Abstract
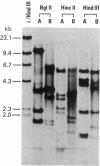
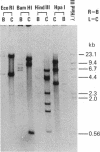
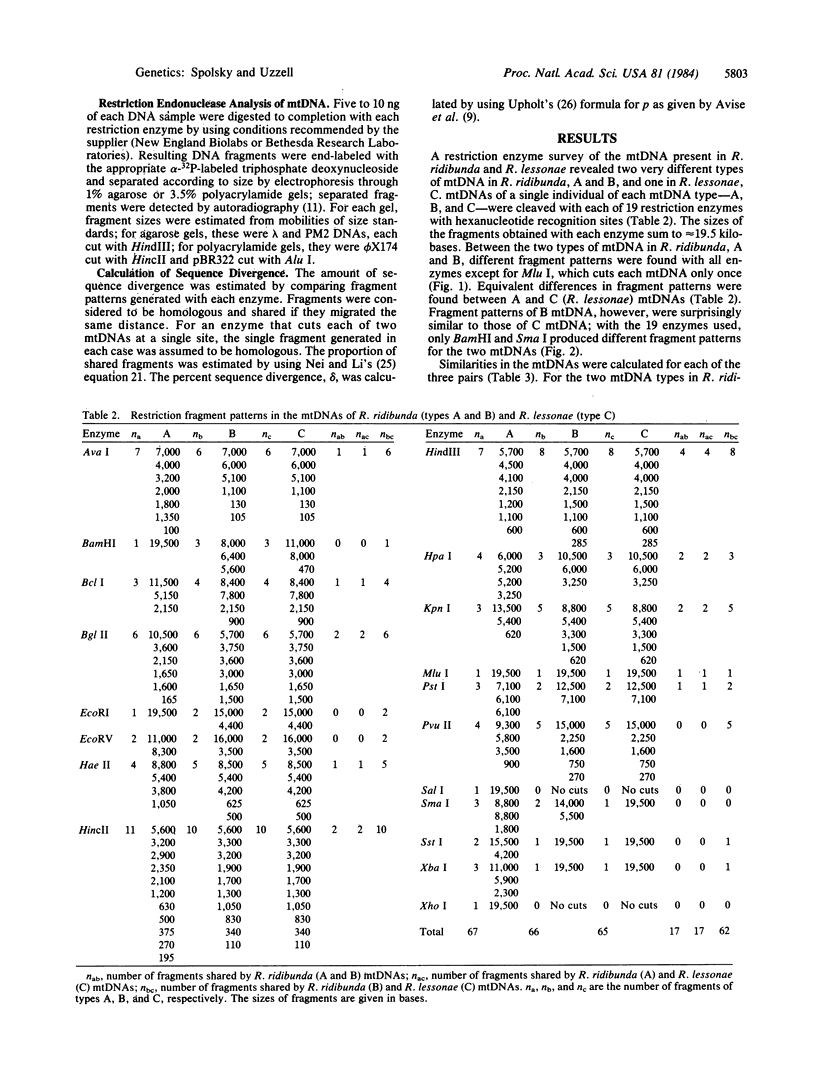
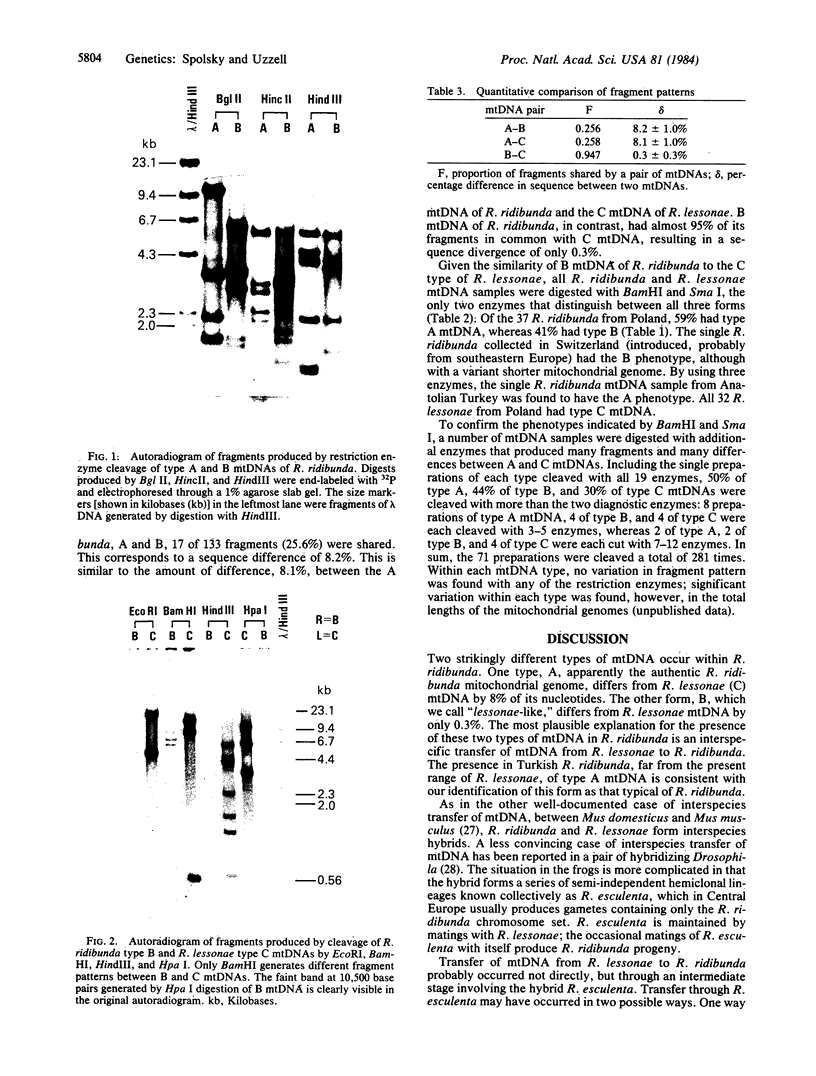
mtDNAs of two Central European water frog species, Rana ridibunda and Rana lessonae, were examined by electrophoresis of restriction enzyme fragments. Two types of mtDNA occur in R. ridibunda. One shares with mtDNA of R. lessonae 25.8% of 132 fragments generated by 19 enzymes, corresponding to a nucleotide sequence divergence of 8.1%; the other has diverged from R. lessonae mtDNA by only 0.3%. This latter type is a variant R. lessonae mtDNA that has been transferred into R. ridibunda; the introgression may have occurred via the hybridogenetic hybrid lineages collectively known as Rana esculenta. Of 37 R. ridibunda from Poland, 59% had the typical R. ridibunda mtDNA; 41% had the modified R. lessonae mtDNA as did a single individual from Switzerland (introduced). A single R. ridibunda from Turkey, outside the present range of R. lessonae, had the typical R. ridibunda mtDNA phenotype. Discordancies between inheritance of mitochondrial and nuclear genomes point up the danger of relying on a single molecular feature in reconstructing phylogeny. In addition, studies of mtDNA provide otherwise inaccessible information on complex evolutionary histories of closely related species. A knowledge of these complexities is important to an understanding of phylogenetic relationships and of the genetic processes that underlie the evolution of clonal taxa.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Giblin-Davidson C., Laerm J., Patton J. C., Lansman R. A. Mitochondrial DNA clones and matriarchal phylogeny within and among geographic populations of the pocket gopher, Geomys pinetis. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6694–6698. doi: 10.1073/pnas.76.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J. C., Lansman R. A., Shade R. O. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics. 1979 May;92(1):279–295. doi: 10.1093/genetics/92.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M. Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3605–3609. doi: 10.1073/pnas.77.6.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Wright J. W. Mitochondrial DNA analyses and the origin and relative age of parthenogenetic lizards (genus Cnemidophorus). Science. 1979 Mar 23;203(4386):1247–1249. doi: 10.1126/science.424751. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Blackler A. W. Maternal and cytoplasmic inheritance of mitochondrial DNA in Xenopus. Dev Biol. 1972 Oct;29(2):152–161. doi: 10.1016/0012-1606(72)90052-8. [DOI] [PubMed] [Google Scholar]
- Dawid I. B. Evidence for the mitochondrial origin of frog egg cytoplasmic DNA. Proc Natl Acad Sci U S A. 1966 Jul;56(1):269–276. doi: 10.1073/pnas.56.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S. D., Brown W. M., Davidson W. S., Wilson A. C. Extensive polymorphism in the mitochondrial DNA of apes. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6319–6323. doi: 10.1073/pnas.78.10.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S. D., Sage R. D., Huang C. M., Nielsen J. T., Ritte U., Wilson A. C. Flow of mitochondrial DNA across a species boundary. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2290–2294. doi: 10.1073/pnas.80.8.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S. D., Wilson A. C., Brown W. M. Evolutionary tree for apes and humans based on cleavage maps of mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2432–2436. doi: 10.1073/pnas.78.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles R. E., Blanc H., Cann H. M., Wallace D. C. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J. D., Karch F., Moreillon M. C. Biochemical variation in the Rana esculenta complex: a new hybrid form related to Rana perezi and Rana ridibunda. Experientia. 1977 Dec 15;33(12):1582–1584. doi: 10.1007/BF01934010. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Laipis P. J. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Newbold J. E., Potter S. S., Edgell M. H. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974 Oct 11;251(5475):536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- Kroon A. M., de Vos W. M., Bakker H. The heterogeneity of rat-liver mitochondrial DNA. Biochim Biophys Acta. 1978 Jun 22;519(1):269–273. doi: 10.1016/0005-2787(78)90079-5. [DOI] [PubMed] [Google Scholar]
- Leslie J. F., Vrijenhoek R. C. Genetic dissection of clonally inherited genomes of poeciliopsis. I. Linkage analysis and preliminary assessment of deleterious gene loads. Genetics. 1978 Dec;90(4):801–811. doi: 10.1093/genetics/90.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MULLER H. J. THE RELATION OF RECOMBINATION TO MUTATIONAL ADVANCE. Mutat Res. 1964 May;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc Natl Acad Sci U S A. 1983 Jan;80(2):492–495. doi: 10.1073/pnas.80.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4(5):1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]