Abstract
Nucleophosmin 1 (NPM1) mutations have been identified in a substantial number of patients with acute myeloid leukemia (AML). Favorable outcomes in AML cases with NPM1 mutations have been previously reported. However, widely differing survival estimates have been indicated. Therefore, a meta-analysis of nine studies including a total of 4509 subjects was performed. The frequency of NPM1 mutations was found to be 6.45–56.08%. NPM1-mutation type (NPM1-mt) patients had >2-fold higher odds of achieving complete remission compared with NPM1-wild-type (NPM1-wt). The summary hazard ratio (HR) of NPM1-mt/NPM1-wt for disease-free survival (DFS) and OS was 0.67 and 0.63, respectively. In conclusion, these findings suggest that the NPM1 mutation has a favorable effect on the outcome for AML. The present meta-analysis was based on data abstracted from observational studies. However, the results obtained may justify the risk-adapted therapeutic strategies for AML according to the NPM1 status.
Keywords: acute myeloid leukemia, nucleophosmin 1, mutation, prognosis, meta-analysis
Introduction
Acute myeloid leukemia (AML) is a heterogeneous clonal disorder characterized by autonomous proliferation and impaired differentiation of hematopoietic progenitor cells (1,2). It is the most common malignant myeloid disorder in adults. Cytogenetic aberrations and molecular genetic alterations provide significant prognostic information for determining the response to chemotherapy and survival outcome (3,4). An increasing number of genetic abnormalities revealed in AML have also contributed to our understanding of the mechanisms and process of leukemogenesis, which leads to an improvement of risk-stratification, and the development of individualized therapies and response assessment (5).
Among these genetic alterations, a potential prognostic genetic marker is the nucleophosmin 1 (NPM1) gene, which is important in many tumor-associated chromosomal translocations (6). NPM1 is an ubiquitously expressed phosphoprotein and continuously shuttles between the cytoplasm and nucleus (7,8). Several functions for this protein have been described, including the binding of p53 (9), the initiation of centrosome duplication (10), and ribosomal protein assembly and transport (11). More recently, NPM1 exon 12 mutations have been reported to be involved in leukemogenesis, and detected in ~35% of AML cases (12,13). However, the prognostic implications of NPM1 mutations are less clear and are notably variable among different institutions (13–18). A study conducted by Konoplev et al including 252 AML patients suggested NPM1 mutations did not impact overall survival (OS) and event-free survival (EFS) (17). However, findings of previous studes have indicated that the NPM1 mutation has a favorable effect on the outcome for AML (13–16,19–22). For this reason, we performed an updated meta-analysis of 9 published studies in order to investigate the prognostic significance of NPM1 mutations for AML.
Materials and methods
Selection of studies
Studies were eligible for inclusion in the meta-analysis if they were: i) original articles written in English and published up to January 2013; ii) dealt only with untreated AML patients; iii) offered survival information based on the NPM1 status, including, NPM1 mutations and NPM1-wild-type (NPM1-wt), and iv) provided survival information on response to induction therapy, including, complete remission (CR), disease-free survival (DFS) and/or OS. Studies were excluded if they focused exclusively on acute promyelocytic leukemia. Multiple reports of a single study were considered as one publication, and only the most recent article was examined.
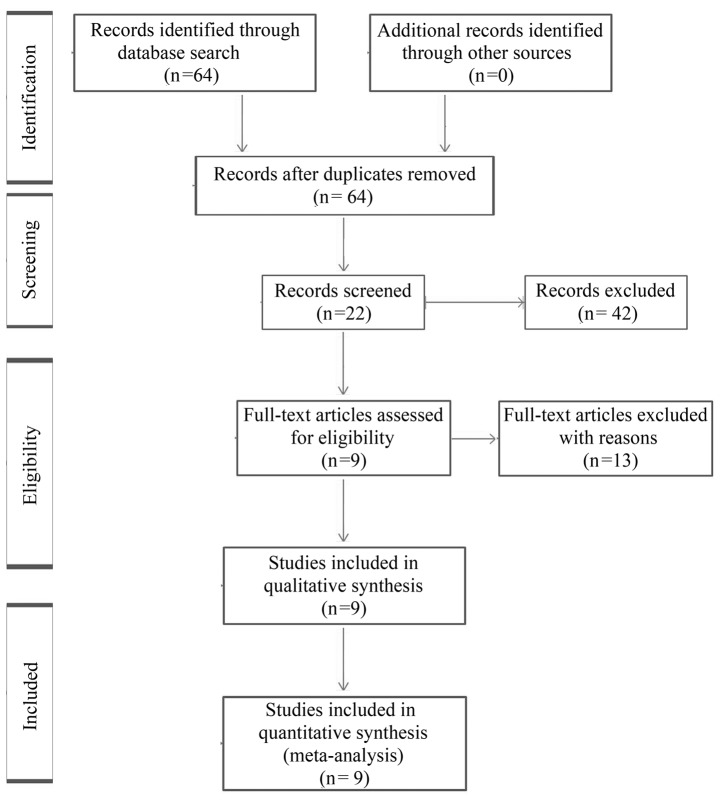
A computerized literature search of the PubMed, Medline and EMBASE databases was conducted using the free text search term AML AND nucleophosmin AND survival, with the publication period limited to prior to January 2013, and the language to English. The initial search yielded a total of 64 articles, and the titles and abstracts of these papers were reviewed, resulting in the exclusion of 42 articles, with 22 candidate articles. Of the 22 articles, 13 full-text articles were excluded as survival information was unavailable. In total, 9 studies satisfied the eligibility criteria and were included in the meta-analysis (Table I). The reasons for excluding articles are shown in Fig. 1 (23–35).
Table I.
List of studies included in the meta-analysis.
| Author (Refs.) | Publication year | Region | Subjects (n) | NPM1 mutation (%) | Normal karyotype (%) |
|---|---|---|---|---|---|
| Thiede et al (14) | 2006 | Germany | 1485 | 27.47 | 79.41 |
| Döhner et al (15) | 2005 | Germany and USA | 300 | 48.33 | NR |
| Schnittger et al (16) | 2005 | Germany and Italy | 401 | 52.90 | 100.00 |
| Suzuki et al (19) | 2005 | Japan | 190 | 30.53 | 63.79 |
| Verhaak et al (13) | 2005 | The Netherlands | 275 | 34.55 | 77.89 |
| Mullighan et al (18) | 2007 | USA | 93 | 6.45 | 50.00 |
| Gale et al (20) | 2008 | United Kingdom | 1217 | 41.33 | 67.59 |
| Becker et al (21) | 2010 | Germany and USA | 148 | 56.08 | NR |
| Boonthimat et al (22) | 2008 | Thailand | 400 | 26.25 | 86.67 |
NPM1, nucleophosmin 1; NR, not reported.
Figure 1.
Flow diagram showing the process of identifying and selecting relevant studies.
Data extraction and quality assessment
In order to avoid bias in the data abstraction process, the reviewers Y.F. Liu and P.C. He independently retrieved the data from the articles and subsequently compared the results. All data were assessed for internal consistency and disagreements were resolved by discussion. Characteristics abstracted from the articles included the name of the first author, year of publication, location of the study, number of subjects, mean or median values of age and median white blood cell (WBC) counts, the incidence of NPM1 mutations, percentage of cases with normal karyotype, outcomes including hematologic CR rate, hazard ratio (HR) and 95% confidence interval (CI) for DFS and OS according to the NPM1 status based on multivariate analysis. When the data required for the analysis could not be abstracted, attempts were made to contact the investigators who conducted the studies.
The quality of evidence and the strength of recommendations were evaluated by GRADE profiler (version 3.2) (36). Any discrepancies in quality assessments were resolved by consensus among the authors. The overall quality of the data was graded as moderate.
Quantitative data synthesis
HR was used to assess the survival effect of NPM1 mutations compared with wild-type. The natural logarithm of a crude HR and its variance within the study was calculated by using the abstracted survival probabilities at each time point with the methods proposed by Parmar et al (37), and those described previously (38). HR was calculated to show how many times higher the probability of survival failure was for the patients with NPM1 mutations cmpared with those with wild-type, as an HR less than unity suggests that NPM1 mutations yield a better survival rate compared with wild-type.
The odds ratio (OR) was calculated to describe the probability of CR following induction therapy based on NPM1 mutation status. An OR >1 indicates that patients with NPM1 mutations are associated with an improved CR rate compared with those without the mutations.
A DerSimonian Laird random method was used to calculate summary HR or OR and their 95% CI. Initially, the fixed effect and random-effect models were used to calculate summary HRs, but eventually the random-effect model was selected. Begg’s funnel plots (39) and Egger’s test (40) were used to detect possible publication bias. The between-study variation (τ2) from the Q statistic was also calculated (41).
Statistical analysis
Statistical analyses were performed using STATA 12 software (College Station, TX, USA). P<0.05 was considered to indicate a statistically significant test result for summary HR or OR.
Results
Study characteristics
As shown in Table I, 9 studies including a total of 4,509 subjects (2,903 with NPM1-wt, 1,606 with NPM1 mutations) were included in the meta-analysis. Four studies originated from Europe (13,14,16,20), two from Asia (19,22) and three from the USA, two of which contained information pertaining to Germany (15,18,21). The frequency of NPM1 mutations varied between 6.45 and 56.08% for AML patients. NPM1 mutations were associated with a higher frequency of FLT3-ITD mutations in 7 studies (Table II) (13–15,19–22). The frequency of the normal karyotype was higher among NPM1 mutant patients (Table I) (13,14,16,18–20,22). No graphical or statistical evidence of publication bias for DFS or OS was identified.
Table II.
Diagnostic characteristics according to the NPM1 status in the AML patients.
| Author (Refs.) | NPM1 status | Subjects (n) | Age (years) | Median WBC count (109/l) | FLT3-ITD mutation (%) |
|---|---|---|---|---|---|
| Thiede et al (14) | NPM1-wt | 1077 | 58.0 (15.0–87.0) | 0.3–465 | 13.74a |
| NPM1-mt | 408 | 60.0 (18.0–83.0) | 0.5–380 | 40.20 | |
| Döhner et al (5) | NPM1-wt | 155 | 16.0–60.0 | 0.41–369 | 24.52a |
| NPM1-mt | 145 | 18.0–60.0 | 0.2–345 | 40.69 | |
| Schnittger et al (16) | NPM1-wt | 189 | 58.1 | 10.0 (0.1–361) | NR |
| NPM1-mt | 212 | 55.8 | 38.7 (0.2–486) | NR | |
| Suzuki et al (19) | NPM1-wt | 141 | 47.0 (15.0–85.0) | 23.3 (0.9–337.6) | 11.35a |
| NPM1-mt | 49 | 58.0 (15.0–77.0) | 52.2 (1.0–372) | 55.10 | |
| Verhaak et al (13) | NPM1-wt | 180 | 39.7±13.3 | NA | 16.76a |
| NPM1-mt | 95 | 47.3±10.7 | NA | 49.47 | |
| Mullighan et al (18) | NPM1-wt | 87 | 9.7 | 38.0 | NR |
| NPM1-mt | 6 | 14.6 | 35.3 | 16.67 | |
| Gale et al (20) | NPM1-wt | 714 | 41.0 | 18.5 | 19.19a |
| NPM1-mt | 503 | 46.0 | 35.4 | 41.35 | |
| Becker et al (21) | NPM1-wt | 65 | 71.0 (60.0–83.0) | 7.0 (1.0–434.1) | 20.00a |
| NPM1-mt | 83 | 67.0 (60.0–81.0) | 26.2 (1.0–249.3) | 39.76 | |
| Boonthimat et al (22) | NPM1-wt | 295 | 40.0 | 25.4 | 20.68a |
| NPM1-mt | 105 | 51.0 | 47.0 | 43.81 |
NPM1, nucleophosmin 1; AML, acute myeloid leukemia; WBC, white blood cell; NR, not reported; NA, not assessed; NPM1-mt, NPM1-mutation type; NPM1-wt, NPM1-wild-type. Statistically significant difference (aP<0.05).
Treatment outcomes
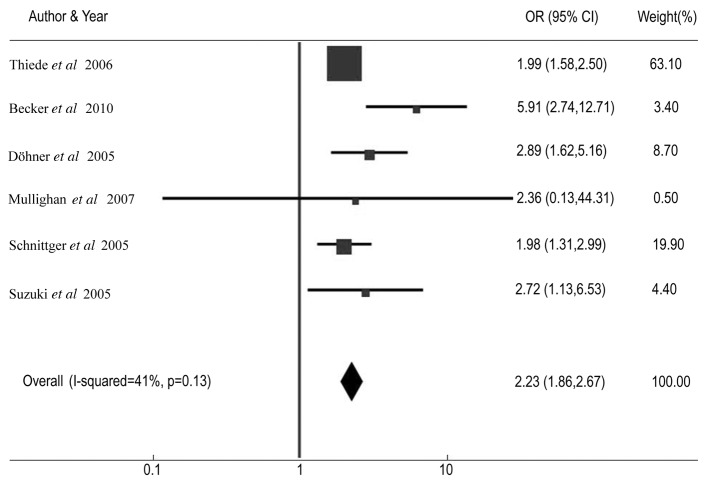
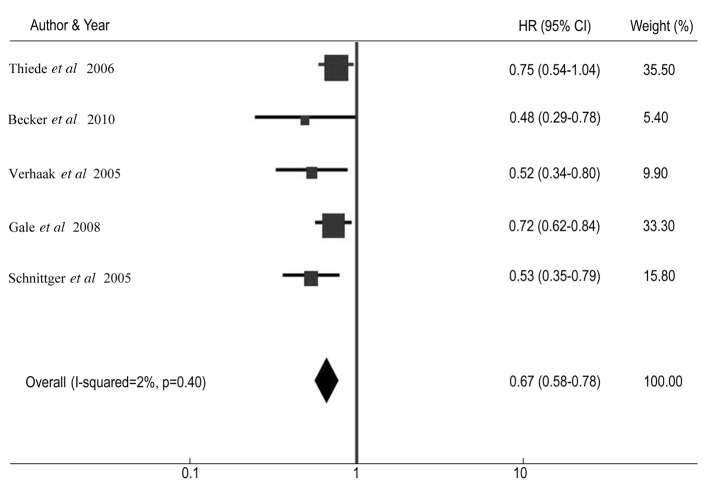
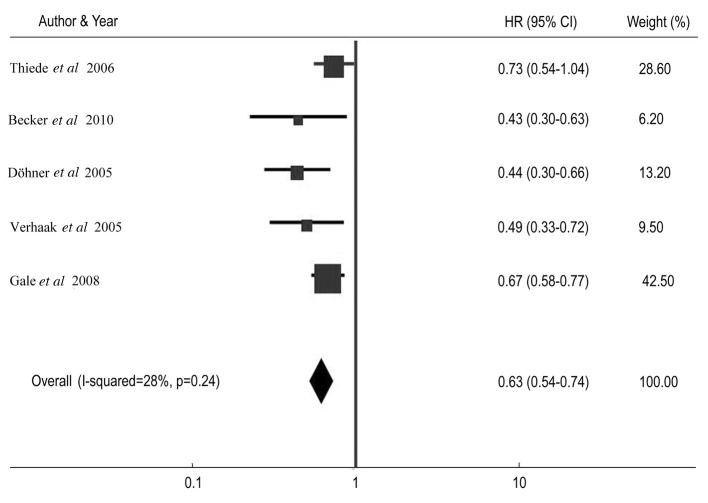
Table III shows the CR rate and HR for DFS and OS among AML patients with NPM1 mutations compared with patients without the mutations in individual studies. The summary OR for CR in the NPM1 mutant group was 2.23 (95% CI: 1.86–2.67, P<0.001) (Fig. 2). The summary HR for DFS of NPM1-mt/NPM1-wt was 0.67 (95% CI: 0.58–0.78; P<0.001) (Fig. 3), and the overall HR for OS of NPM1-mt/NPM1-wt was 0.63 (95% CI: 0.54–0.74; P<0.001) (Fig. 4). The test for heterogeneity, which evaluates the variation in study outcomes between studies in a meta-analysis, showed no significant heterogeneity among studies included in the DFS analysis (Q=4.08, df=4, P=0.40, τ2=2) and OS analysis (Q=5.52, d=4, P=0.24, τ2=28).
Table III.
NPM1 mutations and outcomes in acute myeloid leukemia.
| Author (Refs.) | NPM1 status | Subjects (n) | CR (%) | HR for OS | 95% CI for OS | HR for DFS | 95% CI for DFS |
|---|---|---|---|---|---|---|---|
| Thiede et al (14) | NPM1-wt | 1077 | 41.60a | 1.00 | Reference | 1.00 | Reference |
| NPM1-mt | 408 | 58.60 | 0.73 | 0.58–0.91 | 0.75 | 0.54–1.04 | |
| Döhner et al (15) | NPM1-wt | 155 | 68.50a | 1.00 | Reference | NR | NR |
| NPM1-mt | 145 | 86.00 | 0.44 | 0.30–0.66 | NR | NR | |
| Schnittger et al (16) | NPM1-wt | 189 | 54.70a | NR | NR | 1.00 | Reference |
| NPM1-mt | 212 | 70.50 | NR | NR | 0.53 | 0.35–0.79 | |
| Suzuki et al (19) | NPM1-wt | 141 | 68.80a | NR | NR | NR | NR |
| NPM1-mt | 49 | 85.70 | NR | NR | NR | NR | |
| Verhaak et al (13) | NPM1-wt | 180 | NA | 1.00 | Reference | 1.00 | Reference |
| NPM1-mt | 95 | NA | 0.49 | 0.33–0.72 | 0.52 | 0.34–0.80 | |
| Mullighan et al (18) | NPM1-wt | 87 | 85.06 | NA | NA | NA | NA |
| NPM1-mt | 6 | 100.00 | NA | NA | NA | NA | |
| Gale et al (20) | NPM1-wt | 714 | NA | 1.00 | Reference | 1.00 | Reference |
| NPM1-mt | 503 | NA | 0.67 | 0.58–0.77 | 0.72 | 0.62–0.84 | |
| Becker et al (21) | NPM1-wt | 65 | 47.69a | 1.00 | Reference | 1.00 | Reference |
| NPM1-mt | 83 | 84.34 | 0.43 | 0.30–0.63 | 0.48 | 0.29–0.78 |
NPM1, nucleophosmin 1; CR, complete remission; HR, hazard ratio; 95% CI, 95% confidence interval; OS, overall survival; NR, not reported; NA, not assessed; DFS, disease-free survival; NPM1-mt, NPM1-mutation type; NPM1-wt, NPM1-wild-type. Statistically significant difference (aP<0.05).
Figure 2.
Forest plots of the odds ratio (OR) and 95% confidence intervals for complete remission. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. OR>1 indicates that the presence of NPM1 mutations is associated with a higher complete remission rate.
Figure 3.
Forest plots of the hazard ratio (HR) and 95% confidence intervals (CIs) for disease-free survival. The size of the blocks or diamonds represents the weight for the random-effects model in the meta-analysis. HR<1 indicates that the presence of nucleophosmin 1 (NPM1) mutations is associated with an improved prognosis.
Figure 4.
Forest plots of the hazard ratio (HR) and 95% confidence intervals (CIs) for overall survival. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. HR<1 indicates that the presence of nucleophosmin 1 (NPM1) mutations is associated with an improved prognosis.
Furthermore, we performed a sensitivity test during the meta-analysis. Exclusion of any single study did not affect the overall results.
Discussion
Previous studies have investigated the prognostic significance of NPM1 mutation status in AML patients. Certain studies demonstrated the positive prognostic effect of NPM1 mutations (13–16,19–22), whereas in other studies no clinical outcome difference between patients with and without NPM1 mutations (17,18). The aim of the present meta-analysis was to clarify the prognostic significance of NPM1 mutation status in AML patients. Meta-analysis is a useful statistical method for integrating results from independent studies for a specified outcome. Combining the relevant studies increases statistical power, thus the effects that may be missed by individual studies may be detected (42). The present meta-analysis demonstrated the effects of NPM1 mutations with the summary OR of 2.23 (95% CI: 1.86–2.67) for CR, HR of 0.67 (95% CI: 0.58–0.78) for DFS, and HR of 0.63 (95% CI: 0.54–0.74) for OS. Furthermore, the present study indicated that NPM1 mutations were associated with a higher frequency of normal karyotype and FLT3-ITD mutations.
Notably, FLT3-ITD mutations have been shown to be the most important abnormality in AML patients and correlated with marked poor outcome (high percentage of bone marrow blast cells, increased risk of relapse from CR, and reduced survival) (43,44). However, our study indicated that patients with FLT3-ITD and NPM1 mutations have an improved CR (14,15,19,20), DFS (14,20) and OS (14,20) compared with those who only have the FLT3-ITD mutation, although this result was inferior to only NPM1 mutation cases.
The present study has several limitations. Firstly, the analyses were based on observational studies rather than prospective controlled studies or randomized trials. Secondly, we used abstracted data, while an individual patient data-based meta-analysis would have provided a more robust estimate of the association. The results reported should therefore be interpreted carefully by clinical physicians. Thirdly, as is often the case with meta-analysis, a substantial effect of heterogeneity should be considered. Although median WBC counts at the time of diagnosis were not identified as sources of heterogeneity, we cannot rule out the potential effect of other factors, such as differences in treatment and distinct cytogenetic categories, which were not examined in our analysis. Publication bias, although not directly detected in the present study, may also have had an impact on the accuracy of our study (42,45).
Although these limitations should be considered, the results of our meta-analysis demonstrated that NPM1 mutations have a favorable effect on the outcome for AML. Thus, distinguishing AML with NPM1 mutations from AML without mutations and justifying the risk-adapted therapeutic strategy for AML based on the NPM1 status.
A large number of patients should be prospectively studied in order for definitive conclusions to be reached. In addition to the presence or absence of NPM1 mutations, several factors relevant to NPM1 have been suggested to have a prognostic value, including the expression levels of NPM1 transcripts, mutant/wild-type allelic ratios for NPM1, and certain types of NPM1 exon mutations. These factors should be investigated in order to arrive at a more accurate estimation of the prognosis for AML.
Acknowledgements
The authors would like to thank Dr Di Wu and Dr Jieying Xi for their technological assistance.
References
- 1.Estey E, Döhner H. Acute myeloid leukemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24:35–63. doi: 10.1016/j.hoc.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Harrison CJ, Hills RK, Moorman AV, et al. Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council Treatment trials AML 10 and 12. J Clin Oncol. 2010;28:2674–2681. doi: 10.1200/JCO.2009.24.8997. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:497–529. doi: 10.1053/beha.2001.0152. [DOI] [PubMed] [Google Scholar]
- 5.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 6.Naoe T, Suzuki T, Kiyoi H, Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97:963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 8.Chan WY, Liu QR, Borjigin J, et al. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989;28:1033–1039. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- 9.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 10.Okuda M, Horn HF, Tarapore P, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 11.Lindstrom MS. NPM1/B23: A multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 13.Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 14.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 15.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 16.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 17.Konoplev S, Huang X, Drabkin HA, et al. Cytoplasmic localization of nucleophosmin in bone marrow blasts of acute myeloid leukemia patients is not completely concordant with NPM1 mutation and is not predictive of prognosis. Cancer. 2009;115:4737–4744. doi: 10.1002/cncr.24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullighan CG, Kennedy A, Zhou X, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21:2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T, Kiyoi H, Ozeki K, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106:2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 20.Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 21.Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boonthimat C, Thongnoppakhun W, Auewarakul CU. Nucleophosmin mutation in Southeast Asian acute myeloid leukemia: eight novel variants, FLT3 coexistence and prognostic impact of NPM1/FLT3 mutations. Haematologica. 2008;93:1565–1569. doi: 10.3324/haematol.12937. [DOI] [PubMed] [Google Scholar]
- 23.Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. 2010;116:5486–5496. doi: 10.1182/blood-2010-02-267955. [DOI] [PubMed] [Google Scholar]
- 24.Yan L, Chen S, Liang J, et al. Analysis of NPM1 gene mutations in Chinese adults with acute myeloid leukemia. Int J Hematol. 2007;86:143–146. doi: 10.1532/IJH97.A10620. [DOI] [PubMed] [Google Scholar]
- 25.Haferlach C, Mecucci C, Schnittger S, et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features. Blood. 2009;114:3024–3032. doi: 10.1182/blood-2009-01-197871. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Rassidakis GZ, Li J, et al. High frequency of NPM1 gene mutations in acute myeloid leukemia with prominent nuclear invaginations (‘cuplike’ nuclei) Blood. 2006;108:1783–1784. doi: 10.1182/blood-2006-03-014340. [DOI] [PubMed] [Google Scholar]
- 27.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 28.Koh Y, Park J, Bae EK, et al. Non-A type nucleophosmin 1 gene mutation predicts poor clinical outcome in de novo adult acute myeloid leukemia: differential clinical importance of NPM1 mutation according to subtype. Int J Hematol. 2009;90:1–5. doi: 10.1007/s12185-009-0350-1. [DOI] [PubMed] [Google Scholar]
- 29.Palmisano M, Grafone T, Ottaviani E, Testoni N, Baccarani M, Martinelli G. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukemia. Haematologica. 2007;92:1268–1269. doi: 10.3324/haematol.11202. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhang M, Yang L, Xiao Z. NPM1 mutations in myelodysplastic syndromes and acute myeloid leukemia with normal karyotype. Leuk Res. 2007;31:109–111. doi: 10.1016/j.leukres.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Wertheim G, Bagg A. Nucleophosmin (NPM1) mutations in acute myeloid leukemia: an ongoing (cytoplasmic) tale of dueling mutations and duality of molecular genetic testing methodologies. J Mol Diagn. 2008;10:198–202. doi: 10.2353/jmoldx.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol. 2010;28:2739–2747. doi: 10.1200/JCO.2009.26.2501. [DOI] [PubMed] [Google Scholar]
- 33.Noguera NI, Ammatuna E, Zangrilli D, et al. Simultaneous detection of NPM1 and FLT3-ITD mutations by capillary electrophoresis in acute myeloid leukemia. Leukemia. 2005;19:1479–1482. doi: 10.1038/sj.leu.2403846. [DOI] [PubMed] [Google Scholar]
- 34.Burnett AK, Hills RK, Green C, et al. The impact on outcome of the addition of all-trans retinoic acid to intensive chemotherapy in younger patients with nonacute promyelocytic acute myeloid leukemia: overall results and results in genotypic subgroups defined by mutations in NPM1, FLT3, and CEBPA. Blood. 2010;115:948–956. doi: 10.1182/blood-2009-08-236588. [DOI] [PubMed] [Google Scholar]
- 35.Falini B. Therapy-related acute myeloid leukaemia with mutated NPM1: treatment induced or de novo in origin? Leukemia. 2008;22:891–892. doi: 10.1038/leu.2008.44. [DOI] [PubMed] [Google Scholar]
- 36.Guyatt GH, Oxman AD, Vist GE, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336:924–926. doi: 10.1136/bmj.39489.470347.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Hotta K, Matsuo K, Ueoka H, Kiura K, Tabata M, Tanimoto M. Meta-analysis of randomized clinical trials comparing Cisplatin to Carboplatin in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:3852–3859. doi: 10.1200/JCO.2004.02.109. [DOI] [PubMed] [Google Scholar]
- 39.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 40.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 42.Yanada M, Matsuo K, Suzuki T, Kiyoi H, Naoe T. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: a meta-analysis. Leukemia. 2005;19:1345–1349. doi: 10.1038/sj.leu.2403838. [DOI] [PubMed] [Google Scholar]
- 43.Small D. FLT3 mutations: biology and treatment. Hematol Am Soc Hematol Educ Program. 2006:178–184. doi: 10.1182/asheducation-2006.1.178. [DOI] [PubMed] [Google Scholar]
- 44.Kottaridis PD, Gale RE, Linch DC. Flt3 mutations and leukaemia. Br J Haematol. 2003;122:523–538. doi: 10.1046/j.1365-2141.2003.04500.x. [DOI] [PubMed] [Google Scholar]
- 45.Feng JH, Guo XP, Chen YY, Wang ZJ, Cheng YP, Tang YM. Prognostic significance of IDH1 mutations in acute myeloid leukemia: a meta-analysis. Am J Blood Res. 2012;2:254–264. [PMC free article] [PubMed] [Google Scholar]