Abstract
Patients with HER2 positive breast cancer often exhibit intrinsic or acquired resistance to trastuzumab treatment. The transmembrane MUC1-C oncoprotein is aberrantly overexpressed in breast cancer cells and associates with HER2. The present studies demonstrate that silencing MUC1-C in HER2-overexpressing SKBR3 and BT474 breast cancer cells results in downregulation of constitutive HER2 activation. Moreover, treatment with the MUC1-C inhibitor, GO-203, was associated with disruption of MUC1-C/HER2 complexes and decreases in tyrosine phosphorylated HER2 (p-HER2) levels. In studies of trastuzumab-resistant SKBR3R and BT474R cells, we found that the association between MUC1-C and HER2 is markedly increased (~20-fold) as compared to that in sensitive cells. Additionally, silencing MUC1-C in the trastuzumab-resistant cells or treatment with GO-203 decreased p-HER2 and AKT activation. Moreover, targeting MUC1-C was associated with downregulation of phospho-p27 and cyclin E, which confer trastuzumab resistance. Consistent with these results, targeting MUC1-C inhibited the growth and clonogenic survival of both trastuzumab-resistant cells. Our results further demonstrate that silencing MUC1-C reverses resistance to trastuzumab and that the combination of GO-203 and trastuzumab is highly synergistic. These findings indicate that MUC1-C contributes to constitutive activation of the HER2 pathway and that targeting MUC1-C represents a potential approach to abrogate trastuzumab resistance.
Keywords: MUC1, HER2, HER3, trastuzumab, resistance, breast cancer
Introduction
The HER2/ERBB2 receptor tyrosine kinase (RTK) is overexpressed in approximately 20% of human breast cancers and is associated with aggressive disease and poor survival (1; 2). HER2 forms heterodimers with HER3 and thereby phosphorylates and activates HER3 (3; 4). Trastuzumab is a humanized monoclonal antibody that binds to the HER2 extracellular domain and destabilizes ligand-independent HER2/HER3 complexes (5). Targeting of HER2 with trastuzumab in HER2-overexpressing breast cancer cells also decreases HER2 levels (6; 7) and induces G1 arrest by stabilizing the CDK inhibitor p27 (8). Trastuzumab extends the overall survival of certain patients with HER2-overexpressing breast cancers when used as monotherapy or in combination with chemotherapy (9–11). However, many patients exhibit de novo unresponsiveness to trastuzumab or develop acquired resistance after treatment (11). Trastuzumab resistance has been associated with constitutive activation of the PI3K pathway as a result of phosphatase and tensin homolog (PTEN) deficiency (12) or PIK3CA gene mutations (13). PTEN has also been linked to SRC activation and thereby trastuzumab resistance in breast cancer cells and in breast tumors (14). Additional mechanisms of resistance have included expression of a truncated p95HER2 that lacks the trastuzumab binding domain (15), heterodimerization with other RTKs (16–18) and downregulation of HER2 expression (19). Other studies have shown that resistance of HER2-overexpressing breast cancer cells to trastuzumab is conferred by (i) upregulation of cyclin E and an increase in CDK2 activity (20), and (ii) decreased expression of the PPM1H phosphatase that regulates stability of the CDK inhibitor p27 (21; 22). These findings have provided the experimental basis for designing strategies that target pathways associated with trastuzumab resistance to reverse unresponsiveness to this agent.
MUC1 is a heterodimeric protein that associates with HER2 at the surface of breast cancer cells (23; 24). MUC1 is translated as a single polypeptide that undergoes autocleavage into N-terminal (MUC1-N) and C-terminal (MUC1-C) fragments, which in turn form a stable complex at the cell membrane (25; 24). The MUC1-N/MUC1-C heterodimer is positioned at the apical border of breast epithelial cells and is sequestered from RTKs that are expressed at the baso-lateral membranes (25; 24). However, with loss of apical-basal polarity as a result of stress or transformation, MUC1 is repositioned over the entire cell membrane and interacts with RTKs such as HER2 (23; 25; 24). MUC1-N, the mucin component of the heterodimer, is shed from the cell surface (25; 24). The MUC1-C subunit spans the cell membrane and includes a 58 amino acid (aa) extracellular domain, a 28 aa transmembrane domain and a 72 aa cytoplasmic domain. MUC1-C associates in part with RTKs through extracellular galectin-3 bridges (26). In addition, the MUC1-C cytoplasmic domain functions as a substrate for phosphorylation by RTKs and SRC (25; 24). The MUC1-C cytoplasmic domain also contains a YHPM motif that, when phosphorylated on tyrosine, functions as a binding site for PI3K p85 SH2 domains (27). These findings and the demonstration that MUC1-C overexpression is sufficient to induce anchorage-independent growth and tumorigenicity (28; 29) provided the basis for developing agents that block the MUC1-C transforming function (25). In this way, the MUC1-C cytoplasmic domain contains a CQC motif that is necessary for its homodimerization and function (30). Notably, cell-penetrating peptides that bind to the MUC1-C CQC motif are effective in inhibiting growth and inducing death of human breast cancer cells growing in vitro and as xenografts in mice (31). However, the effects of MUC1-C inhibition on (i) the interaction between MUC1-C and HER2, and (ii) HER2 signaling in breast cancer cells are not known.
The present studies demonstrate that MUC1-C contributes to HER2 activation in HER2-overexpressing breast cancer cells and thereby promotes their growth and clonogenic survival. The results also demonstrate that the formation of MUC1-C/HER2 complexes is substantially increased in the setting of trastuzumab resistance and that targeting MUC1-C in trastuzumab-resistant cells results in downregulation of HER2 activation. In concert with these findings, we show that targeting MUC1-C is effective in reversing trastuzumab resistance.
Results
Silencing MUC1-C suppresses HER2 activation
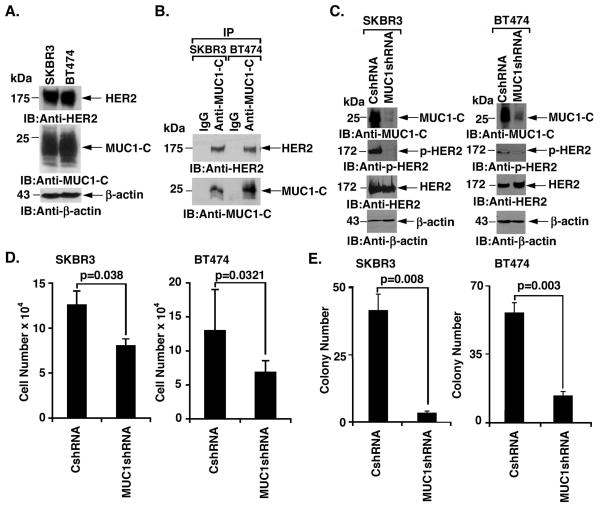
MUC1 associates with HER2 in non-HER2-amplified breast cancer cells and this interaction is increased by heregulin stimulation (23). However, the functional significance of the MUC1-C/HER2 interaction has remained unclear. Accordingly, studies were performed on SKBR3 and BT474 breast cancer cells that overexpress HER2 and are dependent on p-HER2 for growth and survival (3). Levels of HER2 and MUC1-C were found to be similar in these cells (Fig. 1A). Coimmunoprecipitation studies further demonstrated that MUC1-C associates with HER2 in both SKBR3 and BT474 cells (Fig. 1B). To assess the potential effects of MUC1-C on HER2 signaling, we stably silenced MUC1-C in SKBR3 cells (Fig. 1C, left). Notably, MUC1-C silencing was associated with downregulation of p-HER2, but not HER2, abundance, consistent with a decrease in HER2 activation (Fig. 1C, left). In concert with these results, silencing MUC1-C in BT474 cells similarly resulted in suppression of p-HER2 levels (Fig. 1C, right). MUC1-C silencing was also associated with a decrease in SKBR3 (Fig. 1D, left) and BT474 (Fig. 1D, right) cell growth. Moreover, colony formation was substantially decreased by silencing MUC1-C expression in both SKBR3 (Fig. 1E, left) and BT474 (Fig. 1E, right) cells. These findings indicated that MUC1-C contributes to HER2 activation and proliferation of HER2-overexpressing breast cancer cells.
Figure 1. Silencing MUC1-C downregulates HER2 activation and colony formation.
A. Lysates from SKBR3 and BT474 cells were immunoblotted with the indicated antibodies. B. Lysates from SKBR3 (left) and BT474 (right) cells were precipitated with anti-MUC1-C or a control IgG. The precipitates were immunoblotted with the indicated antibodies. C. SKBR3 (left) and BT474 (right) cells were infected with lentiviruses to stably express a control shRNA (CshRNA) or a MUC1shRNA. Lysates were immunoblotted with the indicated antibodies. D. The indicated SKBR3 (left) and BT474 (right) cells were seeded at 5 × 104 cells/well. The results (mean±SD of three replicates) are expressed as the cell number on day 4. E. The indicated SKBR3 (left) and BT474 (right) cells were seeded at 2000 and 1000 cells/well, respectively, and incubated for 12 d. Colony number is expressed as the mean±SD of three replicates.
The MUC1-C inhibitor, GO-203, downregulates HER2 phosphorylation
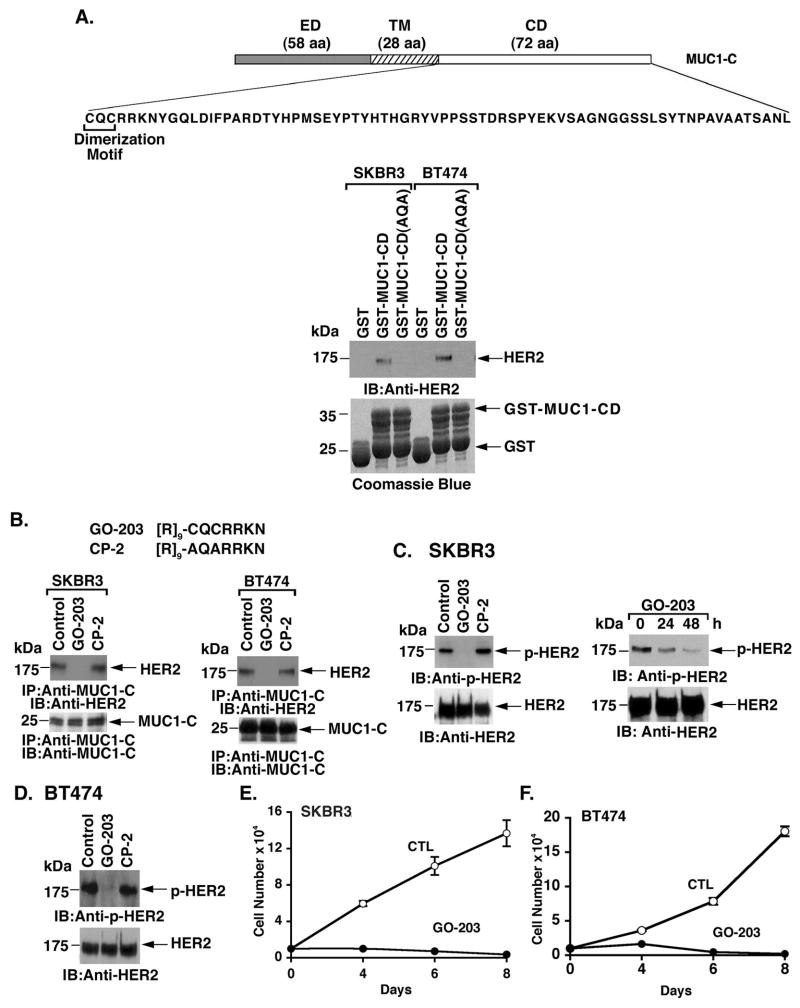
The 72 aa MUC1-C cytoplasmic domain (MUC1-CD) contains a CQC motif that is necessary for its homodimerization (30; 32) (Fig. 2A). The results of pull-down studies using lysates from SKBR3 cells demonstrated that MUC1-CD is sufficient for forming complexes with HER2 (Fig. 2A, left). However, binding to HER2 was not detectable with MUC1-CD in which the CQC motif had been mutated to AQA (Fig. 2A, left). Similar results were obtained when pull-down experiments were performed with lysates from BT474 cells (Fig. 2A, right), indicating that the MUC1-C cysteine residues are of importance for forming MUC1-C/HER2 complexes. GO-203 is a cell-penetrating peptide that contains a poly-Arg cell transduction domain linked to CQCRRKN that binds to the MUC1-C cytoplasmic domain at the CQC motif and thereby blocks MUC1-C homodimerization (31; 27; 32) (Fig. 2B). Another cell-penetrating peptide, designated CP-2, was synthesized in which the cysteine residues are altered to alanines, resulting in an inactive control that does not inhibit MUC1-C homodimerization (31; 27; 32) (Fig. 2B). Treatment of SKBR3 cells with GO-203 disrupted the interaction between MUC1-C and HER2 (Fig. 2B, left). By contrast, CP-2 had no apparent effect on the abundance of MUC1-C/HER2 complexes (Fig. 2B, left). GO-203, but not CP-2, also blocked the interaction between MUC1-C and HER2 in BT474 cells (Fig. 2B, right). Analysis of p-HER2 levels further demonstrated that GO-203, but not CP-2, decreases HER2 activation in SKBR3 cells (Fig. 2C, left and right). BT474 cells also responded to GO-203, and not CP-2, exposure with decreased HER2 activation (Fig. 2D). In concert with these results, GO-203 treatment of SKBR3 (Fig. 2E) and BT474 (Fig. 2F) cells was associated with inhibition of growth. These findings and those obtained with MUC1-C silencing provided support for the premise that binding of MUC1-C and HER2 contributes to HER2 activation.
Figure 2. Targeting the MUC1-C cytoplasmic domain with GO-203 blocks the interaction with HER2 and suppresses HER2 activation.
A. Schema of the MUC1-C subunit with the 58 amino acid (aa) extracellular domain (ED), the 28 aa transmembrane domain (TM) and the amino acid sequence of the 72 aa cytoplasmic domain (CD). Highlighted is the CQC motif that is necessary and sufficient for dimerization of the MUC1-C subunit. Lysates from SKBR3 (left) and BT474 (right) cells were incubated with the indicated GST or GST-fusion proteins. The adsorbates were immunoblotted with anti-HER2. Input of the GST proteins was assessed by Coomassie blue staining. B. D-amino acid sequences of the GO-203 and CP-2 peptides. SKBR3 (left) and BT474 (right) cells were treated with 5 μM GO-203 or CP-2 for 48 h. Anti-MUC1-C precipitates were immunoblotted with anti-HER2 and anti-MUC1-C. C. SKBR3 cells were treated with 5 μM GO-203 or CP-2 for 48 h (left) or with 5 μM GO-203 for the indicated times (right). Lysates were immunoblotted with the indicated antibodies. D. BT474 cells were treated with 5 μM GO-203 or CP-2 for 48 h. Lysates were immunoblotted with the indicated antibodies. E and F. SKBR3 (E) and BT474 (F) cells were left untreated (Control; CTL) or treated with 5 μM GO-203 for the indicated times. Cell number (mean±SE of three determinations) was determined by trypan blue staining.
MUC1-C/HER2 complexes are upregulated in association with trastuzumab resistance
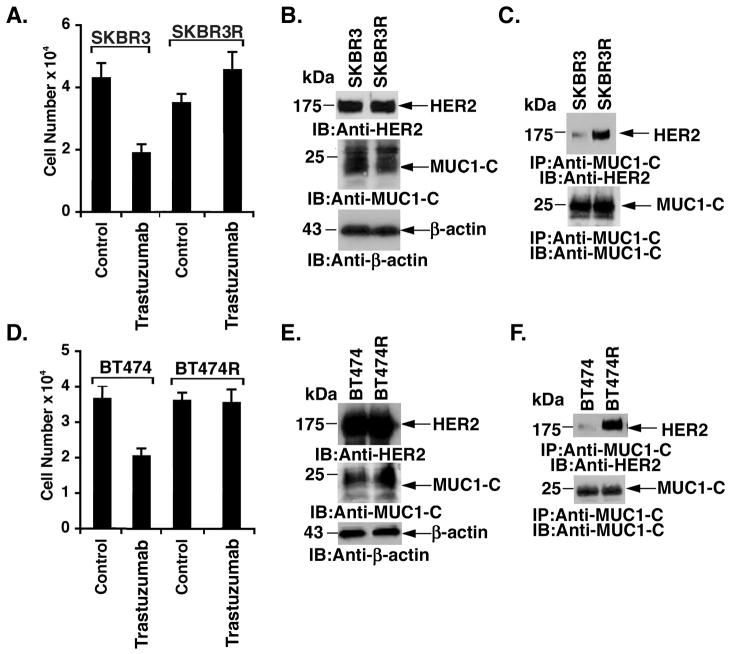
Trastuzumab-resistant SKBR3R cells were generated by chronic exposure to increasing concentrations of trastuzumab for over 18 months in vitro (33). Thus, in contrast to parental SKBR3 cells, growth of the resistant SKBR3R cells was not inhibited by exposure to 80 nM trastuzumab (Fig. 3A). Levels of HER2 and MUC1-C were similar in SKBR3 and SKBR3R cells (Fig. 3B). Strikingly, however, MUC1-C/HER2 complexes were 19-fold higher (as determined by densitometric scanning of the signals) in the resistant SKBR3R cells (Fig. 3C). Analysis of trastuzumab-resistant BT474R cells (33) (Fig. 3D) also demonstrated similar levels of MUC1-C and HER2 as compared to that in the sensitive BT474 cells (Fig. 3E). Moreover, BT474R cells exhibited a substantial increase (28-fold) in MUC1-C/HER2 complexes (Fig. 3F), indicating that the interaction between MUC1-C and HER2 is increased in the setting of trastuzumab resistance.
Figure 3. Interaction between MUC1-C and HER2 is upregulated in trastuzumab-resistant breast cancer cells.
A. SKBR3 (left) and SKBR3R (right) cells (1 × 104/well) were left untreated or treated with 80 nM trastuzaumb for 72 h. Cell number (mean±SE of three determinations) was determined by trypan blue staining. B. Lysates from SKBR3 and SKBR3R cells were immunoblotted with the indicated antibodies. C. Anti-MUC1-C precipitates from SKBR3 and SKBR3R cells were immunoblotted with the indicated antibodies. D. BT474 (left) and BT474R (right) cells (1 × 104/well) were left untreated or treated with 80 nM trastuzumab for 72 h. Cell number (mean±SE of three determinations) was determined by trypan blue staining. E. Lysates from BT474 and BT474R cells were immunoblotted with the indicated antibodies. F. Anti-MUC1-C precipitates from BT474 and BT474R cells were immunoblotted with the indicated antibodies.
Targeting MUC1-C suppresses HER2 activation in trastuzumab-resistant cells
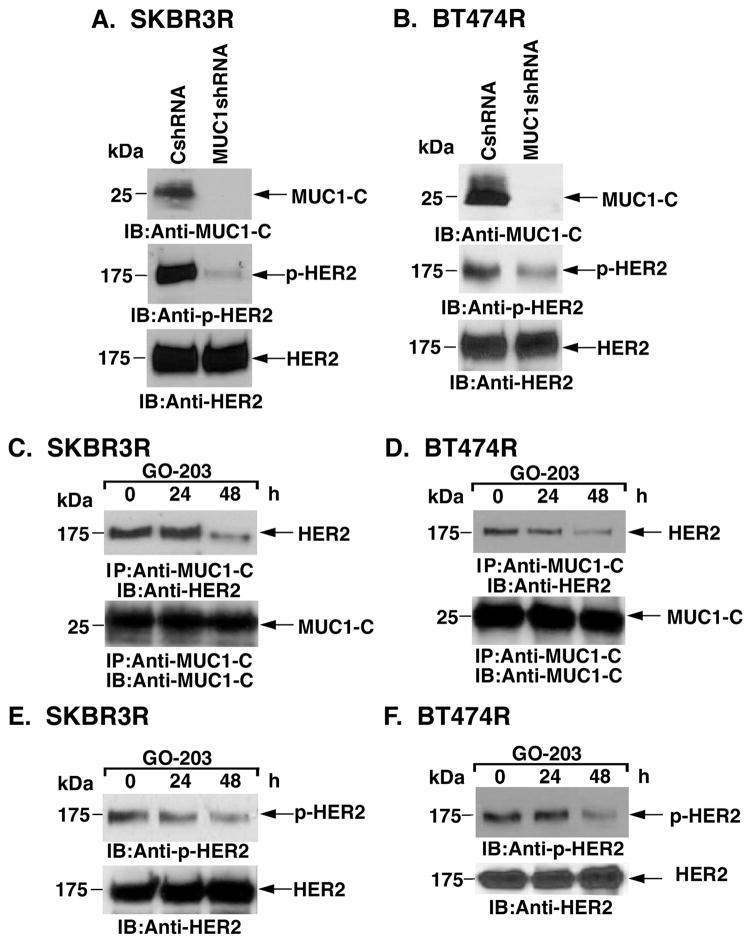
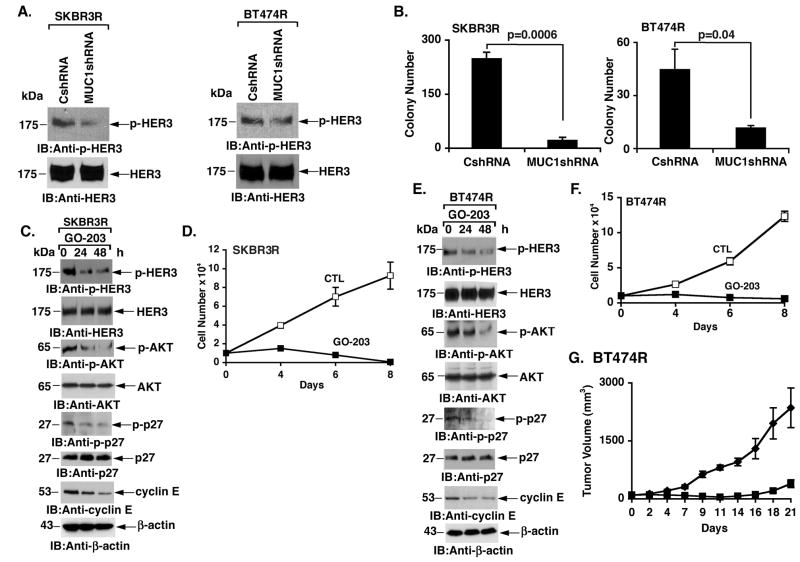
As found in wild-type SKBR3 cells, stable silencing of MUC1-C in SKBR3R (Fig. 4A) and BT474R (Fig. 4B) cells was associated with decreases in p-HER2 levels, indicating that MUC1-C also contributes to HER2 activation in trastuzumab-resistant cells. Densitometric scanning of the p-HER2 signals from multiple experiments demonstrated a decrease in p-HER2 abundance of 74% and 45% in the SKBR3R and BT474R cells, respectively (Supplemental Figs. S1A and S1B). In studies with SKBR3R cells, we found that treatment with GO-203 is associated with disruption of MUC1-C/HER2 complexes (Fig. 4C). Similar results were obtained when BT474R cells were treated with the MUC1-C inhibitor (Fig. 4D). GO-203 also suppressed HER2 activation in both SKBR3R (Fig. 4E) and BT474R (Fig. 4F) cells. These results indicate that targeting MUC1-C with silencing or GO-203 treatment results in suppression of HER2 activation in trastuzumab-resistant cells.
Figure 4. Downregulation of MUC1-C suppresses HER2 activation in trastuzumab-resistant cells.
A and B. SKBR3R (A) and BT474R (B) cells were infected with lentiviruses expressing a control shRNA (CshRNA) or MUC1shRNA. Lysates were immunoblotted with the indicated antibodies. C and D. SKBR3R (C) and BT474R (D) cells were treated with 5 μM GO-203 for the indicated times. Anti-MUC1-C precipitates were immunoblotted with the indicated antibodies. E and F. SKBR3R (E) and BT474R (F) cells were treated with 5 μM GO-203 for the indicated times. Lysates were immunoblotted with the indicated antibodies.
Targeting MUC1-C suppresses HER3 phosphorylation
HER2 phosphorylates HER3 and disruption of the HER2/HER3 interaction is associated with HER3 dephosphorylation (5). These findings and the demonstration that targeting MUC1-C downregulates HER2 invoked the possibility that MUC1-C inhibition could also affect HER3 activation. Indeed, silencing MUC1-C in SKBR3R and BT474R cells was associated with decreases in p-HER3 levels (Fig. 5A, left and right). Scanning of the p-HER3 signals from multiple experiments demonstrated a decrease in p-HER3 abundance of 55% and 52% in the SKBR3R and BT474R cells, respectively (Supplemental Figs. S1C and S1D). HER3 is as crucial as HER2 in promoting proliferation of breast cancer cells that overexpress HER2 (34). In that sense, the suppression of both phosphorylated HER2 (Figs. 4A and B) and HER3 (Fig. 5A) was associated with a marked decrease in colony formation (Fig. 5B, left and right). GO-203 treatment of SKBR3R cells also suppressed p-HER3 abundance and activation of the downstream effector AKT (Fig. 5C). AKT phosphorylates the CDK inhibitor p27 on T198 and thereby inactivates p27 by preventing its localization to the nucleus (35; 36). In concert with the GO-203-induced decreases in AKT activation, phosphorylation of p27 was also decreased in the absence of apparent changes in p27 abundance (Fig. 5C). Trastuzumab resistance is associated with increases in p27 phosphorylation (22) and upregulation of cyclin E (20). In addition to the decreases in phospho-p27 levels, we also found that GO-203 treatment is associated with downregulation of cyclin E abundance (Fig. 5C). Moreover, GO-203 treatment of SKBR3R cells was associated with inhibition of growth (Fig. 5D). Similar effects of GO-203 treatment were obtained in BT474R cells with decreases in phospho-p27 and cyclin E levels (Fig. 5E) and an arrest of growth in vitro (Fig. 5F) and as tumor xenografts in nude mice (Fig. 5G). These findings thus indicate that silencing MUC1-C or inhibition of MUC1-C with GO-203 results in downregulation of HER3 activation and suppression of cell growth and survival.
Figure 5. Targeting MUC1-C in trastuzumab-resistant cells suppresses HER3 activation and inhibits growth and clonogenic survival.
A. Lysates from the indicated SKBR3R (left) and BT474R (right) cells were immunoblotted with anti-p-HER3 and anti-HER3. B. The indicated SKBR3R (left) and BT474R (right) cells were seeded at 4000 and 1000 cells/well, respectively, and incubated for 12 d. The colony number is expressed as the mean±SE of three replicates. C. SKBR3R cells were treated with 5 μM GO-203 for the indicated times. Lysates were immunoblotted with the indicated antibodies. D. SKBR3R cells were left untreated (Control; CTL) or treated with 5 μM GO-203 for the indicated times. Cell number (mean±SE of three determinations) was determined by trypan blue staining. E. BT474R cells were treated with 5 μM GO-203 for the indicated times. Lysates were immunoblotted with the indicated antibodies. F. BT474R cells were left untreated (Control; CTL) or treated with 5 μM GO-203 for the indicated times. Cell number (mean±SE of three determinations) was determined by trypan blue staining. G. BALB/c nu/nu mice were injected subcutaneously in the flank with 1 × 107 BT474R cells. The mice were pair-matched when the tumors were 80–100 mm3. Treatment groups consisted of 8 mice injected intravenously with PBS (vehicle control; diamonds) or 7.5 mg/kg GO-203 (squares) each day for 21 days. There was no weight loss in the two groups. The results are expressed as tumor volume (mean±SE for the 8 mice in each group).
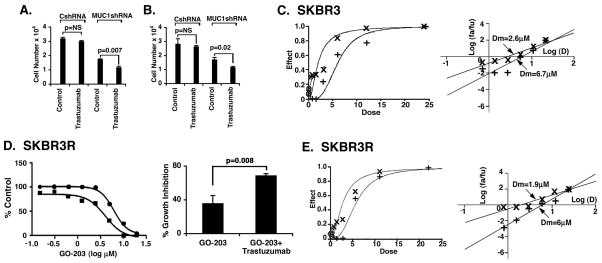
Targeting MUC1-C with silencing or GO-203 treatment reverses trastuzumab resistance
The demonstration that MUC1-C promotes HER2 signaling invoked the possibility that targeting MUC1-C might affect the response to trastuzumab. In this context, analysis of SKBR3R cells demonstrated that silencing MUC1-C is associated with increased sensitivity to the growth inhibitory effects of trastuzumab (Fig. 6A). Trastuzumab resistance of BT474R cells was also reversed in part by silencing MUC1-C (Fig. 6B), indicating that targeting MUC1-C might be effective in combination with trastuzumab. To assess the effects of combining GO-203 and trastuzumab, we first identified the IC50s for each agent against SKBR3 and BT474 cells. In studies with SKBR3 cells, the IC50s for GO-203 and trastuzumab were 5.9 μM and 93 nM, respectively (Supplemental Table 1A). Based on the Chou-Talalay method, we then evaluated the effects of combining GO-203 and trastuzumab. As shown in the dose-effect curves, treatment of SKBR3 cells with the combination was more effective in inhibiting growth and survival than that obtained with either agent alone (Fig. 6C, left). The median-effect analysis and calculation of the median dose (Dm) values further showed a ~3-fold reduction in the Dm for GO-203 in the presence of trastuzumab as compared to GO-203 alone (Fig. 6C, right). Calculation of the combination index (CI) at the ED50, ED75 and ED90 for these agents demonstrated a high degree of synergy with the values less than 1 (Supplemental Table 1B). Comparable results were obtained when BT474 cells were treated with GO-203 and trastuzumab as shown in the dose-effect curves (Supplemental Fig. S2A, left) and the median-effect plots (Supplemental Fig. S2A, right), and as supported by CI values ranging from 0.47 to 0.68 (Supplemental Table 1B). IC50s for GO-203 were also generated for the SKBR3R and BT474R cells (Supplemental Table 1A). For these trastuzumab-resistant cells, there is no definable IC50 for trastuzumab (Supplemental Table 1A); accordingly, we used the Bliss independence method for assessing interactions between two agents when one of the agents is inactive (37; 38). As expected, treatment with trastuzumab alone at 40 nM had no effect on SKBR3R cell growth. However, GO-203 alone was effective in inhibiting growth, and combining GO-203 with 40 nM trastuzumab was more effective than either agent alone (Fig. 6D, left and right), indicating that GO-203 reverses resistance to trastuzumab. An observed BI score of 0.84 as compared to 0.46 for the predicted BI supported a synergistic interaction (Supplemental Table 2). Based on these findings, we used the Chou-Talalay method to further analyze the interaction between GO-203 and trastuzumab. Under these experimental conditions, we confirmed that the combination of GO-203 and trastuzumab is synergistic in the treatment of SKBR3R cells (Fig. 6E) with the CI<1 (Supplemental Table 1B). GO-203 was also highly effective in combination with trastuzumab in the treatment of trastuzumab-resistant BT474R cells using the BI (Supplemental Fig. S2B and Supplemental Table 2) and Chou-Talalay (Supplemental Fig. S2C and Supplemental Table 1B) methods. These findings indicate that GO-203 is synergistic with trastuzumab in the treatment of trastuzumab-resistant cells.
Figure 6. Targeting MUC1-C reverses trastuzaumab resistance.
A and B. The indicated SKBR3R (A) and BT474R (B) cells (1 × 104/well) were left untreated or treated with 80 nM trastuzumab for 72 h. Cell number (mean+SE of three determinations) was assessed by trypan blue staining. C. SKBR3 cells were treated with GO-203 alone (+), trastuzumab alone (⊙), and the combination (X) based on the IC50 values listed in Supplemental Table 1A. Growth inhibition data were analyzed by the method of Chou and Talalay using the CalcuSyn program. The dose-effect curves (left) and median-effect plots (right) are shown using μM concentrations for GO-203 and trastuzumab. The CI values are listed in Supplemental Table 1B. D. SKBR3R cells were treated with GO-203 alone (circles) and in combination with 40 nM trastuzumab (squares) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software and the BI score was calculated as described in the Materials and Methods (Supplemental Table 2). The percentage growth inhibition (mean±SE of three determinations) obtained with 5 μM GO-203 was significantly different from that with 5 μM GO-203+40 nM trastuzumab (right). E. SKBR3R cells were treated with GO-203 alone (+), trastuzumab alone (⊙), and the combination (X). Growth inhibition data were analyzed by the method of Chou and Talalay using the CalcuSyn program. The dose-effect curves (left) and median-effect plots (right) are shown using μM concentrations for GO-203 and trastuzumab. The CI values are listed in Supplemental Table 1B.
Effects of targeting MUC1-C on lapatinib treatment of trastuzumab-resistant cells
Lapatinib is a small molecule ATP competitor that inhibits phosphorylation of the HER2 intracellular kinase domain and has reported activity in trastuzumab-resistant cells (39–41; 7). In the present studies with SKBR3R cells, the combination of lapatinib and GO-203 was more effective in inhibiting growth than lapatinib alone (Supplemental Fig. S3A, left and right). Synergy between lapatinib and GO-203 was supported by an observed BI score of 0.85 as compared to 0.58 for the predicted BI (Supplemental Table 3). In addition, lapatinib was synergistically active with GO-203 in inhibiting growth of BT474R cells (Supplemental Fig. S3B, left and right; Supplemental Table 3). Based on these observations, we assessed the effects of (i) lapatinib alone, (ii) lapatinib+trastuzumab, and (iii) lapatinib+trastuzumab+GO-203. Using SKBR3R cells, the lapatinib+trastuzumab combination was not significantly different in inhibiting growth as compared to lapatinib alone (Supplemental Fig. S4A, left and right). However, the triple lapatinib+trastuzumab+GO-203 combination was significantly more effective than lapatinib alone or lapatinib+trastuzumab (Supplemental Fig. S4A, left and right) and as evidenced by an observed BI of 0.81 compared to a predicted BI of 0.59 (Supplemental Table 4). These findings with the lapatinib+trastuzumab+GO-203 combination were further supported in studies of BT474R cells (Supplemental Fig. S4B, left and right); Supplemental Table 4), indicating that GO-203 potentiates the effects of both lapatinib and trastuzumab in trastuzumab-resistant cells.
Discussion
MUC1-C is a previously unrecognized effector of HER2 activation
Previous work demonstrated that MUC1 associates with HER2 in the mammary glands of a MUC1 transgenic mouse model and in human non-HER2-amplified breast cancer cell lines (24). To define the functional significance of this association, the present studies were performed on SKBR3 and BT474 breast cancer cells that overexpress HER2 and are dependent on HER2 for proliferation (3). As anticipated, we found that MUC1-C associates with HER2 in these HER2-overexpressing cells. Surprisingly, however, our results showed that silencing MUC1-C is associated with downregulation of HER2 activation and loss of clonogenic survival, supporting the contention that MUC1-C is of importance to HER2 signaling. The MUC1-C subunit includes a cytoplasmic domain that contains a CQC motif, which is necessary for the formation of MUC1-C homodimers (30). Our results show that the MUC1-C cytoplasmic domain is sufficient to form complexes with HER2 and that this association is abrogated by altering the CQC motif to AQA, indicating that MUC1-C homodimerization is necessary for the MUC1-C/HER2 interaction. Cell membrane-penetrating peptides, such as GO-203, block homodimerization of endogenous MUC1-C through binding to the CQC motif (31; 32). The present results demonstrating that treatment of HER2-overexpressing breast cancer cells with GO-203 blocks the interaction between MUC1-C and HER2 provided further support for the premise that MUC1-C homodimerization is necessary for forming complexes with HER2. In addition and as found with MUC1-C silencing, targeting MUC1-C with GO-203 treatment was associated with marked decreases in HER2 phosphorylation. Overexpression of HER2 in human breast cancer cells facilitates the formation of HER2/HER3 heterodimers, activation of HER2 and HER2-mediated phosphorylation of HER3 (4). The importance of HER3 in the HER2/HER3 heterodimer is supported by the demonstration that loss of HER3 function attenuates HER2-mediated transformation (3). In addition to downregulation of HER2, we found that targeting MUC1-C is associated with suppression of HER3 activation. These findings support a previously unrecognized model in which MUC1-C homodimerization contributes to HER2 activation and HER3 phosphorylation.
Targeting MUC1-C is a potential strategy for circumventing trastuzumab resistance
Patients with HER2-overexpressing breast cancers frequently display primary unresponsiveness or develop acquired resistance to trastuzumab therapy. Trastuzumab resistance has been attributed to number of mechanisms, including activation of the PI3K and SRC pathways (12; 13). Selection for growth of BT474 cells in the presence of trastuzumab for 8 weeks has been associated with upregulation of MUC1 expression (42). However, the present results show that the development of trastuzumab resistance over 18 months has little if any effect on MUC1-C abundance in SKBR3R and BT474R cells. The basis for this discrepancy in findings is not clear. Nonetheless, the present results demonstrate that trastuzumab resistance is associated with a substantial (~20-fold) increase in the association of MUC1-C and HER2. Trastuzumab disrupts ligand-independent HER2/HER3 interactions in HER2-overexpressing cells (5). Our results indicate that targeting MUC1-C attenuates HER2 activation in trastuzumab-resistant cells. These findings raised the possibility that MUC1-C may contribute to trastuzumab resistance by promoting HER2-mediated signaling. Indeed, targeting MUC1-C was associated with downregulation of (i) p-HER3 and p-AKT levels, and (ii) phosphorylation of p27, an AKT substrate. In addition, targeting MUC1-C resulted in decreases in cyclin E abundance, a finding consistent with the demonstration that cyclin E levels decrease upon HER2 inhibition (43). The downregulation of p27 phosphorylation could also be linked to decreases in cyclin E expression as a result of increased localization of p27 to the nucleus and thereby inhibition of CDK2, which in turn results in cyclin E degradation (35; 36; 44). Further studies will be needed to address the mechanistic basis for cyclin E downregulation in GO-203-treated trastuzumab-resistant cells. Nonetheless, since both phospho-p27 and cyclin E have been linked to trastuzumab resistance (21; 22; 20), we treated trastuzumab-resistant cells with GO-203 in combination with trastuzumab. The observation that GO-203 and trastuzumab are highly synergistic lends further support to the premise that MUC1-C contributes to the trastuzumab-resistant phenotype by promoting HER2/HER3→AKT activation. These findings thus indicate that targeting MUC1-C can reverse trastuzumab resistance. In addition, our results indicate that GO-203 can potentiate the effects of lapatinib and the lapatinib+trastuzumab combination in the SKBR3R and BT474R models. Translation of these findings with regard to the potential effectiveness of combining GO-203 with trastuzumab and/or lapatinib in the clinic will therefore require further study. In this respect, a Phase I trial of GO-203 is presently being completed for patients with refractory solid tumors and, based on the present results, this agent may be effective in combination with HER2 inhibitors in the setting of trastuzumab-resistant breast cancer.
Why would MUC1-C contribute to HER2 signaling in breast cancer cells?
Epithelia are single cell layers with apical-basal polarity that separate metazoans from the external environment. As such, robust defense mechanisms emerged during evolution to protect epithelial integrity. The mucins contribute in part to that defense by forming a protective physical barrier (25). The MUC1-N subunit is shed from the epithelial cell surface into this barrier as a first line of defense. In turn, the transmembrane MUC1-C subunit functions in a subsequent line of defense by signaling stress to the interior of the epithelial cell to promote repair, growth and survival (25). Importantly in this regard, the response of epithelial cells to stress is associated with loss of apical-basal polarity and activation of a HER2-mediated proliferation and survival program (45). With loss of polarity, the apical MUC1-C protein is transiently repositioned over the cell membrane, allowing it to interact with HER2, which is normally sequestered at the basolateral membrane (25; 24). The present results provide evidence that, in associating with HER2, MUC1-C contributes to HER2 activation and HER3 phosphorylation. Thus, a potential advantage of this interaction with MUC1-C could conceivably be prolonged HER2/HER3 signaling in the absence of ligand. The epithelial stress response is reversible such that the interaction between MUC1-C and HER2 is transient with return of epithelial cell polarity. By contrast, in carcinoma cells with sustained activation of the epithelial-mesenchymal transition, MUC1-C is positioned to constitutively interact with the HER2 complex and promote activation of the HER2 pathway. In this way, breast cancer cells would appear to have appropriated and subverted a physiologic stress response to support their own growth and survival. Importantly, the interaction between MUC1-C and HER2 has been further increased and subverted to circumvent the growth inhibitory effects of trastuzumab treatment, such that targeting MUC1-C reverses trastuzumab resistance.
Materials and Methods
Cell culture
SKBR3 and BT474 breast cancer cells (ATCC) were maintained in McCoy’s 5a modified medium and Dulbecco’s modified Eagle medium/Ham F12 (1:1) supplemented with 10% heat-inactivated FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine. SKBR3R and BT474R cells were selected for resistance to trastuzumab as described (33). Cells were infected with lentiviral vectors expressing a MUC1 shRNA (Sigma) or a control scrambled shRNA (CshRNA; Sigma). Cells were treated with GO-203 and CP-2 peptides (AnaSpec) as described (27). Trastuzumab (Genentech) was dissolved in sterile apyrogen water (stock solution of 21 mg/ml) and stored at 4°C. Lapatinib (Selleck Chemicals) was dissolved in DMSO. Viability was determined by trypan blue exclusion.
Immunoprecipitation and immunoblot analysis
Cell lysates were prepared as described (31). Soluble proteins were immunoprecipitated with anti-MUC1-C (Ab5; Neomarkers) or a control IgG. The precipitates and lysates not subjected to immunoprecipitation were immunoblotted with anti-MUC1-C, anti-β-actin (Sigma-Aldrich), anti-p-HER2(Tyr1221/1222), anti-HER2, anti-p-HER3(Tyr1289), anti-HER3, anti-p-AKT(S473), anti-AKT (Cell Signaling Technology), anti-p-p27(T198), anti-p27 and anti-cyclin E (Santa Cruz Biotechnology). Reactivity was detected with horseradish peroxidase-conjugated secondary antibodies and chemiluminescence.
In vitro binding assay
GST and GST-MUC1-CD fusion proteins were prepared and incubated with cell lysates as described (27). Adsorbates to glutathione-conjugated beads were analyzed by immunoblotting.
Colony formation assays
Cells were seeded on 6-well plates and incubated for 10–14 d. The cells were then washed and stained with 0.5% crystal violet in 25% methanol. Colonies >25 cells were counted in triplicate wells.
Trastuzumab-resistant BT474R tumor xenograft model
Four- to 6-week-old BALB/c nu/nu female mice were injected subcutaneously with 1 × 107 BT474R cells in the flank. When tumors were ~80–100 mm3, the mice were pair-matched into control and treatment groups of 8 mice each. PBS (control vehicle) or GO-203 at 7.5 mg/kg body weight were administered daily by intravenous injection for 21 days. Tumor volumes were calculated as described (27).
Determination of IC50 and synergism
Cells were seeded on 96-well plates in 100 μl of growth medium at a density of 3000 cells per well. After 24 h, the cells were exposed to drugs in treatment medium for an additional 72 h. Cell viability was assessed using the alamar blue viability assay (Invitrogen). Triplicate wells of each treatment were analyzed and each experiment was done three times. The IC50 values were determined by nonlinear regression of the dose-response data using Prism 5.0 for Mac OSX (GraphPad Software). The presence or absence of synergism between GO-203 and trastuzumab was determined by the method of Chou and Talalay (46; 47). Briefly, cells were exposed to 1:1 ratios of the respective IC50 values for GO-203 and trastuzumab at (i) ¼ x IC50, (ii) ½ x IC50, (iii) IC50, (iv) 2 x IC50, and (v) 4 x IC50. Cell viability was determined after treatment for 72 h. The combination index (CI) was calculated to determine the presence of synergism (CI<1) or antagonism (CI>1) using CalcuSyn software (Biosoft).
Assessment of drug interaction by Bliss independence
The Bliss independence (BI) model was also used to assess the interaction between GO-203 and trastuzumab in studies of trastuzumab-resistant cells. BI was determined using the equation (37; 38), where EA is the fractional inhibition for GO-203 alone and EB is that for trastuzumab alone. Using this equation, if the experimentally measured inhibition is greater than Ei, then the combination is synergistic (38). Plots were generated by Prism GraphPad software.
Supplementary Material
A and B. Densitometric scanning of the p-HER2 signals in SKBR3R (A) and BT474R (B) cells demonstrated that silencing MUC1 is associated with a 74% and 45% decrease in p-HER2 levels (mean±SE of three determinations), respectively. C and D. Densitometric scanning of the p-HER3 signals in SKBR3R (C) and BT474R (D) cells demonstrated that silencing MUC1 is associated with a 55% and 52% decrease in p-HER3 levels (mean±SE of three determinations), respectively.
A. BT474 cells were treated with GO-203 alone (+), trastuzumab alone (⊙), and the combination (X) based on the IC50 values listed in Supplemental Table 1A. Growth inhibition data were analyzed by the method of Chou and Talalay using the CalcuSyn program. The dose-effect curves (left) and median-effect plots (right) are shown using μM concentrations for GO-203 and trastuzumab. The CI values are listed in Supplemental Table 1B. B. BT474R cells were treated with GO-203 alone (circles) and in combination with 40 nM trastuzumab (squares) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software and the BI score was calculated as described in the Materials and Methods (Supplemental Table 2). The percentage growth inhibition (mean±SE of three determinations) obtained with 5 μM GO-203 was significantly different from that with 5 μM GO-203+40 nM trastuzumab (right). C. BT474R cells were treated with GO-203 alone (+), trastuzumab alone (⊙), and the combination (X). Growth inhibition data were analyzed by the method of Chou and Talalay using the CalcuSyn program. The dose-effect curves (left) and median-effect plots (right) are shown using μM concentrations for GO-203 and trastuzumab. The CI values are listed in Supplemental Table 1B.
Effects of combining lapatinib with GO-203 in the treatment of trastuzumab-resistant cells. A and B. SKBR3R (A) and BT474R (B) cells were treated with the indicated lapatinib concentrations alone (circles) or in combination with 4 μM GO-203 (squares) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software (left) and the BI score was calculated as described in the Materials and Methods (Supplemental Table 3). The percentage growth inhibition (mean±SE of three determinations) obtained with 0.75 μM lapatinib was significantly different from that with 0.75 μM lapatinib+4 μM GO-203 (right).
A and B. SKBR3R (A) and BT474R (B) cells were treated with the indicated lapatinib concentrations alone (circles), in combination with 40 nM trastuzumab (squares) or in combination with 40 nM trastuzumab+4 μM GO-203 (triangles) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software (left) and the BI score was calculated as described in the Materials and Methods (Supplemental Table 4). The percentage growth inhibition (mean±SE of three determinations) obtained with 1.5 μM lapatinib was significantly different from that with 1.5 μM lapatinib+40 nM trastuzumab+4 μM GO-203 (right).
Acknowledgments
Grant Support
This work was supported by Grants CA97098 and CA166480 awarded by the National Cancer Institute, and by the Susan G. Komen for the Cure grant SAC110046 (M.S. and J.B.).
Abbreviations
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- MUC1-CD
MUC1 cytoplasmic domain
- PI3K
phosphatidylinositol-3 kinase
- RTK
receptor tyrosine kinase
- Dm
median dose
- CI
combination index
Footnotes
Conflict of Interest: D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
References
- 1.Slamon D, Clark G, Wong S, Levin W, Ullrich A, Mcguire W. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 3.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 5.Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 7.Scaltriti M, Verma C, Guzman M, Jimenez J, Parra JL, Pedersen K, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28:803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 8.Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J, et al. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med. 2002;8:1145–1152. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 9.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 10.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 11.Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2009;27:5838–5847. doi: 10.1200/JCO.2009.22.1507. [DOI] [PubMed] [Google Scholar]
- 12.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 16.Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 17.Shattuck DL, Miller JK, Carraway KL, 3rd, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68:1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Gao L, Wang S, McManaman JL, Thor AD, Yang X, et al. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-1 receptor in breast cancer cells resistant to herceptin. Cancer Res. 2010;70:1204–1214. doi: 10.1158/0008-5472.CAN-09-3321. [DOI] [PubMed] [Google Scholar]
- 19.Mittendorf EA, Wu Y, Scaltriti M, Meric-Bernstam F, Hunt KK, Dawood S, et al. Loss of HER2 amplification following trastuzumab-based neoadjuvant systemic therapy and survival outcomes. Clin Cancer Res. 2009;15:7381–7388. doi: 10.1158/1078-0432.CCR-09-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scaltriti M, Eichhorn PJ, Cortes J, Prudkin L, Aura C, Jimenez J, et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Natl Acad Sci U S A. 2011;108:3761–3766. doi: 10.1073/pnas.1014835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res. 2004;64:3981–3986. doi: 10.1158/0008-5472.CAN-03-3900. [DOI] [PubMed] [Google Scholar]
- 22.Lee-Hoeflich ST, Pham TQ, Dowbenko D, Munroe X, Lee J, Li L, et al. PPM1H Is a p27 Phosphatase Implicated in Trastuzumab Resistance. Cancer Discov. 2011;1:326–337. doi: 10.1158/2159-8290.CD-11-0062. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Yu W-H, Ren J, Huang L, Kharbanda S, Loda M, et al. Heregulin targets γ-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 protein. Mol Cancer Res. 2003;1:765–775. [PubMed] [Google Scholar]
- 24.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duraisamy S, Kufe T, Ramasamy S, Kufe D. Evolution of the human MUC1 oncoprotein. Int J Oncology. 2007;31:671–677. [PubMed] [Google Scholar]
- 27.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Therapeutics. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 30.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 31.Raina D, Ahmad R, Joshi M, Yin L, Wu Z, Kawano T, et al. Direct targeting of the MUC1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scaltriti M, Serra V, Normant E, Guzman M, Rodriguez O, Lim AR, et al. Antitumor activity of the Hsp90 inhibitor IPI-504 in HER2-positive trastuzumab-resistant breast cancer. Mol Cancer Ther. 2011;10:817–24. doi: 10.1158/1535-7163.MCT-10-0966. [DOI] [PubMed] [Google Scholar]
- 34.Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 35.Viglietto G, Motti ML, Bruni P, Melillo RM, D’Alessio A, Califano D, et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med. 2002;8:1136–44. doi: 10.1038/nm762. [DOI] [PubMed] [Google Scholar]
- 36.Motti ML, De Marco C, Califano D, Fusco A, Viglietto G. Akt-dependent T198 phosphorylation of cyclin-dependent kinase inhibitor p27kip1 in breast cancer. Cell Cycle. 2004;3:1074–1080. [PubMed] [Google Scholar]
- 37.Buck E, Eyzaguirre A, Brown E, Petti F, McCormack S, Haley JD, et al. Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors. Mol Cancer Ther. 2006;5:2676–2684. doi: 10.1158/1535-7163.MCT-06-0166. [DOI] [PubMed] [Google Scholar]
- 38.Guertin AD, Martin MM, Roberts B, Hurd M, Qu X, Miselis NR, et al. Unique functions of CHK1 and WEE1 underlie synergistic anti-tumor activity upon pharmacologic inhibition. Cancer Cell Int. 2012;12:45. doi: 10.1186/1475-2867-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 40.Konecny GE, Pegram MD, Venkatesan N, Finn R, Yang G, Rahmeh M, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 41.Nahta R, Yuan LX, Du Y, Esteva FJ. Lapatinib induces apoptosis in trastuzumab-resistant breast cancer cells: effects on insulin-like growth factor I signaling. Mol Cancer Ther. 2007;6:667–674. doi: 10.1158/1535-7163.MCT-06-0423. [DOI] [PubMed] [Google Scholar]
- 42.Fessler S, Wotkowicz M, Mahanta S, Bamdad C. MUC1* is a determinant of trastuzumab (Herceptin) resistance in breast cancer cells. Breast Cancer Res Treat. 2009;118:113–124. doi: 10.1007/s10549-009-0412-3. [DOI] [PubMed] [Google Scholar]
- 43.Mittendorf EA, Liu Y, Tucker SL, McKenzie T, Qiao N, Akli S, et al. A novel interaction between HER2/neu and cyclin E in breast cancer. Oncogene. 2010;29:3896–3907. doi: 10.1038/onc.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li P, Li C, Zhao X, Zhang X, Nicosia SV, Bai W. p27(Kip1) stabilization and G(1) arrest by 1,25-dihydroxyvitamin D(3) in ovarian cancer cells mediated through down-regulation of cyclin E/cyclin-dependent kinase 2 and Skp1-Cullin-F-box protein/Skp2 ubiquitin ligase. J Biol Chem. 2004;279:25260–25267. doi: 10.1074/jbc.M311052200. [DOI] [PubMed] [Google Scholar]
- 45.Vermeer PD, Einwalter LA, Moninger TO, Rokhlina T, Kern JA, Zabner J, et al. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature. 2003;422:322–326. doi: 10.1038/nature01440. [DOI] [PubMed] [Google Scholar]
- 46.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 47.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A and B. Densitometric scanning of the p-HER2 signals in SKBR3R (A) and BT474R (B) cells demonstrated that silencing MUC1 is associated with a 74% and 45% decrease in p-HER2 levels (mean±SE of three determinations), respectively. C and D. Densitometric scanning of the p-HER3 signals in SKBR3R (C) and BT474R (D) cells demonstrated that silencing MUC1 is associated with a 55% and 52% decrease in p-HER3 levels (mean±SE of three determinations), respectively.
A. BT474 cells were treated with GO-203 alone (+), trastuzumab alone (⊙), and the combination (X) based on the IC50 values listed in Supplemental Table 1A. Growth inhibition data were analyzed by the method of Chou and Talalay using the CalcuSyn program. The dose-effect curves (left) and median-effect plots (right) are shown using μM concentrations for GO-203 and trastuzumab. The CI values are listed in Supplemental Table 1B. B. BT474R cells were treated with GO-203 alone (circles) and in combination with 40 nM trastuzumab (squares) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software and the BI score was calculated as described in the Materials and Methods (Supplemental Table 2). The percentage growth inhibition (mean±SE of three determinations) obtained with 5 μM GO-203 was significantly different from that with 5 μM GO-203+40 nM trastuzumab (right). C. BT474R cells were treated with GO-203 alone (+), trastuzumab alone (⊙), and the combination (X). Growth inhibition data were analyzed by the method of Chou and Talalay using the CalcuSyn program. The dose-effect curves (left) and median-effect plots (right) are shown using μM concentrations for GO-203 and trastuzumab. The CI values are listed in Supplemental Table 1B.
Effects of combining lapatinib with GO-203 in the treatment of trastuzumab-resistant cells. A and B. SKBR3R (A) and BT474R (B) cells were treated with the indicated lapatinib concentrations alone (circles) or in combination with 4 μM GO-203 (squares) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software (left) and the BI score was calculated as described in the Materials and Methods (Supplemental Table 3). The percentage growth inhibition (mean±SE of three determinations) obtained with 0.75 μM lapatinib was significantly different from that with 0.75 μM lapatinib+4 μM GO-203 (right).
A and B. SKBR3R (A) and BT474R (B) cells were treated with the indicated lapatinib concentrations alone (circles), in combination with 40 nM trastuzumab (squares) or in combination with 40 nM trastuzumab+4 μM GO-203 (triangles) for 72 h. Growth inhibition plots (left) were generated by Prism GraphPad software (left) and the BI score was calculated as described in the Materials and Methods (Supplemental Table 4). The percentage growth inhibition (mean±SE of three determinations) obtained with 1.5 μM lapatinib was significantly different from that with 1.5 μM lapatinib+40 nM trastuzumab+4 μM GO-203 (right).