Abstract
The mechanisms involved in progression from monoclonal gammopathy of undetermined significance (MGUS) and smoldering myeloma (SMM) to malignant multiple myeloma (MM) and plasma cell leukemia (PCL) are poorly understood but believed to involve the sequential acquisition of genetic hits. We performed exome and whole genome sequencing on a series of MGUS (n=4), high risk (HR)-SMM (n=4), MM (n=26) and PCL (n=2) samples, including four cases who transformed from HR-SMM to MM, to determine the genetic factors which drive progression of disease. The pattern and number of non-synonymous mutations show that the MGUS disease stage is less genetically complex than MM, and HR-SMM is similar to presenting MM. Intraclonal heterogeneity is present at all stages and using cases of HR-SMM, which transformed to MM, we show that intraclonal heterogeneity is a typical feature of the disease. At the HR-SMM stage of disease the majority of the genetic changes necessary to give rise to MM are already present. These data suggest that clonal progression is the key feature of transformation of HR-SMM to MM and as such the invasive clinically predominant clone typical of MM is already present at the SMM stage and would be amenable to therapeutic intervention at that stage.
Keywords: myeloma, smoldering, progression, genome, sequencing
Introduction
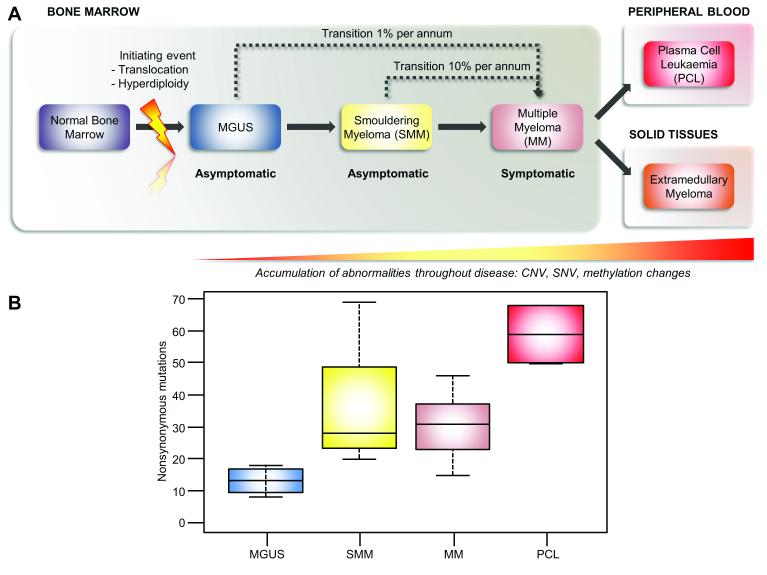
A key question in cancer biology is how a cell transforms from an essentially benign premalignant condition to a malignant invasive state. Some hints as to how this happens can come from the study of myeloma (MM) which is characterized by the proliferation of abnormal plasma cells1 that have fewer mutations than the more complex solid tumors and as such provides a good model in which to address the question of cancer progression. Current understanding of MM development is thought to involve a multistep transformation process, occurring as a consequence of the sequential acquisition of genetic hits that deregulate the behavior of a normal plasma cell, Figure 1A. The first stage of this process is a benign premalignant condition, known as Monoclonal Gammopathy of Undetermined Significance (MGUS) that transforms to MM at a rate of 1% per annum.2, 3 The next step of MM evolution is the development of Smoldering Myeloma (SMM), that shares the same morphologic features as symptomatic MM, but lacks evidence of end-organ damage4 and progresses to MM at a rate of 10% per annum.2 In the final stage of the transformation process, the malignant plasma cell clone gains independence from the bone marrow microenvironment, presenting either as plasma cell leukemia (PCL) or as extra-medullary myeloma (EMM).5
Figure 1. The number of acquired mutations increases as disease progresses.
A, a schematic of disease progression from MGUS through to PCL. B, Boxplot showing the median number of acquired NS-SNVs increases as the disease progresses from MGUS through HR-SMM and MM to PCL.
Translocations, involving the IGH locus, or hyperdiploidy are seen as primary events in myeloma that are present in 100% of cells but these alone are not sufficient to result in clinically invasive myeloma, given that they can be found in patients with MGUS and may be stable for many years.6 Therefore, there must be additional genetic events, which contribute towards disease progression leading to the invasive stages of the disease. There is significant interest in defining these driver mutations that push the malignant clone towards more invasive behavior, as they constitute good therapeutic targets. The main approach to identifying these variables has been through comparing the genetics of samples taken from different individuals who have MGUS, SMM or MM. As a result of such studies we have identified a number of lesions, which are known to contribute to disease progression including an increased frequency of copy number abnormalities,7, 8 genome-wide hypomethylation and gene-specific hypermethylation,9, 10 secondary translocations (often involving MYC)11 and altered gene expression.12, 13 A more elegant way of accurately defining the genetic factors associated with disease progression is to study sequential samples from the same patient as they progress from one clinical stage to another, yet few such studies have done this because of the difficulty in obtained paired samples from the same individual. The limited number of studies that have taken this approach, however, have been very informative showing that the frequency of genomic alterations, such as translocations and copy number alterations, increases with disease progression but as a rule they are generally present during the early stages of disease.14-18 Since such studies have been performed technology has moved on and the availability of genome-wide analyses, such as massively parallel sequencing, provides a tool able to deliver an unbiased genome wide assessment of the changes present during the transition from one clinical state to another.
Understanding the biology underlying cancer progression is important for therapeutic strategies aimed at cancer control and is being studied intensively. The results of recent analyses have suggested that instead of following a linear multistep pathway through this transformation process, progression is more likely to develop via a branching evolutionary process, as first used by Darwin to explain the evolution of species.19, 20 This model of cancer evolution requires the coexistence of multiple diverging clones, the essential substrate for evolution, and has been recently demonstrated in a number of different malignancies, including multiple myeloma (MM).21-23 Even within such Darwinian models it remains important to understand what drives the competition between clones in order to effectively manipulate it therapeutically. Therefore, investigating the relationship between HR-SMM and MM in paired samples that have undergone the SMM-MM transformation provides the opportunity to define novel genetic hits that contribute to sub-clonal growth and survival advantage that mediates the development of end organ damage, typical of MM that requires treatment and is the myeloma equivalent of invasive cancer. In this study we have used massively parallel sequencing to study the genetic makeup of the stages of MM and in particular to investigate the genetic relationship of the transition from HR-SMM to MM.
Materials and methods
Samples were derived from the QUIREDEX study (NCT00480363) where high risk (HR) SMM cases were randomized to receive either therapy with lenalidomide and dexamethasone or to undergo longitudinal follow up only. High-risk SMM was defined as the presence of >10% plasma cells (PC) in the bone marrow (BM) and a monoclonal component IgG >= 3 g/dL, IgA >=2 g/dL or Bence Jones proteinuria >= 1 g/24 h together with absence of CRAB (Calcium increase, Renal Insufficiency, Anemia, Bone lesions).24 Patients meeting either but not both of these two criteria were also included in the study if they met the additional criteria of having ≥95% phenotypically aberrant plasma cells (aPC) in the BM PC compartment (aPC/BMPC) and immunoparesis.25 BM samples were collected at the time of entering the study and at disease progression to symptomatic myeloma. Patient characteristics are shown in Supplementary Table 1. In all the BM samples CD138-positive PC isolation was carried out using the AutoMACs automated separation system (Miltenyi-Biotec, Auburn, CA, USA). Purity was >95% in all SMM cases. The systematic screening for genomic aberrations includes interphase fluorescence in situ hybridization (FISH) studies for detecting IGH rearrangements (LSI IGH dual-color, break-apart rearrangement probe; Abbott Molecular/Vysis, Des Plaines, IL, USA), 13q (LSI 13, RB1 13q14) and 17p deletions (LSI p53, 17p13.1) (Abbott Molecular/Vysis) as previously described, and 1q gains (ON 1q21/SRD 1p36, Kreatech Diagnostics, Amsterdam). Those SMM samples with IGH translocations were explored for t(11;14)(q13;q32), t(4;14)(p16;q32) and t(14;16)(q32;q23) with the corresponding dual-color, dual-fusion translocation probes from Abbott Molecular/Vysis. The interphase FISH procedure has been described previously in detail.26 A total of 200 interphase nuclei were analyzed using the scoring criteria recommended by the manufacturer. The cut-off level for the identification of IGH translocations (fusion/break-apart probes) and 1q gains was set at 10% and at 20% for numerical abnormalities, as recommended by the European Myeloma Network FISH workshop. All tests were performed on the SMM sample and positive tests were repeated on the progression MM sample.
Approximately 100 ng of DNA underwent whole genome sequencing using 120 bp paired-end reads on a GAIIx (Illumina) to a median depth of 44x, with 99% of the genome covered at >1x and 96% >20x coverage. From each patient the non-involved peripheral white blood cells were sequenced as well as bone marrow-derived CD138-selected cells from the HR-SMM diagnosis sample and the sample following progression to MM. Two patients were also sequenced to a higher depth by exome sequencing (see Supplementary Table 2 for metrics). Additionally, whole exome sequencing was performed in an identical manner following exome capture using the Agilent SureSelect Human All Exon 50 Mb system on 4 MGUS, 22 MM (previously published)27 and 2 PCL paired tumor/normal samples (see Supplementary Table 2 for metrics). These additional MM, MGUS and PCL samples were acquired at the Royal Marsden Hospital, London.
Paired reads were aligned to the human genome (GRCh37) using Stampy28 and BWA29 and duplicate reads were removed using Picard. Sequence recalibration, local realignment and single nucleotide variant (SNV) calling were all undertaken using the GATK.30 SnpEff was used to functionally annotate all variants. Further filtering of variants and comparisons between samples were performed using code written in R. Variants present in both the peripheral blood and tumor samples were discarded. Only variants sequenced to a minimum depth of 10x in both the tumor and matched normal sample, as well as having a minimum genotype score of greater than 50 (representing a 1 in 100 000 error rate) and no more than one variant read in the normal sample were retained. Variants were determined to be unique to a disease stage or present in both stages by comparing the base calls at the location of the SNV in both samples. A high-level overview of the analysis pipeline is given in supplementary figure 1.
Whole genome copy number was called using a combination of the R package BICseq to perform segmentation of the log ratio of the read depth between matched peripheral blood and tumor samples and the R package CGHcall to produce integer copy calls within the segmented regions. Copy number in exome samples was similarly determined using the R package ExomeCNV.
The proportion of cells containing a variant was estimated using the following equation:
Where p is the proportion of cells containing the variant, C is the integer copy number at that position, r is the number of reads containing the variant and R is the total number of reads. Note that this method assumes that the tumor cell population is free from normal cell contamination and that detected copy number changes are not subclonal.
Results
Mutation characteristics
We performed whole exome sequencing of DNA derived from a series of samples with MGUS (n=4), MM (n=22) and PCL (n=2), as well as four HR-SMM and MM paired samples from the same patients, in order to characterize the changes present in MGUS, HR-SMM, MM and finally in PCL. The tumor acquired SNVs and copy number variants in each sample were defined and the number and nature of mutations in each stage of disease are tabulated, Supplementary Table 3 and Supplementary Table 4. The median number of non-synonymous (NS)-SNVs in the MGUS exomes was 13 (range 8-18), in HR-SMM 28 (range 20-69), in MM 31 (range 15-46), and in the PCL samples is 59 (50-68). We, therefore, show that the number of NS-SNVs increases with disease progression from MGUS to PCL, Figure 1B.
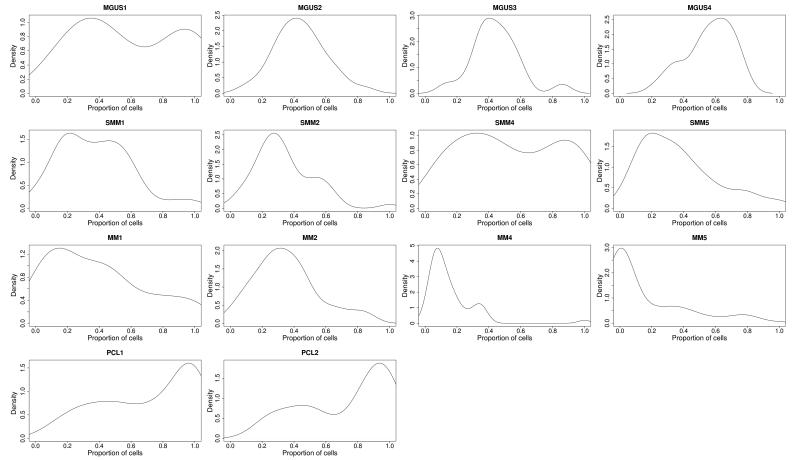
We have previously shown that there is intraclonal heterogeneity, at the level of SNVs, in presenting myeloma.27 We used a similar approach to analyze the data generated in this study to determine if there is clonal heterogeneity present at the early premalignant stages of disease (MGUS and HR-SMM) as well as the advanced stage of disease (PCL). The kernel density plots in Figure 2 show several distinct peaks indicating a similar level of heterogeneity exists at all stages of disease from MGUS to PCL. This intraclonal heterogeneity at the MGUS stage is consistent with clonal diversity arising early on in the process of myelomagenesis and with clonal competition being an essential requirement from the earliest phases of disease. The rate of transformation to MM from MGUS is slow which is consistent with the requirement for the acquisition of multiple mutations able to deregulate driver genes but that these constitute rare events.
Figure 2. Clonal heterogeneity is present in all disease states.
Gaussian kernel density plots indicating the frequency of cells carrying all acquired exonic mutations. Frequency is calculated by adjusting mutant allele burden by copy number of the loci mutated. Top row, MGUS samples; Middle rows, matched HR-SMM/MM samples; Bottom row, PCL samples.
Having shown the presence of intraclonal heterogeneity at each stage of the disease we were keen to understand how the relationship between subclones changes with the development of clinical symptoms and whether this relationship followed Darwinian principles. In order to accurately determine the changes occurring in the transition from HR-SMM to MM we analyzed three sets of patients with paired HR-SMM and MM samples taken at least 21 months apart. SNVs were called for each of the genomes sequenced and in the order of 19 000 acquired variants were detected, Supplementary Table 4. Of the total acquired SNVs, 93% (range 91-95%) were detected in both the HR-SMM and MM samples, with 81% (range 64-94%) of the variants being present in less than half of the tumor cells at both stages. In only 3·3% (range 0·3-8·1%) of cases were variants present in greater than 90% of MM cells. This observation suggests that both HR-SMM and MM contain many sub-clones at low frequencies, a feature that would be anticipated if disease progression was the result of clonal competition.
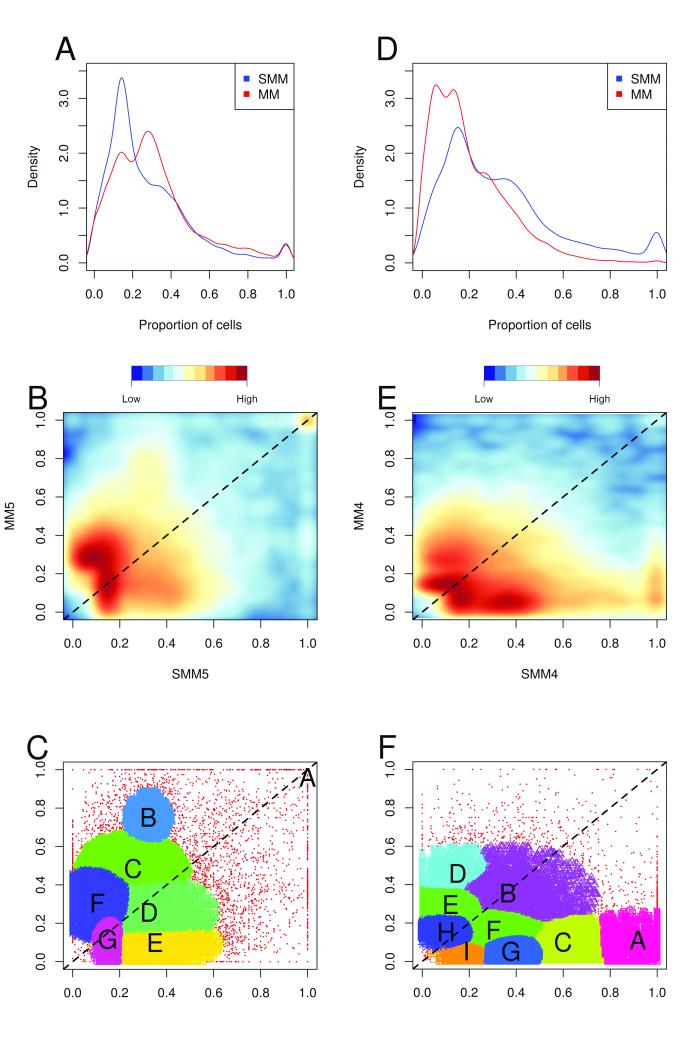
After calculating the proportion of cells that contained each variant, by combining base calls and copy number data, it was possible to define the sub-clonal composition of each disease stage. The relationship between the various sub-clones was best illustrated when kernel density plots were created for each patient, Figure 3A, and these clearly show changes in clonal composition over time. The same data are displayed as a 2-dimensional scatterplot with kernel density estimates represented as a color scale, Figure 3B. In order to identify clusters in these plots an expectation-maximization based model was used to group the data, Figure 3C (and Supplementary Figure 2). Typically, around six clusters were identified per sample pair and although these variant clusters do not directly represent clones they can be used to demonstrate clonal evolution over time.
Figure 3. Clonal evolution of paired HR-SMM and MM samples from an untreated (patient 5; A-C) and a treated patient (patient 4; D-F).
A and D: Kernel density plots of the proportion of cells containing each variant from whole genome sequencing in HR-SMM (blue) and MM (red); B and E: Comparison of proportion of cells containing each variant in HR-SMM and MM samples. Positive and negative vertical deviations from the main diagonal (marked at a dashed line) indicate an increase or decrease in the variant from the HR-SMM to MM stage respectively. C and F: After estimating noise using a nearest-neighbor based classifier, an EM based clustering strategy was employed to define clusters of variants. Most of the variants in HR-SMM occur at low frequency, but a distinct increase in frequency for a large set of variants is seen in the MM sample.
One paired set of samples was sourced from a patient who was treated with lenalidomide and dexamethasone and clearly demonstrates that chemotherapy results in a reduction in clonal complexity, Figure 3D-F. For example, cluster A was dominant at the HR-SMM stage, with mutations present in 80-100% of cells, but after treatment it has decreased to 0-20% of cells. Some clusters were largely unaffected by treatment (clusters B, E, F, H and I which stay around the diagonal). Conversely, cluster D has gone from 0-20% of cells at HR-SMM and risen to 40-60% at progression to MM, perhaps due to therapy resistance.
Acquired genetic change in the progression from HR-SMM to MM
Having shown changes in intraclonal composition during disease progression we sought to define the genetic changes mediating the differences in sub-clonal behavior facilitating the growth and survival advantage for the clone mediating disease progression.
There were on average 433 additional novel and unique mutations (range 341-517) gained per sample during the transition from HR-SMM to symptomatic MM, few (mean 2·3) were present within coding regions and only one was non-synonymous, Table 1. The one case with the acquired NS-mutation was characterized by the acquisition of a novel acquired stop mutation at amino acid position 31 (c.G92A, p.W31*) in RUNX2, located on chromosome 6, which has features of an important driver mutation and could mediate the changes in biology that could cause the transition. There was no copy number change at this locus, indicating that haplo-insufficiency may a mechanism at work. Understanding the rate of mutation acquisition is important and as we know the time to progression for the patients studied we used the number of variants unique to each HRSMM and MM sample to calculate the rate at which new mutations were gained and existing mutations lost, or at least became undetectable at the level of sensitivity of the test used. The mean across the three cases was 19 mutations gained and 36 mutations lost per month.
Table 1. Shared and unique variants in HR-SMM and MM samples.
| Sample | Variant Type | Total SNV | Coding SNV | NS SNV |
|---|---|---|---|---|
| Patient 1 | HR-SMM unique | 1 213 | 9 | 3 |
| MM unique | 431 | 0 | 0 | |
| Shared | 16 061 | 56 | 32 | |
| Patient 2 | HR-SMM unique | 645 | 2 | 2 |
| MM unique | 341 | 1 | 1 | |
| Shared | 20 433 | 46 | 32 | |
| Patient 4 | HR-SMM unique | 1 042 | 2 | 2 |
| MM unique | 443 | 0 | 0 | |
| Shared | 18 227 | 51 | 32 | |
| Patient 5 | HR-SMM unique | 637 | 51 | 39 |
| MM unique | 517 | 2 | 0 | |
| Shared | 17 645 | 48 | 38 |
Importantly from a mechanistic perspective it seems that copy number change occurs relatively early in these cases as using FISH, at a macroscopic level, we could not see significant copy number change during the transition, a feature that was recapitulated when the sequencing data were used to generate copy number data across the genome, Supplementary Figure 3. These data indicate that in these three cases the acquisition of copy number abnormalities does not facilitate the transformation between the two disease stages and was a feature of earlier stages of the disease.
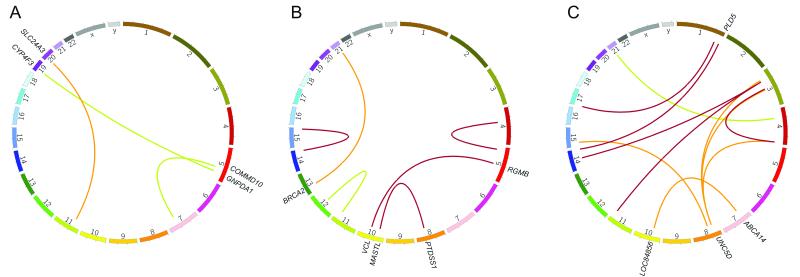
Progression could also be mediated by chromosomal rearrangement and we used the data generated here to describe a number of translocations, finding events that exist in both the HR-SMM and MM samples, as well as those that are either gained or lost in the HR-SMM to MM transition, Figure 4 and Supplementary Table 7. Translocations acquired in the transition from HR-SMM to symptomatic MM could readily mediate disease progression and we describe a number of these. These include a translocation in patient two, involving BRCA2 which was disrupted by a t(13;21). Also, in patient five several translocations involving UNC5D were seen which was notable for its complexity, involving three separate translocation events.
Figure 4. Circos plots of translocations detected in SMM-MM paired samples.
A, patient 1. B, patient 2. C, patient 5. Red lines indicate translocations present in both SMM and MM samples, orange in only the MM sample, and green in only the SMM sample. No translocations were identified in patient 4.
The limited number of novel NS-SNVs, indels and translocations which characterize the transformed MM samples indicates that by the time a case has evolved to the HR-SMM stage that the majority, if not all, of the exonic genetic diversity necessary to give rise to an aggressive clinical state is already present. We show that 1 732 mutations were acquired in the MM samples, of which only one was non-synonymous. However, there is substantial intronic diversity, the significance of which has not previously been demonstrated.
Discussion
It has been thought that the acquisition of mutations over time leads to the progression of cancer from essentially benign to clinically aggressive behavioral states. We have used massively parallel sequencing to study the multistep progression from a normal plasma cell to one which has leukemic properties, with the aim of describing the genetic and sub-clonal events which characterize disease progression. We show that the number of NS mutations defining cases of symptomatic MM is approximately 23, a number intermediate between simple cancers, such as AML,31 and the more complex solid cancers.32 Based on the data generated here and single cell data analyzed previously we suggest that there are approximately 6 predominant clones with much greater complexity existing below the sensitivity of the sequencing approach we have used. We go on to show that MGUS has fewer NS mutations than the later stages of disease including HR-SMM, MM and PCL. However, when we look at the nature and sites of mutation present at each disease stage, they do not differ, an observation that is consistent with the same molecular mechanisms being active throughout the course of the disease.
To date, the genomes of the premalignant stages of MM have not been sequenced and compared to those of symptomatic MM or PCL. Here we demonstrate that there is an increase in the number of NS-mutations as the disease progresses from MGUS to HR-SMM and MM. This is in line with previous copy number abnormality data which indicate that the genetic complexity of the disease increases towards MM.14-17 MGUS is known to take >25 years to progress to symptomatic MM, whereas SMM takes <5 years to progress. In this context it is interesting to note that the MGUS samples are very much less complex compared to symptomatic MM, having approximately half the number of NS-mutations, whereas the HR-SMM samples have an equal number of mutations compared to symptomatic MM.
We demonstrate clear evidence of sub-clonal heterogeneity in all stages of disease from MGUS to PCL, which is an essential substrate for Darwinian type evolution, suggesting that disease progression is mediated via competition between sub-clones and outgrowth of the fittest. In this report, for the first time, we demonstrate that intra-clonal heterogeneity is present in MGUS, the earliest clinically recognizable stage of MM, and that it is also present in the later stages of disease including HR-SMM and PCL. These observations are consistent with intra-clonal heterogeneity being a critical and consistent feature of both the early and late stages of myeloma. It is also consistent with clonal competition being active from the earliest phases of disease until its highly aggressive late stages and provides direct evidence to support the hypothesis that Darwinian evolution mediates progression through the multiple steps of the disease model of MM. The development of PCL via this complex evolutionary process characterized by extreme deregulation of the behavior of the normal cellular counterpart is consistent with the poor results of treatment associated with this stage of disease. This situation can be contrasted with the results of treatment of de-novo acute leukemia which can be cured probably because of the significantly less number of genetic hits required to deregulate a hematopoietic stem cell.
In order to directly demonstrate the sub-clonal relationships underlying the transitions between the disease phases of myeloma we paid particular attention to the HR-SMM/MM transition using serial samples from the same individuals. This is a critical transition where an essentially benign state, that it not treated, undergoes a change that results in clinical symptoms consistent with clonally destructive behavior and the development of end organ damage. Using kernel plot analysis of these paired samples we can see changes in clonal composition, mediated via expansion of specific sub-clones, associated with the transition an observation that is consistent with previous observations concerning the clonal expansion of genetically abnormal cells.14 Further, looking at the clonal composition of the paired samples we show that the transformation of HR-SMM to MM is not the result of the outgrowth of a single clone but results from the outgrowth of a number of sub-clones, already present in the HR-SMM sample. It seems that, in MM, up to six clones change during the HR-SMM-MM transition, which is more than has been suggested previously in pediatric acute lymphoblastic leukemia,23 reflecting the increased genetic and sub-clonal heterogeneity seen in MM compared to this rather simple cancer model.
The question then arises as to the nature of the molecular mechanisms driving the sub-clonal changes. At the HR-SMM time point we show that the majority of the non-synonymous exonic changes are already present and that only a few additional changes are seen following transformation. We describe one case, patient two, in which an acquired NS-SNV variant in RUNX2 is seen following progression to MM, which could plausibly drive the expansion of the clone carrying it. Consistent with this RUNX2 is expressed in myeloma cells and regulates osteopontin (OPN), a multifunctional bone matrix glycoprotein that is involved in angiogenesis, cell survival and proliferation.33, 34 RUNX2 mutations are also seen in a range of cancers with three stop mutations, 17 non-synonymous and six synonymous mutations having been described.35 These features are consistent with inactivating mutations in RUNX2 having a credible role in the progression of myeloma.
We also describe a number of translocations, three of which are only present in the MM sample. Of interest, also in patient two, we describe a translocation disrupting one allele of BRCA2, which could mediate clonal behavior. The other interesting feature is several translocations into UNC5D, in patient five, which is very complex and could be an interesting mechanism mediating disease progression and is a known feature of myeloma.36, 37 UNC5D has been implicated in predisposition to colon cancer and is mutated in 9% of diffuse large B-cell lymphomas as well as being involved in translocations in breast cancer.35, 38, 39 It has a role as a netrin-1 receptor, is transcriptionally controlled by p53, and induces apoptosis making it a good candidate for tumor progression in myeloma.40
Apart from inactivating mutations in RUNX2 and acquired translocations into BRCA2 and UNC5D we could not identify any truly acquired genetic abnormalities between the paired samples despite thoroughly checking for coding SNVs, indels and copy number abnormalities. Having exhaustively searched for acquired mutational change to explain the transition and having found only limited numbers of potential molecular drivers to explain the development of clinical symptoms and how disease progression could be mediated alternative explanations should be considered. There are a number of non-mutational changes which could mediate the transition including epigenetic changes, which we have not looked for in this study but have been described previously.9, 10 Alternatively, based on the changes in sub-clonal composition, as a consequence of an increase in tumor bulk there is a threshold effect above which the MM cells modify the microenvironment such that they favor increased proliferation and expansion of a sub-clone best suited to that environment, the net consequence of which is sub-clonal expansion and the development of end organ damage.
When these observations based on the experimental characterization of the multiple disease phases of myeloma and the transition of HR-SMM to MM in particular are considered it seems likely that HR-SMM is not a distinct disease entity but is rather a transition state between MGUS and MM where the sub-clonal structure is evolving. MGUS is a benign clinical state which is stable with no clinical symptoms which contrasts with presenting myeloma when tumor bulk has reached a critical threshold and clinical damage has developed. The in-between state, SMM, seems to represent a phase characterized by ongoing competition between sub-clones with the rate of transformation depending upon the nature and rate of mutations which drive the process.
We present a coherent hypothesis based on the data we find in myeloma tumour sequencing data. In this hypothesis, adaptation is driven by genetic variation in the tumour cells. At presentation the only selective pressures are those which occur naturally and would involve competition for the myeloma cell niche in the bone marrow and the avoidance of selective pressures exerted by the immune system. Once treatment has been initiated the selective pressures change and possibly the evolution of the tumour is enhanced by exposure to treatment.
Supplementary Material
Acknowlegements
We gratefully acknowledge The Institute of Cancer Research Tumour Profiling Unit for their support and technical expertise in this study.
Grant Funding: This study was supported by grants from Myeloma UK, Cancer Research UK and the National Institutes of Health Biomedical Research Centre at the Royal Marsden Hospital.
Footnotes
Conflicts of Interest: SH, LM, MR and DB are employed by Illumina Cambridge Ltd.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF, et al. A Long-Term Study of Prognosis in Monoclonal Gammopathy of Undetermined Significance. New England Journal of Medicine. 2002;346(8):564–569. doi: 10.1056/NEJMoa01133202. [DOI] [PubMed] [Google Scholar]
- 3.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009;113(22):5418–5422. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. British journal of haematology. 2003;121(5):749–57. [PubMed] [Google Scholar]
- 5.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–48. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 6.Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA, et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood. 2002;100(4):1417–1424. [PubMed] [Google Scholar]
- 7.Chiecchio L, Dagrada GP, Ibrahim AH, Dachs Cabanas E, Protheroe RK, Stockley DM, et al. Timing of acquisition of deletion 13 in plasma cell dyscrasias is dependent on genetic context. Haematologica. 2009;94(12):1708–13. doi: 10.3324/haematol.2009.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magrangeas F, Lode L, Wuilleme S, Minvielle S, Avet-Loiseau H. Genetic heterogeneity in multiple myeloma. Leukemia. 2005;19(2):191–194. doi: 10.1038/sj.leu.2403555. [DOI] [PubMed] [Google Scholar]
- 9.Salhia B, Baker A, Ahmann G, Auclair D, Fonseca R, Carpten J. DNA methylation analysis determines the high frequency of genic hypomethylation and low frequency of hypermethylation events in plasma cell tumors. Cancer Res. 2010;70(17):6934–44. doi: 10.1158/0008-5472.CAN-10-0282. [DOI] [PubMed] [Google Scholar]
- 10.Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011;117(2):553–62. doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- 11.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. Journal of the National Cancer Institute. Monographs. 2008;(39):25–31. doi: 10.1093/jncimonographs/lgn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies FE, Dring AM, Li C, Rawstron AC, Shammas MA, O’Connor SM, et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003;102(13):4504–4511. doi: 10.1182/blood-2003-01-0016. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser MF, Walker BA, Hockley SL, Begum DB, Wardell CP, Gonzalez D, et al. A TC classification based predictor for multiple myeloma using multiplexed real-time quantitative PCR. Leukemia. 2013 doi: 10.1038/leu.2013.12. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Corral L, Gutierrez NC, Vidriales MB, Mateos MV, Rasillo A, Garcia-Sanz R, et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res. 2011;17(7):1692–700. doi: 10.1158/1078-0432.CCR-10-1066. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Corral L, Mateos MV, Corchete LA, Sarasquete ME, de la Rubia J, de Arriba F, et al. Genomic analysis of high-risk smoldering multiple myeloma. Haematologica. 2012;97(9):1439–43. doi: 10.3324/haematol.2011.060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Corral L, Sarasquete ME, Bea S, Garcia-Sanz R, Mateos MV, Corchete LA, et al. SNP-based mapping arrays reveal high genomic complexity in monoclonal gammopathies, from MGUS to myeloma status. Leukemia. 2012;26(12):2521–9. doi: 10.1038/leu.2012.128. [DOI] [PubMed] [Google Scholar]
- 17.Chiecchio L, Dagrada GP, Protheroe RK, Stockley DM, Smith AG, Orchard KH, et al. Loss of 1p and rearrangement of MYC are associated with progression of smouldering myeloma to myeloma: sequential analysis of a single case. Haematologica. 2009;94(7):1024–8. doi: 10.3324/haematol.2008.004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weston-Bell N, Gibson J, John M, Ennis S, Pfeifer S, Cezard T, et al. Exome sequencing in tracking clonal evolution in multiple myeloma following therapy. Leukemia. 2013;27(5):1188–91. doi: 10.1038/leu.2012.287. [DOI] [PubMed] [Google Scholar]
- 19.Swanton C. Intratumor Heterogeneity: Evolution through Space and Time. Cancer research. 2012;72(19):4875–82. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S, Van Loo P, Wedge David C, Alexandrov Ludmil B, Greenman Christopher D, Lau King W, et al. The Life History of 21 Breast Cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–61. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 24.Kyle RA, Remstein ED, Therneau TM, Dispenzieri A, Kurtin PJ, Hodnefield JM, et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. The New England journal of medicine. 2007;356(25):2582–90. doi: 10.1056/NEJMoa070389. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Persona E, Vidriales MB, Mateo G, Garcia-Sanz R, Mateos MV, de Coca AG, et al. New criteria to identify risk of progression in monoclonal gammopathy of uncertain significance and smoldering multiple myeloma based on multiparameter flow cytometry analysis of bone marrow plasma cells. Blood. 2007;110(7):2586–92. doi: 10.1182/blood-2007-05-088443. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez NC, Castellanos MV, Martin ML, Mateos MV, Hernandez JM, Fernandez M, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia. 2007;21(1):143–50. doi: 10.1038/sj.leu.2404413. [DOI] [PubMed] [Google Scholar]
- 27.Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120(5):1077–86. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 28.Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21(6):936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465(7297):473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 33.Colla S, Morandi F, Lazzaretti M, Rizzato R, Lunghi P, Bonomini S, et al. Human myeloma cells express the bone regulating gene Runx2/Cbfa1 and produce osteopontin that is involved in angiogenesis in multiple myeloma patients. Leukemia. 2005;19(12):2166–76. doi: 10.1038/sj.leu.2403976. [DOI] [PubMed] [Google Scholar]
- 34.Giuliani N, Colla S, Morandi F, Lazzaretti M, Sala R, Bonomini S, et al. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood. 2005;106(7):2472–83. doi: 10.1182/blood-2004-12-4986. [DOI] [PubMed] [Google Scholar]
- 35.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118(3):675–8. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P, Lu Y, Liu H, Wen W, Jia D, Wang Y, et al. Genome-wide association and fine mapping of genetic loci predisposing to colon carcinogenesis in mice. Mol Cancer Res. 2012;10(1):66–74. doi: 10.1158/1541-7786.MCR-10-0540. [DOI] [PubMed] [Google Scholar]
- 39.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Ozaki T, Shamim Hossain M, Nakamura Y, Kamijo T, Xue X, et al. A newly identified dependence receptor UNC5H4 is induced during DNA damage-mediated apoptosis and transcriptional target of tumor suppressor p53. Biochem Biophys Res Commun. 2008;370(4):594–8. doi: 10.1016/j.bbrc.2008.03.152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.