Abstract
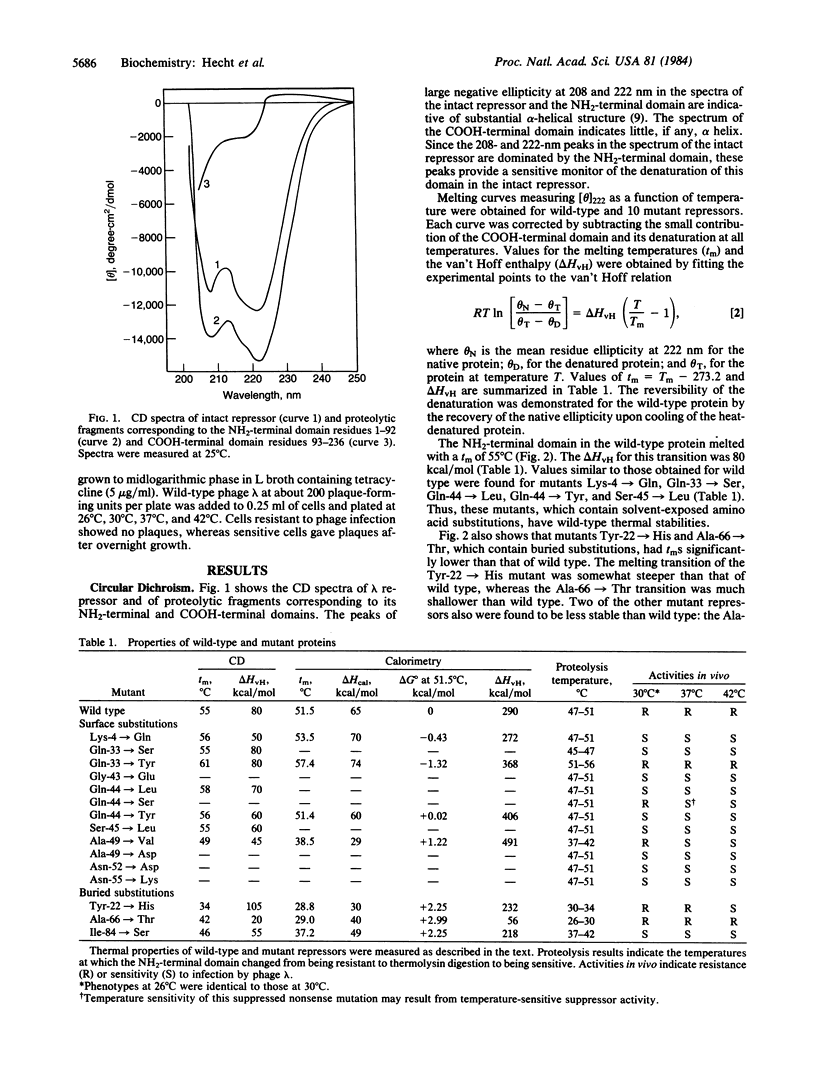
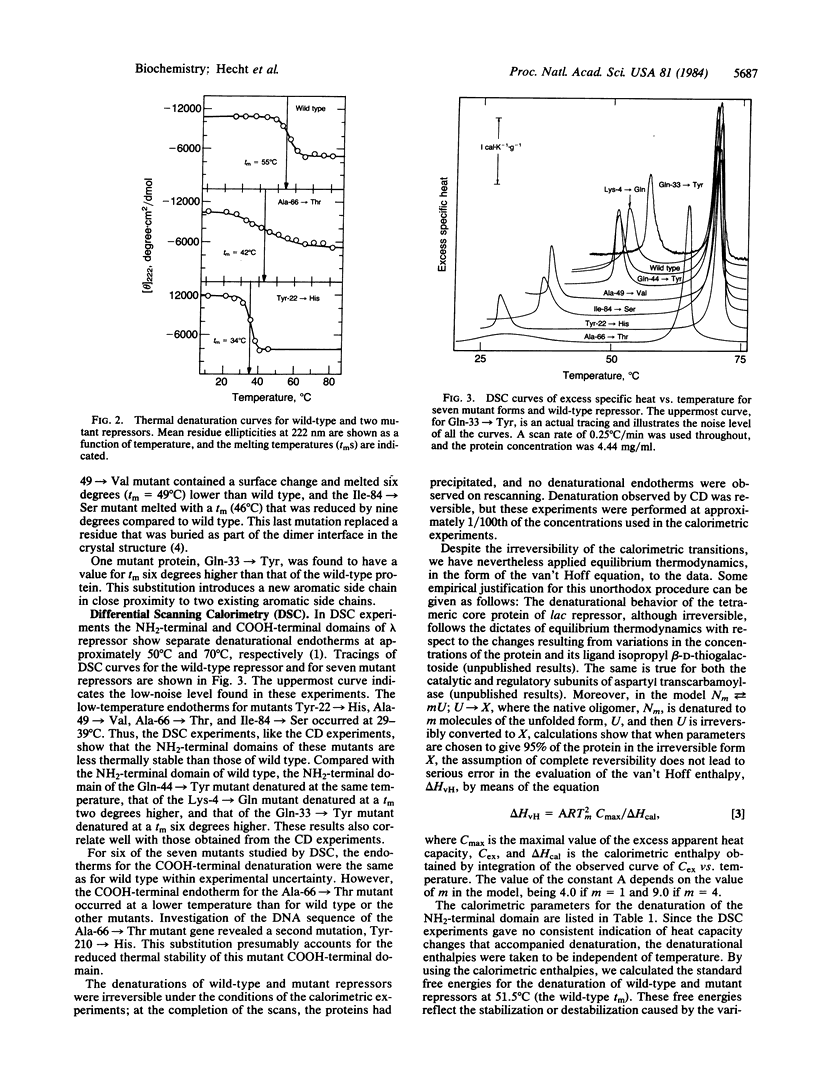
The thermal stabilities of mutant phage lambda repressors that have single amino acid replacements in the NH2-terminal domain have been studied by means of circular dichroism and differential scanning calorimetry. The variations in stability determined by these physical methods correlate with the resistance to proteolysis at various temperatures and can be compared with the temperature-sensitive activity of the mutants in vivo. In general, mutant proteins bearing solvent-exposed substitutions have thermal stabilities identical to wild type, whereas buried substitutions reduce stability. In one case, a single amino acid replacement increases the thermal stability of the repressor.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Argos P., Rossman M. G., Grau U. M., Zuber H., Frank G., Tratschin J. D. Thermal stability and protein structure. Biochemistry. 1979 Dec 11;18(25):5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978 Jan;13(1):65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Hecht M. H., Sauer R. T. Mutations defining the operator-binding sites of bacteriophage lambda repressor. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):441–449. doi: 10.1101/sqb.1983.047.01.052. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T., Sturtevant J. M., Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz M. F., Raidt H. Stereochemical basis of heat stability in bacterial ferredoxins and in haemoglobin A2. Nature. 1975 May 15;255(5505):256–259. doi: 10.1038/255256a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Karplus M., Patel D. J., Sauer R. T. Solution NMR studies of intact lambda repressor. J Biomol Struct Dyn. 1983 Oct;1(1):151–157. doi: 10.1080/07391102.1983.10507431. [DOI] [PubMed] [Google Scholar]



