glucuronidase (Sigma, St. Louis, MO) and 0.4 mg/ml lysing enzyme (L2265, Sigma).
Cellular Metabolic Activity Assay
Colorimetric tetrazolium assay using the dye XTT {2,3-bis (2-methoxy-4-nitro-5- sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide} was conducted to compare the WT and Sec cell metabolic activities as previously described (39). In brief, the WT spores and Sec mycelia were inoculated into SDB for 12 h and then transferred into 1 L flasks containing 300 ml SDB for 48 h at 28 °C. The mycelial balls of both cultures were dispersed mechanically and equal amounts were grown in 25 ml SDB for another 4, 5, 6, 7, and 8 h before measurement. XTT was diluted to 5.75 mg/ml in PBS. Menadione (Sigma) was prepared in acetone at a concentration of 0.52 mg/ml. For activity assays, five ml culture broth was transferred to a 15 ml tube and incubated with 250 μl of XTT-menadione solution (200 μl of 5.75 mg/ml XTT in PBS and 50 μl of 0.52 mg/ml menadione in acetone) at 31 °C for 2 h. The absorbance of samples was measured at 460 nm against the blank (five ml SDB treated with XTT-menadione solution). Following the assays, mycelia were harvested from the parallel treatments and heat dried at 75 °C for 1 day to determine the dry cell weight (DCW). Relative metabolic activity was determined as the unit absorbance value of DCW. Each sample had three replicates and the experiment was repeated three times.
Measurement of ATP Turnover
ATP production by the WT and Sec cells was quantified using an ATPlite bioluminescence assay kit according to manufacturer's instructions (Vigorous Biotechnology Ltd, Beijing, China). The mycelia were grown in SDB at 28 °C for 20 h, harvested and equal amounts disrupted with silica beads (one mm in diameter) by FastPrep (MP Biomedicals, Solo, OH) in one ml 0.02% trichloroacetic acid. The homogenates were centrifuged at 5,000 g for 5 min and supernatants were collected for protein quantification by Bradford protein assay. Twenty μl of supernatant was used for ATP measurement and the ATP turnover was determined as relative luminescence units of 1 mg protein.
Quantification of Different Metal Ions
For measurement of metal ions in mitochondria and whole cells, the WT and Sec cells were grown in SDB at 28 °C for 48 h and one part of the harvested mycelia were heat dried, weighed, and digested in concentrated nitric acid. The remaining mycelia were used for isolation of mitochondria as described above and the purified mitochondria were solubilized in a lysis buffer (8 m urea, 150 mm Tris-HCl pH 8.5, 10 mm DTT, 1 mm PMSF). Protein concentration was determined as indicated above. The concentrations of Ca2+, Fe2+/3+, Zn2+, Cu2+, and Mn2+ ions were quantified using an inductively coupled plasma-atomic emission spectroscopy (ICP-AES, PerkinElmer, Waltham, MAs) (40).
RESULTS
Association Between Oxidative Stress and Culture Degeneration
Phenotypic degeneration of filamentous fungi occurs spontaneously when maintained on artificial medium (1, 14, 15). This was also the case for the model fungus A. nidulans with around 8% of the cultures producing fluffy mycelium-type non-sporulation sectors on PDA plates (Figs. 1A, 1B, 1G). By inoculating the culture of A. nidulans into the plates amended with H2O2, we found a significant increase in sectorization frequency (Figs. 1C and 1G). The Sec cultures failed to produce asexual spores and their irreversible phenotype could be sustained for many generations (Figs. 1D and 1E). However, addition of an antioxidant agent, NAC, only partially restored the ability of Sec cultures to form conidiopheres and conidial spores (Fig. 1F). In contrast to the WT, the Sec cells accumulated higher levels of ROS as indicated by NBT staining assay (Fig. 2A). Thus, similar to the observations in Metarhizium (14, 15), a correlation was evident between fungal culture degeneration and cellular oxidative stress in A. nidulans.
Fig. 2.
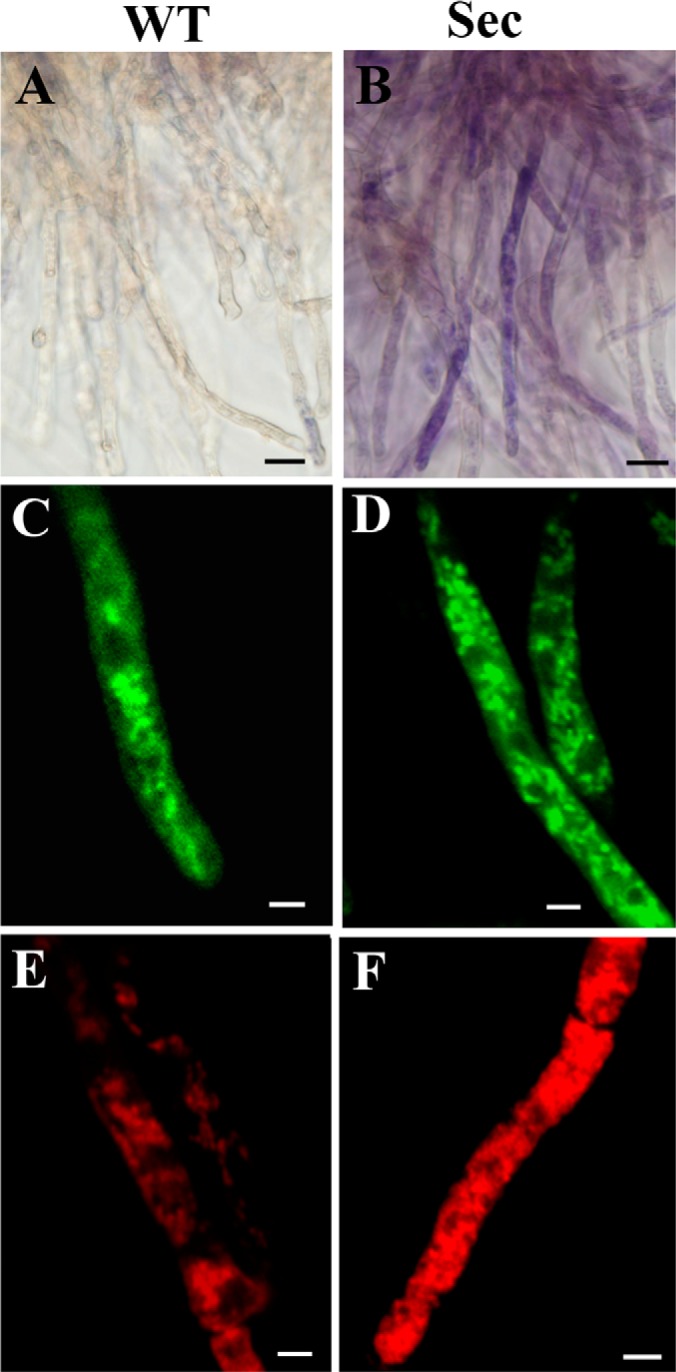
Microscopic examinations of the wild-type (WT) and sector (Sec) cells. Nitroblue tetrazolium staining showed that in contrast to WT (A), blue/purple formazan precipitate formation was observed in sector mycelia (B), an indication of cellular oxidative stress. MitoTracker Green staining and confocal microscopic observations showed mitochondrial phenotypic differences between the WT (C) and Sec (D) cells. MitoTracker Red staining revealed the differences of mitochondrial morphology and membrane potential between the WT (E) and Sec (F) cells. Bar, 5 μm.
Mitochondria are one of the primary targets of ROS-induced organelle damages (19, 41). Therefore, mitochondrial morphologies were compared between the WT and Sec cells by fluorescent staining with different dyes. Similar to previous observations (42), the WT cells of A. nidulans harbored thread-like and spherical mitochondria and these were contrast to the smaller and densely distributed mitochondria in sector mycelia when stained with MitoTracker Green (Figs. 2C and 2D). Staining with the membrane potential-dependent MitoTracker Red dye indicated a higher membrane potential in Sec cells than the WT (Figs. 2E and 2F) suggesting a higher rate of ATP synthesis in Sec mitochondria.
Accelerated Autophagic and Apoptotic Processes in Sector Cells
Oxidative stress triggers autophagy, apoptosis and cell aging (19). In filamentous fungi, increased volume of vacuoles was observed in cells undergoing autophagy that resulted in degradation and recycling of cytoplasmic and organelle components that are damaged under stress conditions (43). Higher levels of ROS in the Sec cells (Fig. 2B) led us to speculate that the sector cells may have enlarged vacuoles when compared with WT. Indeed, a neutral red staining indicated that the sector cells contained many more large vacuoles than WT (Figs. 3A, 3B). Large vacuoles were evident in WT cells when treated with H2O2 (Fig. 3A), which was a reminiscent of oxidative stress in Sec cells. These observations indicated that the appearance of large vacuoles was because of oxidative stress. Consistent with neutral red staining, large vacuoles were also evident in Sec but not WT cells when examined under TEM (Figs. 3C, 3D). In addition, unlike the WT, autophagic bodies were found exclusively in the vacuoles of sector cells (Fig. 3D). Moreover, when compared with WT much denser melanin pigment layers were deposited outside the Sec cell wall (Figs. 3E, 3F). Fungal melanin pigments act as traps for oxidative radicals and thus have high antioxidant activities (44). In this respect, production of higher amounts of melanin by the Sec mycelia could be an adaptive response against cellular oxidative stress. To further determine whether oxidative stress could cause cell apoptosis, we counterstained the sector cells with Hoechst/PI and compared with the WT. The results indicated that, in contrast to the WT (Figs. 4A–4C) significantly increased numbers of Sec cells contained condensed chromatins and were also positive for PI staining (Figs. 4D–4F). Thus, the Sec cells of A. nidulans appeared to undergo apoptosis.
Fig. 3.
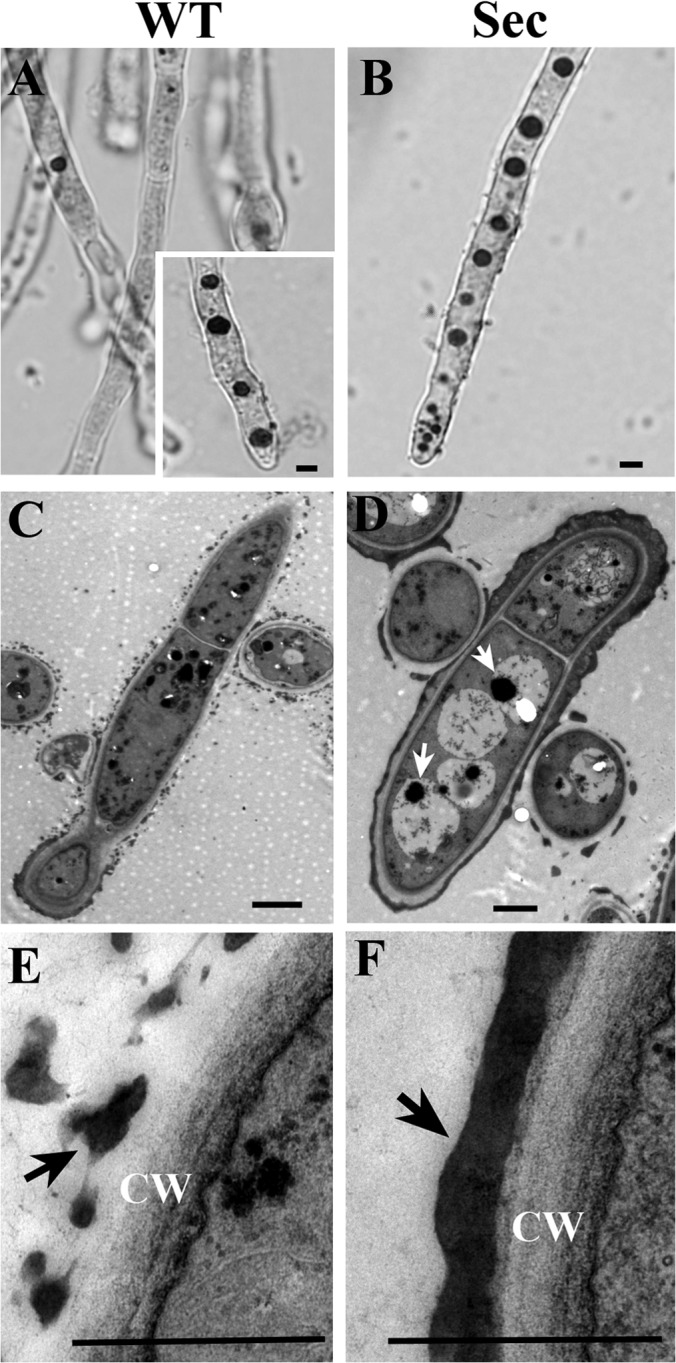
Neutral red (NR) staining and transmission electron microscope (TEM) observations. NR staining demonstrated the difference in vacuole formation between the wild-type (WT) (A, inset shows WT cells challenged with H2O2 for two hours and then stained with NR) and sector (Sec) (B) cells. TEM analysis confirmed that in contrast to WT (C), large vacuoles were formed in Sec cells (D, arrows indicate autophagic bodies). Ultrastructure TEM analysis also revealed the differences in melanin (arrowed) formation on cell wall (CW) surface between the WT (E) and Sec (F) cells. Bar, 2 μm.
Fig. 4.
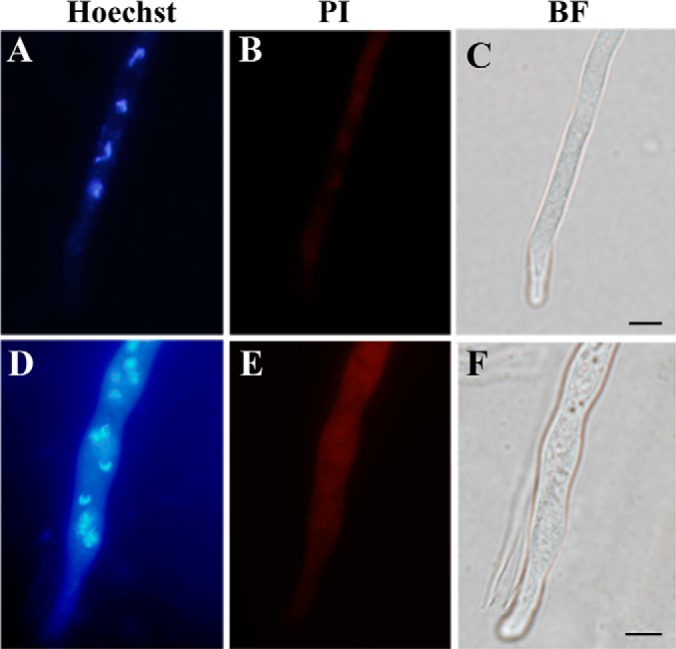
Assessment of cell apoptosis. Both the wild-type (A–C) and sector (D–F) mycelia were counterstained using Hoechst and propidium iodide (PI), which demonstrated the differences between the wild-type and sector cells in nuclear phenotype and the ability to uptake PI. BF, bright field. Bar, 5 μm.
General Features of Mitochondrial Proteomic Profiling
To better understand the mechanisms of fungal culture degeneration, we conducted a high throughput label-free proteomic analysis to profile and compare the WT and Sec mt proteomes of A. nidulans. The mt proteins extracted from highly purified mitochondria of two independent experiments demonstrated the reproducible patterns and the differences in intensity and composition of protein bands between the WT and Sec samples (Fig. 5A). The protein samples were further subjected to trypsin digestion and LC-MS/MS analysis. Both spectra profiles (supplemental Fig. S1) and regression analysis of the WT (supplemental Fig. S2A, r = 0.7895, p < 0.05) and Sec (supplemental Fig. S2B, r = 0.7243, p < 0.05) indicated the high reproducibility of the three technical repeats. By mapping the fragmented peptides on protein sequences, a total of 858 proteins were detected from two samples (supplemental Tables S1–S3). Among these, 703 were in both WT and Sec samples whereas 53 and 102 proteins were detected only in the WT and Sec, respectively. Consistent with SDS-PAGE analysis (Fig. 5A), considerable differences were also observed in mt protein expressions between the WT and Sec (Fig. 5B). A statistical analysis identified 260 differentially expressed proteins (cutoff values at p < 0.05; FDR < 0.05), which included 148 up-regulated and 112 down-regulated proteins in Sec when compared with WT (supplemental Table S1).
Fig. 5.
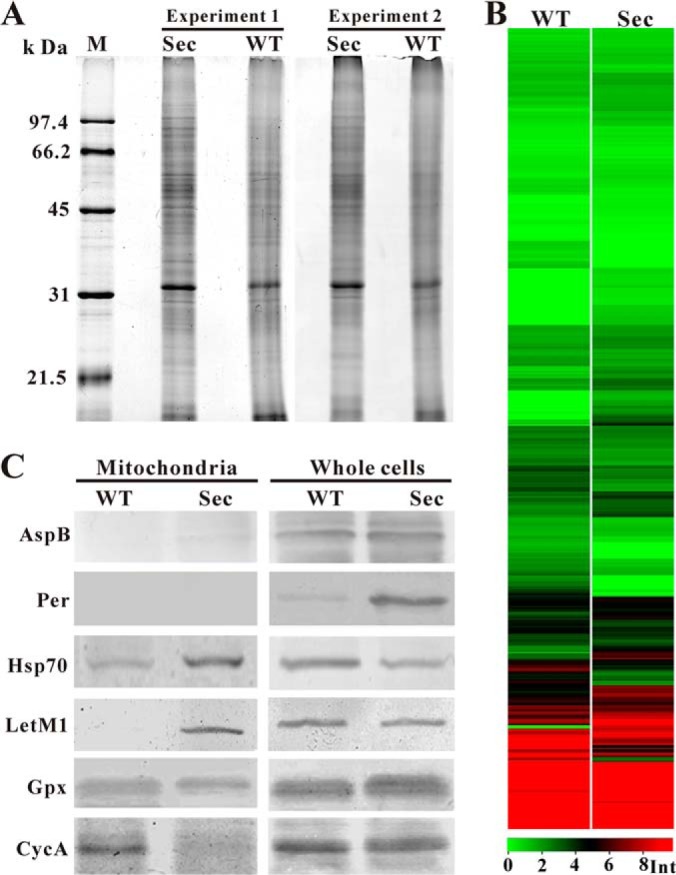
Proteome profiling and Western blot analyses. A, Mitochondrial proteins of the wild-type (WT) and sector (Sec) prepared from two independent experiments were analyzed by SDS-PAGE and silver staining. B, Heat map analysis of the WT and Sec mitochondrial protein expression profiles. Int, intensity of protein peak. C, Western blot analysis of the WT and Sec mitochondrial and whole cell protein samples using different antibodies. Asp B, septum-specific protein septin B; Per, lipid droplet-specific perilipin-like protein; Hsp70, heat shock protein Hsp70; LetM1, calcium antiporter LetM1; Gpx, glutathione perioxidases; CycA, cytochrome c protein.
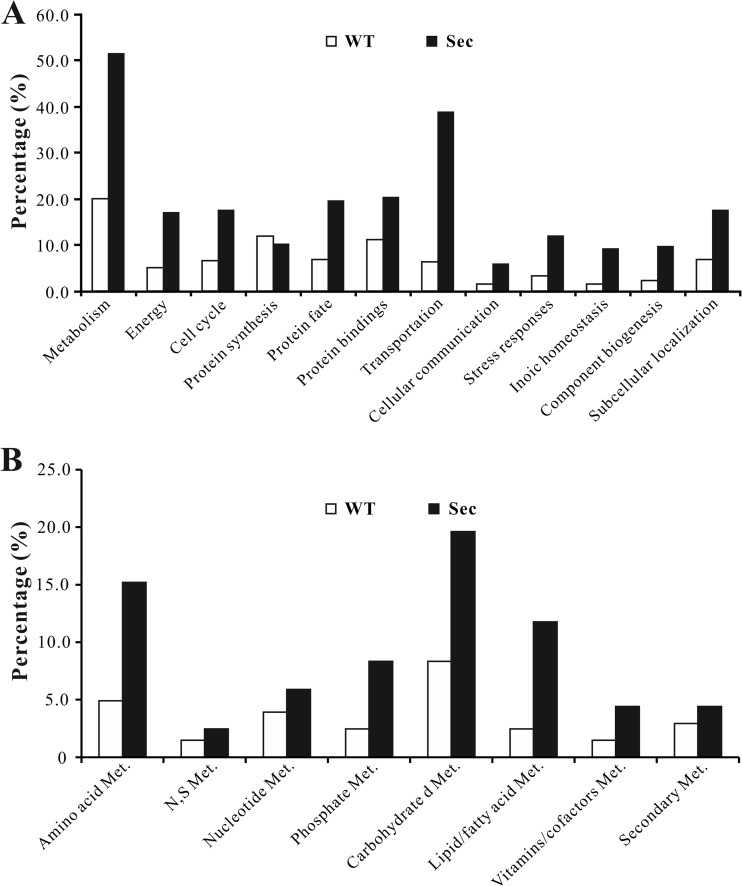
An InterProScan analysis indicated that 778 out of 858 proteins detected contained conserved domains. Functional characterization of differentially expressed proteins as well as those specifically expressed either in the WT or Sec mitochondria revealed that expect for those involved in protein synthesis, more proteins were up-regulated in different functional categories in the Sec mitochondria, especially those involved in cell metabolisms and transportation (Fig. 6A). For those involved in cell metabolisms, the proteins involved in carbohydrate, amino acid and lipid/fatty acid metabolism were more highly up-regulated in sector mitochondria than in WT (Fig. 6B). In addition, proteins associated with the energy metabolic pathways, such as tricarboxylic acid cycle (TCA cycle, supplemental Fig. S3), oxidative phosphorylation and β-oxidation of fatty acids, were also mostly up-regulated in sector cells.
Fig. 6.
Functional classification analysis. A, Functional category analysis of the mitochondrial proteins differentially expressed by the wild-type (WT) and sector (Sec). B, Functional category analysis of differentially expressed mitochondrial proteins involved in cell metabolisms (Met.) in WT and Sec.
Verification of Protein Localization
Based on LC-MS/MS analysis, varied numbers of mt proteins were identified in the WT (756 proteins) and Sec (805 proteins) mitochondria. However, in silico analysis of these proteins with the program ProtComp only predicted 209 and 229 mt proteins in the WT and Sec, respectively. And, 233 (WT) and 244 (Sec) mt proteins were predicted above the confidence level by the program WoLF PSORT. To validate our proteome data, Western blot analyses were performed for a few selected proteins (Fig. 5C). As expected, antibodies against the septum protein, AspB, and lipid droplet surface protein perilipin (Per) did not bind to any mt protein from WT or Sec mitochondria as shown by the lack of signals, but did bind to proteins from whole cell samples (Fig. 5C). The heat shock protein Hsp70 (AN5129) was 1.7-fold up-regulated in the Sec mitochondria (p = 0.0014; FDR = 0.0043) (supplemental Table S1). Consistent with the MS data, a stronger signal was detected in the Sec mt sample than the WT. The mitochondrial H+/Ca2+ antiporter, Letm1 (AN2939), was only detected in the Sec mitochondria by MS analysis (supplemental Table S2) and this was confirmed by Western blot analysis (Fig. 5C). Relative to the WT, the antioxidant mt enzyme, glutathione peroxidase (Gpx, AN2846), was up-regulated (1.7-fold) in Sec although not significantly (p = 0.1599; FDR = 0.0972) (supplemental Table S1). This was also evident in Western blot analysis (Fig. 5C). Release of cytochrome c from mitochondria is a hallmark of cell apoptosis (45). Cytochrome c (CycA, AN6246) was not identified in our MS data, however, the protein was detected in the WT but not Sec mitochondria by Western blot assay, i.e. an indicative of CycA release from sector mitochondria (Fig. 5C). Overall, the MS data were supported by Western blot analyses.
In our LC-MS/MS analysis, six of 13 genome-encoded Hsp70s were detected in the WT and Sec mt proteins. Besides AN5129 mentioned above, three other Hsp70s, i.e. AN6010 (2.1-fold, p = 0.0504; FDR = 0.0443), AN1047 (2.0-fold, p = 0.0009; FDR = 0.0036), and AN4616 (2.1-fold, p = 0.005; FDR = 0.0091) were significantly up-regulated in Sec when compared with WT (supplemental Table S1). We were interested to determine whether these molecular chaperones were “real” mt proteins. Previous studies have shown that yeast Ssc1 (P0CS90) and Ssg1 (Q9UXR0) are mitochondrial chaperones of the Hsp70 class (46). The ortholog of these Hsp70s in A. nidulans is AN6010 (76% identity with Ssc1 and 53% with Ssg1), which was also predicted as a mitochondrial protein by both programs in in silico analyses. To further verify the mt localization of other Hsp70s, AN5129 was selected and its C terminus was fused with a GFP protein for fungal transformation of the WT strain (supplemental Fig. S4). The acquired transformants were challenged with H2O2 to mimic oxidative stress in the sector cells. As shown in Fig. 7, the occurrence of AN5129-GFP translocation into mitochondria was confirmed, which supported again the MS data.
Fig. 7.
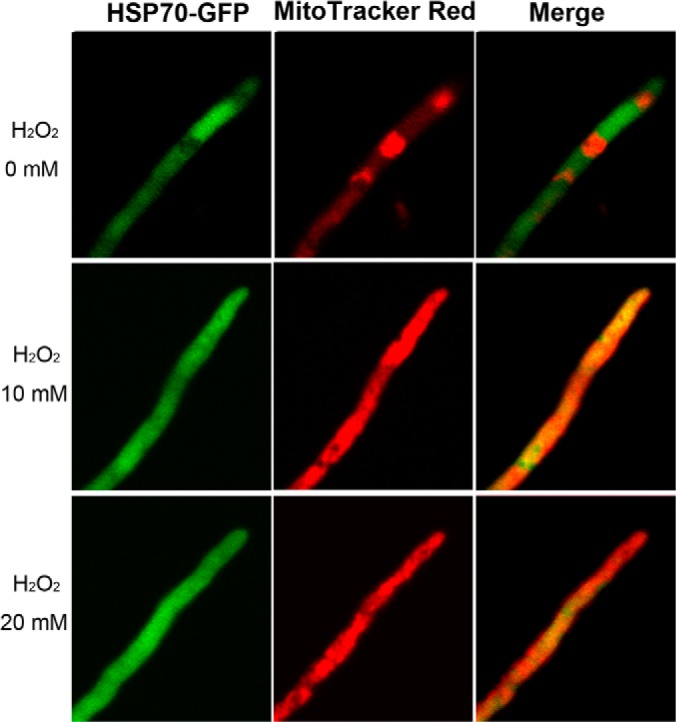
Translocation of Hsp70 from cytosol to mitochondria. The wild-type strains were transformed with an Hsp70-Gfp fusion gene and the acquired transformants were grown in a medium amended with different concentrations of H2O2 for 4 h and then stained with the mitochondrial dye MitoTracker Red for microscopic examination.
Proteins Responsible for Enhanced Metabolic Activities and ATP Production in Sector Cells
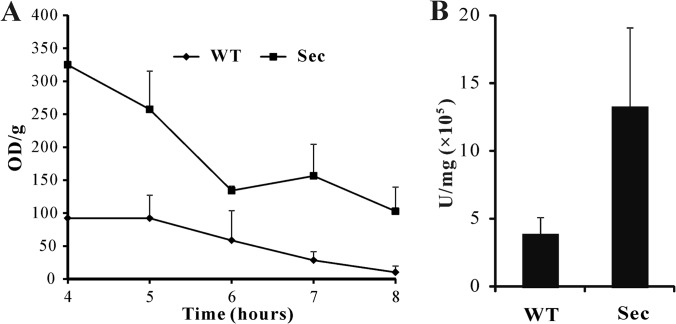
Proteomics data suggested an enhancement of the overall metabolic activities in sector cells (Fig. 6; supplemental Fig. S3). To confirm these experimentally, we quantified the overall metabolic activity using an XTT assay. The results indicated a higher metabolic activity in sector cells when compared with WT at all-time points examined (Fig. 8A). Consistent with the observation of a higher mt membrane potential in Sec than WT (Fig. 2F), the detected mt proteins involved in TCA cycle (supplemental Fig. S3), β-oxidation (AN2072, AN0023, and AN1078) and oxidative phosphorylation for ATP synthesis (i.e. ATP synthases including AN11303, AN11565, AN10032, AN1523, AN6232, AN5713, and AN8674) were up-regulated in sector cells when compared with WT (supplemental Tables S1 and S2). Besides peroxisomal β-oxidation of fatty acids, mt β-oxidation was evident in A. nidulans (47). The up-regulated activities of 3-hydroxyacyl-CoA dehydrogenase (AN0023, 1.5-fold), acyl-CoA dehydrogenase (AN0272, 1.4-fold) and D3, D2-enoyl-CoA isomerase (AN1078, 1.6-fold) could be related to the production of acetyl-CoA to enter mitochondria for the TCA cycle (supplemental Table S1; supplemental Fig. S3). In addition, a long chain fatty acid CoA ligase (AN6014) was only detected in the Sec mitochondria (supplemental Table S2). This enzyme catalyzes the formation of fatty acyl-CoA, which is the prestep reaction for β-oxidation of fatty acids (48). These data suggested the production of higher amounts of unit ATP in the Sec cells than the WT. This speculation was verified by measurement of ATP turnover with an ATPlite assay kit, which showed larger amounts of ATP (p = 0.0062) in sector cells when compared with WT (Fig. 8B).
Fig. 8.
Cellular metabolic activity and ATP production assays. A, An XTT assay revealing the differences in the overall cellular metabolic activities between the wild type (WT) and sector (Sec) cells. B, Bioluminescence assay showing the differences in ATP production between the WT and Sec cells.
Up-regulated Transporters to Elevate Mitochondrial Ca2+ Level in Sector Cells
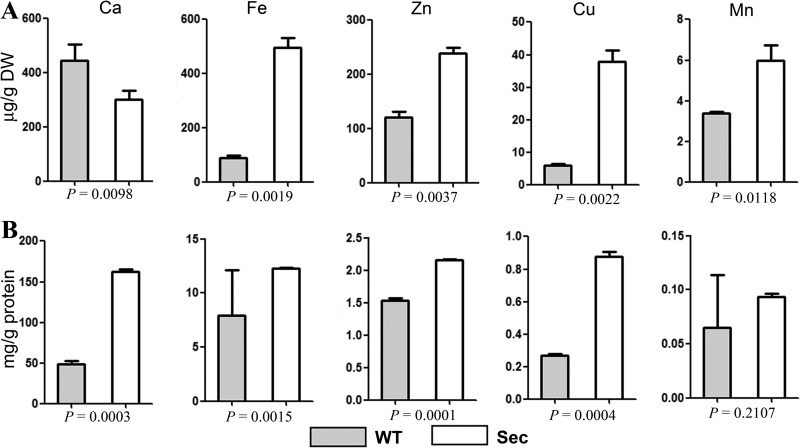
Mitochondrial Ca2+ stimulates TCA cycle and oxidative phosphorylation for energy metabolisms (49). Mitochondrial Ca2+ uptake occurs through the mt Ca2+ uniporter (Mcu) and H+/Ca2+ antiporter Letm1 (34). Mcu homologs have not been characterized in yeasts and A. nidulans (50). In this study, we found a P-type ATPase (AN10367, i.e. ion transporter) being significantly up-regulated (1.6-fold, p = 0.0013; FDR = 0.0042) in Sec mitochondria when compared with WT (Tables S1). This protein is a homolog of yeast Spf1 (52% identity), which is implicated in Ca2+ homeostasis (51). Antiporter Letm1 was specifically expressed in Sec mitochondria (Fig. 5B; supplemental Table S2). Up-regulation of these transporters suggested an accumulation of more Ca2+ cations in sector mitochondria than in WT. To test this, we measured the metal ion contents in whole cells and mitochondria, respectively, by ICP-AES spectroscopy analysis. The results demonstrated a lesser amount of Ca2+ ion in sector whole cells than in WT (Fig. 9A). However, a 3-fold increase in Ca2+ content accumulation was observed in the Sec mitochondria when compared with WT (Fig. 9B). This is in contrast to other detectable metal ions (Fe, Zn, Cu, and Mn), which were highly accumulated in whole sector cells and mt than those in WT (Fig. 9).
Fig. 9.
Quantification of ion concentrations. A, Different ion concentrations in the whole cell samples of wild-type (WT) and sector (Sec). B, Different ion concentrations in the WT and Sec mitochondria. p values under each panel show the significance level of differences between WT and Sec.
Proteins Involved in Controlling Mitochondrial Morphology
Mitochondria have a dynamic network and its morphology within a cell is precisely controlled by a set of proteins that regulate organelle fusion and fission (52). Consistent with the report that oxidative stress results in mt fragmentation (53, 54), much denser and smaller type of mitochondria were observed in Sec cells than those in WT (Figs. 2C–2F). In A. nidulans, very few proteins involved in controlling mt morphology have been characterized. By examining the MS data set, we found that the orthologs of yeast Mdv1 (AN6867, 32% identity; 1.5-fod, p = 0.004; FDR = 0.0008), Tim11 (AN11303, 32% identity; 2.2-fold, p = 0.0015; FDR = 0.0044), and Num1 (AN7757, 20% identity; 2.77-fold, p = 0; FDR = 0.0006) (supplemental Table S1), which are involved in mt fission (54), were significantly up-regulated in Sec mitochondria. In addition, the yeast dynamin (Dm1) homolog AN8874 (69% identity), which is implicated in mt fission (54), was only detected in the Sec mt protein sample (supplemental Table S2). It has also been reported that calcium overload in mt is one of the factors that induces mt-network fragmentation (55). Taken together, more frequent events of fission than fusion would be speculated to occur in sector cells and thereby resulted in mt fragmentation (Fig. 2D).
Proteins Involved in Anti-oxidative Stress and Anti-apoptosis
As indicated above, Sec culture demonstrated the characteristic features of apoptosis. This was consistent with the up-regulation of a putative apoptosis-inducing factor (Aif) (AN0394, 1.3-fold, p = 0.0181; FDR = 0.0208) in the sector sample (supplemental Table S1). In yeast, strong stimulation of apoptotic cell death by overexpression of Aif-1 (24% identity with AN0394) has been evident (56). A putative nucleoside diphosphate kinase (AN8216), involved in the negative regulation of apoptosis (57), was down-regulated in sector (1.8-fold, p = 0.0227; FDR = 0.0244). Down-regulation of a superoxide dismutase homolog (AN0785) (1.8-fold, p = 0.0006; FDR = 0.0028) in sector mitochondria and the presence of the homologs of anti-oxidant enzymes, thioredoxins (AN0170 and AN8571) and peroxiredoxin (AN8692) only in WT but not in Sec (supplemental Table S3) could contribute, at least in part, to the accumulation of ROS in Sec cells. However, unlike the senescence observed in P. anserina cultures (13), A. nidulans sector culture could be subsequently maintained on media, that is, without the signs of cell death. Apart from the massive biosynthesis of melanin to combat oxidative stress, a few mt proteins were found to be up-regulated in Sec, which may be responsible for anti-oxidative stress and/or anti-apoptosis. For example, besides the up-regulation of Gpx enzyme mentioned above, an anti-oxidant enzyme glutathione S-transferase (AN10273) was only detected in the Sec mitochondria (supplemental Table S2). A putative glutamine amidotransferase (AN6141) involved in the biosynthesis of pyridoxine (vitamin B6, an inhibitor of cell apoptosis) (58), was up-regulated 2.1-fold in Sec (p = 0.0303; FDR = 0.031) (supplemental Table S1). In yeasts, plants and animals, two copies of mt inner membrane prohibitins (Phb) form a complex and play active roles in response to oxidative stress and mt dysfunction (59). Similarly, two prohibitins were detected in our MS data, i.e. AN0686 (yeast Phb1-like, 71% identity) only expressed in Sec and AN6072 (yeast Phb2-like, 66% identity) up-regulated in Sec (1.3-fold) (supplemental Tables S1 and S2). In addition, it has been well established that Hsp70 can efficiently inhibit cell apoptosis (60). Thus, the up-regulations of different Hsp70s could additionally contribute to the anti-apoptotic process in sector. The up-regulations of ABC-type multidrug transporters in Sec could be also involved in stress defenses to maintain Sec culture viability (Fig. 6A; supplemental Tables S1 and S2).
Oxidative stress also induces mutation and degradation of mtDNA (61) and mtDNA glycation is evident in the degenerated culture of Metarhizium (15). In light of these, failure to maintain mtDNA integrity could result in cell death rather than apoptosis of A. nidulans sector culture. By examining the MS data, we observed up-regulation of different proteins involved in DNA repair in Sec including AN10677 (Rad54, 1.7-fold, p = 0.0444; FDR = 0.0399), AN0855 (Rad54b, 3-fold, p = 0.0006; FDR = 0.0028), AN10415 (Rad18, 3.4-fold, p = 0.0028; FDR = 0.0066), and AN10757 (RecQ, 2.3-fold, p = 0.0002; FDR = 0.0017) (supplemental Table S2). RecQ AN10757 is a homolog of yeast Hrq1, which is reported to support the repair of excised nucleotides (62). The putative DNA helicase AN5278 and DNA repair nuclease AN3118 were only detected in sector (supplemental Table S2) and are the homologs of yeast Pif1 (26%) and Mus81 (36% identity), respectively. Pif1 helicase has been shown to play important roles in the maintenance of both nuclear and mt genome integrity by interacting with a single-strand binding protein (63). Mus81 is shown to serve as a subunit of a heterodimeric nucleases complex that efficiently repairs DNA by resolving DNA intermediates (64). Taken together, it could be reasonable to speculate that oxidative damages of mtDNA in sector were the stimuli to trigger the up-regulation or translocation of DNA repair proteins to sustain mt genome stability and thus, the viability of sector culture.
DISCUSSION
In this article, by using the filamentous fungus A. nidulans as a model, we establish that spontaneous fungal culture degeneration is linked to cellular accumulation of ROS and mitochondrial dysfunctions. Comparative mt proteome analyses of the WT and nonsporulation sector cultures further helped understand the mechanisms of spontaneous degeneration of fungal cultures. Most significantly, we found that, in contrast to WT, the degenerated culture demonstrated the characteristics of cell apoptosis, including the release of CycA, calcium overload and up-regulations of apoptosis inducing factors in sector mitochondria (Fig. 10). Elevated accumulation of Ca2+ was evident in the Sec mitochondria which could accelerate the release of CycA as well as the productions of ROS to trigger cell apoptosis (65, 66). However, unlike cell senescence (13), the Sec culture did not show signs of growth arrest during maintenance, which could be caused by the up-regulations of the Hsp70 chaperones, anti-apoptotic factors and mtDNA repair proteins (Fig. 10).
Fig. 10.
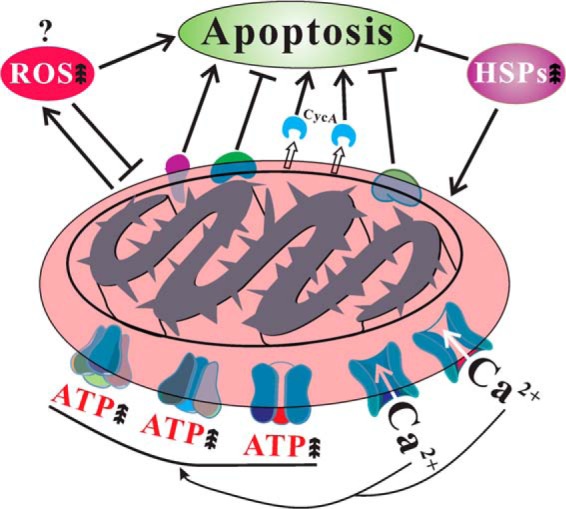
Schematic representation of mitochondrial dysfunctions in the degenerated sector of A. nidulans. The factor(s) that triggers cellular accumulation of reactive oxygen species (ROS) is not known. However, accumulation of ROS, release of cytochrome c (CycA), calcium overload and up-regulation of apoptosis inducing factors will induce sector cell apoptosis. On the other hand, up-regulation of anti-apoptotic factors, mtDNA repair proteins, heat shock proteins (HSPs) and translocation of HSPs to mitochondria will suppress sector cell apoptosis. Up arrows represents increase.
Phenotypic features of degenerated fungal cultures include non-sporulation and impaired abilities of sexuality, fruiting, and production of secondary metabolites (6, 7, 15). However, mutation or deletion of different genes involved in controlling sporulation could also result in a fluffy phenotype in A. nidulans and other fungi. For example, the zinc finger region-containing transcription factor BrlA (AN0973) controls conidiophore development and its proper expression is crucial for regulating conidiation in A. nidulans (67). Deletion of a Zn2Cys6-type transcription factor Fl (for fluffy) led to the failure of macroconidia production in N. crassa and thereby the fluffy phenotype (8, 68). The RGS (regulator of G-protein signaling) domain-containing protein FlbA (fluffy low BrlA expression, AN5893) has been verified in different fungi in controlling spore production and deletion of this gene could result in aerial hyphal growth, that is, the fluffy phenotype (9). The genes FlbB (AN7542), FlbC (AN2421), and FlbD (AN0279) encoding transcription factors are also important for regulating similar phenotypes featured by a failure in conidiophore development and asexual spore formation (10). Global regulators like LaeA (AN0807, methyltransferase) and VeA (AN1052, velvet protein) coupled with the G-protein signaling pathway have been functionally demonstrated to coordinate the regulation of asexual sporulation, sexuality and secondary metabolism in A. nidulans (69, 70). Not surprisingly, none of these transcription factors or RGS protein was detected in our MS data. In addition, it has not been reported that deletion of any of these genes could lead to cellular ROS accumulation, the feature essentially different from spontaneous culture degeneration. However, the characteristic fluffy phenotype of the A. nidulans sector suggests that it would be worthwhile to determine in the future whether or not these global regulators participate in fungal culture degeneration.
Mitochondrial proteins are also directly or indirectly involved in controlling fungal developments. For example, mitochondrial ATP synthases have been found to be involved in regulations of ascospore germination and sexual reproduction in P. anserina (71). In this study, we detected a high expression of ATP-dependent protease (AN0122) only in the WT mitochondria (supplemental Table S3). This protein is a homolog of Lon protease, which shows 33% identity to PaLon1 from P. anserina mitochondria. Deletion of PaLon1 leads to the defects in fungal developments including sexuality and a decrease in fungal life span (72). Down-regulation of the Lon protease in human cells also impairs mitochondrial structure and functions, and lead to extensive apoptotic cell death (73). Thus, loss of Lon protease in sector mitochondria further supports the conclusion of apoptotic process occurring in the degenerated cultures of A. nidulans.
Mitochondrial theory of aging is based on the vicious cycle of ROS generation and damages (18, 19). We found the causal link among oxidative stress, mt dysfunctions, and culture degeneration in A. nidulans. Besides the up-regulation of apoptosis-inducing factors, the cytochrome c release and calcium overload were evident in the sector mitochondria. The former is the hallmark of cell apoptosis and aging across the kingdom (18, 45). And, the latter is also one of the apoptotic ways to perturb or rupture mt membrane permeability and in turn the release of apoptotic factors including cytochrome c and Aif factor into the cytosol (54, 74). In addition, it has been established in P. anserina and yeast that reducing mt fission could increase cellular resistance to the induction of apoptosis. Relative to the wild type, the up-regulations of proteins involved in mt fission, and the smaller/denser-type of mitochondria in sector cells also suggested the occurrence of cell apoptosis in sector associated with oxidative stress (53, 75). Thus, it could be reasonably concluded that fungal culture degeneration is linked to cell apoptosis. Nevertheless, in contrast to the aging yeast, Podospora or mammalian cells, which show decreased levels of mt membrane potential (Δψm) and ATP production (76, 77), the levels of Δψm and ATP turnover were higher in sector than the wild type. Provided that there are emerging data with contrary results based on different models of apoptosis and different systems (78, 79), the up-regulations of anti-oxidant and anti-stress enzymes in the sector could contribute to the differences between this study and previous reports on other systems. For example, relative to the wild type, a glutathione peroxidase was up-regulated in the sector mitochondria, the enzyme could protect mitochondrial ATP generation against oxidative damage (80). Further studies are still required to determine the similar and dissimilar mechanisms between the cell apoptosis and fungal culture degeneration.
The mt genome of A. nidulans (NC_017896) contains 20 protein coding genes, which is close to the numbers in other fungi (81). However, the exact numbers of mt proteome in different organisms including A. nidulans is still not clear. Based on mass spectrometry and cellular localization studies the mt proteome of the budding yeast has been estimated to be ∼800 proteins (81). In addition, based on a GFP-tagged library, about 500 yeast proteins are observed to be localized in mitochondria (82). However, the MitoP2 mitochondrial proteome database catalogues 601 proteins (83) and the MitoMiner database (84) includes more than 1000 proteins as yeast mt proteins. Likewise, the MitoP2 database shows 442 mt proteins for the model fungus N. crassa whereas at least 738 genes encoding proteins are estimated in the Neurospora mt proteome (85). In this study, different numbers of mt proteins were detected in the WT and Sec mitochondria, respectively. The data suggested that mt proteome changed dynamically in association with cell physiological states. However, both numbers are considerably higher than those predicted in silico by different algorithms. For example, the predicted “none” Hsp70 AN5129 could be evidently translocated into mitochondria from cytosol when the fungus was challenged with oxidative stress (Fig. 7). The proteins involved in DNA repairs are all predicted as nuclear proteins in in silico analysis. However, different DNA repair proteins have been shown to target both nuclear and mitochondrial compartments (86). A recent study has shown that more than one third of yeast mt proteome has dual localizations (87). Thus, it is not uncommon to detect these proteins in the mitochondria of A. nidulans. There are also well-known mt proteins that were not detected in our MS spectra. Besides cytochrome c, the MdmB (AN6901) protein of A. nidulans has been shown to regulate mt morphology and localize within mitochondria (88). This protein was also not detected in our MS data. Thus, the analyses with combined proteomic techniques to determine the mt proteins isolated from the cells under different physiological stages could account for a more reliable mt proteome.
Overall, based on cell biology, biochemical and comparative mt proteomic analyses, we establish the causal link among oxidative stress, mitochondrial dysfunctions and fungal culture degeneration with the latter showing the signs of cell apoptosis and aging. Besides the cellular accumulation of ROS and over-deposition of melanin on the cell wall of sector culture, the findings of cytochrome c release, calcium overload, accelerated rate of ATP production, down-regulation of Lon protease, and protein translocations could be potentially used as “biomarkers” to monitor fungal culture degeneration to prevent or reduce economic losses during mass production of valued economic fungi. Mechanistically, many issues still remain elusive, for example, the cause and effect relationships between frequent culture degeneration and cellular oxidative stress and between mitochondrial dysfunction and fungal developments. Apart from the involvement of mitochondrial proteins, cytosolic and other organelle proteins, especially the downstream transcription factors, are likely to be implicated in physiological responses and adaptations of fungal culture deterioration. Future functional studies of differentially expressed mt proteins as well as related non-mitochondrial proteins will help identify the “trigger(s)” of fungal culture degeneration and elucidate the associated signaling networks.
Supplementary Material
Acknowledgments
We thank Shanghai Applied Protein Technology Co. Ltd for proteome analysis service, Prof. Lin Lu at Nanjing Normal University for technical supports for fungal transformation, Prof. Jiming Gong for help quantify metal ions and Mr. Xiaoyan Gao for helps with the transmission electron microscopy analysis. We also than the PRIDE team for help with the MS data deposition.
Footnotes
Author contributions: C.W. designed research; L.L. performed research; Y.X. and P.Z. contributed new reagents or analytic tools; L.L., X.H., Y.X., G.X., P.Z., and C.W. analyzed data; L.L. and C.W. wrote the paper.
* This study is supported by the National Natural Science Foundation of China (Grant Nos. 30970034 and 31225023).
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S3.
1 The abbreviations used are:
- ROS
- reactive oxygen species
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- WT
- wild type
- Sec
- Sector
- mt
- mitochondrial
- mtDNA
- mitochondrial DNA
- NBT
- nitroblue tetrazolium
- FDR
- false discovery rate
- Hsp
- heat shock protein
- CycA
- Cytochrome c
- XTT
- 2,3-bis (2-methoxy-4-nitro-5- sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide
- ICP-AES
- Inductively Coupled Plasma-Atomic Emission Spectroscopy
- TEM
- transmission electron microscopy.
REFERENCES
- 1. Ryan M. J., Bridge P. D., Smith D., Jeffries P. (2002) Phenotypic degeneration occurs during sector formation in Metarhizium anisopliae. J. Appl. Microbiol. 93, 163–168 [DOI] [PubMed] [Google Scholar]
- 2. Wang C. S., Skrobek A., Butt T. M. (2004) Investigations on the destruxin production of the entomopathogenic fungus Metarhizium anisopliae. J. Invertebr. Pathol. 85, 168–174 [DOI] [PubMed] [Google Scholar]
- 3. Sheng T. C. (1951) The genetic basis of woolly degeneration in Neurospora crassa. Bot. Gaz. 113, 203–206 [Google Scholar]
- 4. Li A., Begin M., Kokurewicz K., Bowden C., Horgen P. A. (1994) Inheritance of strain instability (sectoring) in the commercial button mushroom, Agaricus bisporus. Appl. Environ. Microbiol. 60, 2384–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Magae Y., Akahane K., Nakamura K., Tsunoda S. (2005) Simple colorimetric method for detecting degenerate strains of the cultivated basidiomycete Flammulina velutipes (Enokitake). Appl. Environ. Microbiol. 71, 6388–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Douma R. D., Batista J. M., Touw K. M., Kiel J. A., Krikken A. M., Zhao Z., Veiga T., Klaassen P., Bovenberg R. A., Daran J. M., Heijnen J. J., van Gulik W. M. (2011) Degeneration of penicillin production in ethanol-limited chemostat cultivations of Penicillium chrysogenum: A systems biology approach. BMC Syst. Biol. 5, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiong C. H, Xia Y. L., Zheng P., Wang C. S. (2013) Increasing oxidative stress tolerance and subculturing stability of Cordyceps militaris by overexpression of a glutathione peroxidase gene. Appl. Microbiol. Biotechnol. 97, 2009–2015 [DOI] [PubMed] [Google Scholar]
- 8. Bailey L. A., Ebbole D. J. (1998) The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 148, 1813–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu J. H., Wieser J., Adams T. H. (1996) The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15, 5184–5190 [PMC free article] [PubMed] [Google Scholar]
- 10. Arratia-Quijada J., Sánchez O., Scazzocchio C., Aguirre J. (2012) FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 11, 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horn B. W., Dorner J. W. (2002) Effect of competition and adverse culture conditions on aflatoxin production by Aspergillus flavus through successive generations. Mycologia 94, 741–751 [PubMed] [Google Scholar]
- 12. Shah F. A., Wang C. S., Butt T. M. (2005) Nutrition influences growth and virulence of the insect-pathogenic fungus Metarhizium anisopliae. FEMS Microbiol. Lett. 251, 259–266 [DOI] [PubMed] [Google Scholar]
- 13. Osiewacz H. D. (2011) Mitochondrial quality control in aging and lifespan control of the fungal aging model Podospora anserina. Biochem. Soc. Trans. 39, 1488–1492 [DOI] [PubMed] [Google Scholar]
- 14. Wang C. S., Butt T. M., St. Leger R. J. (2005) Colony sectorization of Metarhizium anisopliae is a sign of ageing. Microbiology 151, 3223–3236 [DOI] [PubMed] [Google Scholar]
- 15. Li L., Pischetsrieder M., St. Leger R. J., Wang C. S. (2008) Associated links among mtDNA glycation, oxidative stress and colony sectorization in Metarhizium anisopliae. Fungal Genet. Biol. 45, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 16. Kim D. H. (1997) Induced change in DNA methylation of Fusarium oxysporum f. sp. niveum due to successive transfer. J. Biochem. Mol. Biol. 30, 216–221 [Google Scholar]
- 17. Wang C. S., Skrobek A., Butt T. M. (2003) Concurrence of losing a chromosome and the ability to produce destruxins in a mutant of Metarhizium anisopliae. FEMS Microbiol. Lett. 226, 373–378 [DOI] [PubMed] [Google Scholar]
- 18. Balaban R. S., Nemoto S., Finkel T. (2005) Mitochondria, oxidants, and aging. Cell 120, 483–495 [DOI] [PubMed] [Google Scholar]
- 19. Pan Y. (2011) Mitochondria, reactive oxygen species, and chronological aging: a message from yeast. Exp. Gerontol. 46, 847–852 [DOI] [PubMed] [Google Scholar]
- 20. Gianazza E., Eberini I., Sensi C., Barile M., Vergani L., Vanoni M. A. (2011) Energy matters: mitochondrial proteomics for biomedicine. Proteomics 11, 657–674 [DOI] [PubMed] [Google Scholar]
- 21. Jiang Y. J., Wang X. (2012) Comparative mitochondrial proteomics: perspective in human diseases. J. Hematol. Oncol. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braun R. J., Westermann B. (2011) Mitochondrial dynamics in yeast cell death and aging. Biochem. Soc. Trans. 39, 1520–1526 [DOI] [PubMed] [Google Scholar]
- 23. Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., Murray S.L., Hynes M. J., Osmani S. A., Oakley B. R. (2006) A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172, 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimamoto K., Hayashi H., Taniai E., Morita R., Imaoka M., Ishii Y., Suzuki K., Shibutani M., Mitsumori K. (2011) Antioxidant N-acetyl-L-cysteine (NAC) supplementation reduces reactive oxygen species (ROS)-mediated hepatocellular tumor promotion of indole-3-carbinol (I3C) in rats. J. Toxicol. Sci. 36, 775–786 [DOI] [PubMed] [Google Scholar]
- 25. Wang J., Hu H., Wang S., Shi J., Chen S., Wei H., Xu X., Lu L. (2009) The important role of actinin-like protein (AcnA) in cytokinesis and apical dominance of hyphal cells in Aspergillus nidulans. Microbiology 155, 2714–2725 [DOI] [PubMed] [Google Scholar]
- 26. Sugui J. A., Kim H. S., Zarember K. A., Chang Y. C., Gallin J. I., Nierman W. C., Kwon-Chung H. K. (2008) Genes differentially expressed in conidia and hyphae of Aspergillus fumigatus upon exposure to human neutrophils. PLoS One 3, e2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duan Z. B., Chen Y. X., Huang W., Shang Y. F., Chen P. L., Wang C. S. (2013) Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy 9, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wright C. A., Howles S., Trudgian D. C., Kessler B. M., Reynard J. M., Noble J. G., Hamdy F. C., Turney B. W. (2011) Label-free quantitative proteomics reveals differentially regulated proteins influencing urolithiasis. Mol. Cell. Proteomics 10, M110.005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta N., Pevzner P. A. (2009) False discovery rates of protein identifications: a strike against the two-peptide rule. J. Proteome Res. 8, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vizcaíno J. A, Côté R. G, Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Griffin N. M., Yu J. Y., Long F., Oh P., Shore S., Li Y., Koziol J. A., Schnitzer J. E. (2010) Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Efron B. (2007) Size, power and false discovery rates. Ann. Stat. 35, 1351–1377 [Google Scholar]
- 33. Benjamini Y., Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 [Google Scholar]
- 34. Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Güldener U., Mannhaupt G., Münsterkötter M., Mewes H. W. (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32, 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westfall P. J., Momany M. (2002) Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patergnani S., Suski J. M., Agnoletto C., Bononi A., Bonora M., De Marchi E., Giorgi C., Marchi S., Missiroli S., Poletti F., Rimessi A., Duszynski J., Wieckowski M. R., Pinton P. (2011) Calcium signaling around Mitochondria Associated Membranes (MAMs). Cell Commun. Signal. 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S. A., Oakley B. R. (2006) Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protocols 1, 3111–3120 [DOI] [PubMed] [Google Scholar]
- 38. Zhong G. W., Wei W. F., Guan Q., Ma Z. F., Wei H., Xu X. S., Zhang S. Z., Lu L. (2012) Phosphoribosyl pyrophosphate synthetase, as a suppressor of the sepH mutation in Aspergillus nidulans, is required for the proper timing of septation. Mol. Microbiol. 86, 894–907 [DOI] [PubMed] [Google Scholar]
- 39. Moss B. J., Kim Y., Nandakumar M. P., Marten M. R. (2008) Quantifying metabolic activity of filamentous fungi using a colorimetric XTT assay. Biotechnol. Prog. 24, 780–783 [DOI] [PubMed] [Google Scholar]
- 40. Gong J. M., Lee D. A., Schroeder J. I. (2003) Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 10118–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osiewacz H. D., Brust D., Hamann A., Kunstmann B., Luce K., Müller-Ohldach M., Scheckhuber C. Q., Servos J., Strobel I. (2010) Mitochondrial pathways governing stress resistance, life, and death in the fungal aging model Podospora anserina. Ann. N.Y. Acad. Sci. 1197, 54–66 [DOI] [PubMed] [Google Scholar]
- 42. Suelmann R., Fischer R. (2000) Mitochondrial movement and morphology depend on an intact actin cytoskeleton in Aspergillus nidulans. Cell Motil. Cytoskeleton 45, 42–50 [DOI] [PubMed] [Google Scholar]
- 43. Shoji J. Y., Kikuma T., Arioka M., Kitamoto K. (2010) Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS One 5, e15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eisenman H. C., Casadevall A. (2012) Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suen D. F., Norris K. L., Youle R. J. (2008) Mitochondrial dynamics and apoptosis. Genes Dev. 22, 1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Voisine C., Schilke B., Ohlson M., Beinert H., Marszalek J., Craig E. A. (2000) Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol. Cell. Biol. 20, 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maggio-Hall L. A., Keller N. P. (2004) Mitochondrial beta-oxidation in Aspergillus nidulans. Mol. Microbiol. 54, 1173–1185 [DOI] [PubMed] [Google Scholar]
- 48. Li M., Zhang X. J., Agrawal A., San K. Y. (2012) Effect of acetate formation pathway and long chain fatty acid CoA-ligase on the free fatty acid production in E. coli expressing acy-ACP thioesterase from Ricinus communis. Metab. Eng. 14, 380–387 [DOI] [PubMed] [Google Scholar]
- 49. Das A. M., Harris D. A. (1990) Control of mitochondrial ATP synthase in heart cells: inactive to active transitions caused by beating or positive inotropic agents. Cardiovasc. Res. 24, 411–417 [DOI] [PubMed] [Google Scholar]
- 50. Bick A. G., Calvo S. E., Mootha V. K. (2012) Evolutionary diversity of the mitochondrial calcium uniporter. Science 336, 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cronin S. R., Rao R., Hampton R. Y. (2002) Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 157, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Palmer C. S., Osellame L. D., Stojanovski D., Ryan M. T. (2011) The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell. Signal. 23, 1534–1545 [DOI] [PubMed] [Google Scholar]
- 53. Youle R. J., van der Bliek A. M. (2012) Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karbowski M., Youle R. J. (2003) Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 10, 870–880 [DOI] [PubMed] [Google Scholar]
- 55. Giorgi C., Baldassari F., Bononi A., Bonora M., De Marchi E., Marchi S., Missiroli S., Patergnani S., Rimessi A., Suski J. M., Wieckowski M. R., Pinton P. (2012) Mitochondrial Ca2+ and apoptosis. Cell Calcium 52, 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wissing S., Ludovico P., Herker E., Büttner S., Engelhardt S. M., Decker T., Link A., Proksch A., Rodrigues F., Corte-Real M., Fröhlich K. U., Manns J., Candé C., Sigrist S. J., Kroemer G., Madeo F. (2004) An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 166, 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schlattner U., Tokarska-Schlattner M., Ramirez S., Brückner A., Kay L., Polge C., Epand R. F., Lee R. M., Lacombe M. L., Epand R. M. (2009) Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim. Biophys. Acta 1788, 2032–2047 [DOI] [PubMed] [Google Scholar]
- 58. Endo N., Nishiyama K., Okabe M., Matsumoto M., Kanouchi H., Oka T. (2007) Vitamin B6 suppresses apoptosis of NM-1 bovine endothelial cells induced by homocysteine and copper. Biochim. Biophys. Acta 1770, 571–577 [DOI] [PubMed] [Google Scholar]
- 59. Van Aken O., Whelan J., Van Breusegem F. (2010) Prohibitins: mitochondrial partners in development and stress response. Trends Plant Sci. 15, 275–282 [DOI] [PubMed] [Google Scholar]
- 60. Beere H.M., Wolf B. B., Cain K., Mosser D. D., Mahboubi A., Kuwana T., Tailor P., Morimoto R. I., Cohen G. M., Green D. R. (2000) Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469–475 [DOI] [PubMed] [Google Scholar]
- 61. Wei Y. H., Lee H. C. (2002) Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp. Biol. Med. 227, 671–682 [DOI] [PubMed] [Google Scholar]
- 62. Groocock L. M., Prudden J., Perry J. J., Boddy M. N. (2012) The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast. Mol. Cell. Biol. 32, 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramanagoudr-Bhojappa R., Blair L. P., Tackett A. J., Raney K. D. (2013) Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 41, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gallo-Fernández M., Saugar I., Ortiz-Bazán M. Á., Vázquez M. V., Tercero J. A. (2012) Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res. 40, 8325–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brustovetsky N., Brustovetsky T., Jemmerson R., Dubinsky J.M. (2002) Calcium-induced cytochrome c release from CNS mitochondria is associated with the permeability transition and rupture of the outer membrane. J. Neurochem. 80, 207–218 [DOI] [PubMed] [Google Scholar]
- 66. Brookes P. S., Yoon Y., Robotham J. L., Anders M. W., Sheu S. S. (2004) Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 287, C817–C833 [DOI] [PubMed] [Google Scholar]
- 67. Adams T. H., Boylan M. T., Timberlake W. E. (1988) brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362 [DOI] [PubMed] [Google Scholar]
- 68. Rerngsamran P., Murphy M. B., Doyle S. A., Ebbole D. J. (2005) Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene. Mol. Microbiol. 56, 282–297 [DOI] [PubMed] [Google Scholar]
- 69. Calvo A. M., Wilson R. A., Bok J. W., Keller N. P. (2002) Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bayram Ö., Braus G. H. (2012) Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24 [DOI] [PubMed] [Google Scholar]
- 71. Déquard-Chablat M., Sellem C.H., Golik P., Bidard F., Martos A., Bietenhader M., di Rago J. P., Sainsard-Chanet A., Hermann-Le Denmat S., Contamine V. (2011) Two nuclear life cycle-regulated genes encode interchangeable subunits c of mitochondrial ATP synthase in Podospora anserina. Mol. Biol. Evol. 28, 2063–2075 [DOI] [PubMed] [Google Scholar]
- 72. Adam C., Picard M., Déquard-Chablat M., Sellem C. H., Hermann-Le Denmat S., Contamine V. (2012) Biological roles of the Podospora anserina mitochondrial Lon protease and the importance of its N-domain. PLoS One 7, e38138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bota D. A., Ngo J. K., Davies K. J. (2005) Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic. Biol. Med. 38, 665–677 [DOI] [PubMed] [Google Scholar]
- 74. Lemasters J. J., Theruvath T. P., Zhong Z., Nieminen A. L. (2009) Mitochondrial calcium and the permeability transition in cell death. Biochim. Biophys. Acta 1787, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Scheckhuber C. Q., Erjavec N., Tinazli A., Hamann A., Nystrom T., Osiewacz H. D. (2007) Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9, 99–105 [DOI] [PubMed] [Google Scholar]
- 76. Groebe K., Krause F., Kunstmann B., Unterluggauer H., Reifschneider N. H., Scheckhuber C. Q., Sastri C., Stegmann W., Wozny W., Schwall G. P., Poznanovic S., Dencher N.A., Jansen-Dürr P., Osiewacz H.D., Schrattenholz A. (2007) Differential proteomic profiling of mitochondria from Podospora anserina, rat and human reveals distinct patterns of age-related oxidative changes. Exp. Gerontol. 42, 887–898 [DOI] [PubMed] [Google Scholar]
- 77. Stöckl P., Zankl C., Hutter E., Unterluggauer H., Laun P., Heeren G., Bogengruber E., Breitenbach M., Jansen-Durr P. (2007) Partial uncoupling of oxidative phosphorylation induces premature senescence in human fibroblasts and yeast mother cells. Free Radic Biol. Med. 43, 947–958 [DOI] [PubMed] [Google Scholar]
- 78. Ly J. D., Grubb D. R., Lawen A. (2003) The mitochondrial membrane potential (Δψm) in apoptosis; an update. Apoptosis 8, 115–128 [DOI] [PubMed] [Google Scholar]
- 79. Spencer S. L., Sorger P. K. (2011) Measuring and modeling apoptosis in single cells. Cell 144, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liang H., van Remmen H., Frohlich V., Lechleiter J., Richardson A., Ran Q. (2007) Gpx4 protects mitochondrial ATP generation against oxidative damage. Biochem. Biophys. Res. Commun. 356, 893–898 [DOI] [PubMed] [Google Scholar]
- 81. Bullerwell C. E., Lang B. F. (2005) Fungal evolution: the case of the vanishing mitochondrion. Curr. Opin. Microbiol. 8, 362–369 [DOI] [PubMed] [Google Scholar]
- 82. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 83. Elstner M., Andreoli C., Ahting U., Tetko I., Klopstock T., Meitinger T., Prokisch H. (2008) MitoP2: an integrative tool for the analysis of the mitochondrial proteome. Mol. Biotechnol. 40, 306–315 [DOI] [PubMed] [Google Scholar]
- 84. Smith A. C., Blackshaw J. A., Robinson A. J. (2012) MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 40, D1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Keeping A., DeAbreu D., DiBernardo M., Collins R. A. (2011) Gel-based mass spectrometric and computational approaches to the mitochondrial proteome of Neurospora. Fungal Genet. Biol. 48, 526–536 [DOI] [PubMed] [Google Scholar]
- 86. Holt I. J. (2009) Mitochondrial DNA replication and repair: all a flap. Trends Biochem. Sci. 34, 358–365 [DOI] [PubMed] [Google Scholar]
- 87. Ben-Menachem R., Tal M., Shadur T., Pines O. (2011) A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics 11, 4468–4476 [DOI] [PubMed] [Google Scholar]
- 88. Koch K. V., Suelmann R., Fischer R. (2003) Deletion of mdmB impairs mitochondrial distribution and morphology in Aspergillus nidulans. Cell Motil. Cytoskeleton 55, 114–124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.