Abstract
MicroRNAs are small non-coding RNA molecules that control expression of target genes. Previous studies showed that microRNA-107 (miR-107) is overexpressed in gastric cancer tissues compared with the matched normal tissues. However, it remains largely unclear as to how miR-107 exerts its function and modulates the malignant phenotypes of gastric cancer, because our understanding of miR-107 signalling pathways is limited. In this study, we demonstrate that miR-107 is frequently up-regulated in gastric cancers and its overexpression is significantly associated with gastric cancer metastasis. Furthermore, silencing the expression of miR-107 could inhibit gastric cancer cell migration and invasion in vitro and in vivo. Subsequent investigation characterized DICER1 as a direct target of miR-107. Up-regulation of DICER1 resulted in a dramatic reduction of in vitro migration, invasion, in vivo liver metastasis of nude mice, which is similar to that occurs with the silencing of miR-107, indicating that DICER1 functions as a metastasis suppressor in gastric cancer. Furthermore, the restoration of DICER1 can inhibit miR-107-induced gastric cancer cell invasion and metastasis. In conclusion, our results suggested that miR-107, an oncogene miRNA promoting gastric cancer metastasis through down-regulation of DICER1. Inhibition of miR-107 or restoration of DICER1 may represent a new potential therapeutic target for gastric cancer treatment.
Keywords: gastric cancer, metastasis, miR-107
Introduction
Gastric cancer is one of the most common malignancies in tumours in East Asian countries [1–2]. Like other solid human cancers, the biggest obstacle for the treatment of gastric cancer is recurrence and metastasis. So, seeking for new therapeutic modalities to prevent the metastasis of gastric cancer becomes an urgent issue. But the precise mechanism of this process has not yet been defined, which has confined the improvement of therapeutic strategies.
MicroRNAs (miRNAs) are small (20–24 nucleotides [nt]) non-coding RNA gene products that post-transcriptionally modulate gene expression by negatively regulating the stability or translational efficiency of their target mRNAs [3]. Currently, more than several hundred unique mature human miRNAs are known (http://microrna.sanger.ac.uk). Aberrant expression of miRNAs has been reported to be involved in tumourigenesis, acting, as one might expect, variously as either oncogenes [4] or tumour suppressors [5]. Previously, altered miRNA expression signature is observed in gastric cancers that have been collected from different study cohorts [6–7] and miR-107 was found significantly up-regulated in gastric cancers, compared with the matched normal tissues.
In this study, we demonstrated that the overexpression of miR-107 is significantly correlated with metastasis of gastric cancer. We comprehensively investigate the biological functions and underlying molecular mechanism of miR-107 in gastric carcinogenesis performed with both in vitro and in vivo models. Our results showed that knockdown of miR-107 could inhibit gastric cancer cell invasion and migration in vitro and in vivo. Furthermore, DICER1, which is a putative metastasis suppressor in gastric cancer, is characterized as a bona fide direct and functional target of miR-107, which provides a possible regulation pathway for DICER1 and a candidate target for gastric cancer treatment.
Materials and methods
Patients and tissue specimens
Fifty formalin fixed paraffin-embedded specimens of gastric cancer tissues, the matched normal tissues and information about the patients were collected from Gastrointestinal Surgery in Xijing Hospital, Xi’an, China. Primary gastric cancer in these patients was diagnosed and treated at Xijing Hospital from 1990 to 1997. The matched ‘normal gastric tissue’ were obtained from the 5 cm distant form the tumour margin, which were further confirmed by pathologist that have not tumour cells. Total RNA, with efficient recovery of small RNAs, was isolated from 20-μm sections from formalin-fixed, paraffin-embedded tissue blocks, performed with the Recover All Total Nucleic Acid Isolation Kit (Ambion, Austin, TX, USA). All patients did not perform any therapy before recruitment to this research. In addition, all patients were received FAM plan chemotherapy after surgery during our research. The protocols used in the study were approved by the Hospital’s Protection of Human Subjects Committee.
RNA extraction and quantitative real-time polymerase chain reaction
Stem-loop reverse transcription for mature miRNA was performed as described previously [8]. All reagents were obtained from Applied Biosystems (Foster City, CA, USA). For miRNA detection, mature miR-107 was reverse-transcribed with specific RT primers, quantified with a TaqMan probe. The small nuclear U6 RNA was used as an internal control for normalization and quantification the miRNAs expression as previously reported [9].
Cell lines and cultures
Cell plates were pre-covered with Matrigel (BD Biosciences, Shanghai, China) at 5 μg/cm2. SGC-7901, MKN-45, MKN-28, AGS, BGC-823, SV40-transformed immortal gastric epithelial cell GES and HEK 293T cell lines were maintained in DMEM containing 10% FBS supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C with 5% CO2.
Plasmid construction and cell transfection
Transient CMV-driven miRNA sponge backbones and a control sponge containing tandem non-targeting binding sites was identical to that utilized previously [9]. The miR-107 sponge was constructed by annealing, purifying and cloning oligonucleotides containing seven tandem ‘bulged’ (at positions 9–12) miR-107 binding motifs into the XhoI and ApaI sites of the CMV sponge backbone. The miR-107 sponge contained ‘bulged’ sites of sequence is CGCCAAUAUUCUAUGCUGCUA. For stable expression studies, the fragments of the miR-107 and control CMV sponge constructs were subcloned into the pBABE-puro retroviral vector. All stable cell lines were generated via retroviral infection performed with HEK293T cells, as has been previously described [10].
The DICER1 expression vector pcDNA-3.1-DICER1 was constructed by inserting its ORF sequence into the pcDNA-3.1 vector (Invitrogen Corp., Carlsbad, CA, USA). pSilencer3.0 (Ambion) was used for construction of human DICER1 siRNA vector psiDICER1 according to manufacturer’s protocol. One pairs of specific oligonucleotide (DICER1, DICER1’) was annealed and then subcloned into the BamHI/HindIII cloning site of pSilencer3.0. Cell transfection was performed with Lipofectamine2000 (Invitrogen) as described in manufacturer’s protocol.
MiR-107 expression vector pcDNA-3.1-miR-107 was amplified from normal genomic DNA and cloned into pcDNA-3.1. 3′-UTR segment of DICER1 was amplified from the genomic DNA of normal gastric tissues and subcloned into the pGL3 control vector (Promega, Madison, WI, USA) immediately downstream of the stop codon of the luciferase gene. PCR with the appropriate primers also generated inserts with point substitutions in the miRNA complementary sites. Wild-type and mutant inserts were confirmed by sequencing.
In vitro migration and invasion assays
For transwell migration assays, 1×105 cells were plated in the top chamber with the non-coated membrane (24-well insert, 8-mm pore size; Corning Costar Corp., Cambridge, MA, USA). For invasion assays, 2×105 cells were plated in the top chamber with Matrigel-coated membrane (24-well insert, 8-mm pore size; Corning Costar Corp.). In both assays, cells were plated in medium without serum. Medium supplemented with serum was used as a chemo-attractant in the lower chamber. The cells were incubated for 24 hrs and cells that did not migrate or invade through the pores were removed by a cotton swab. Cells on the lower surface of the membrane were fixed with methanol and stained with haematoxylin. The cell numbers were determined by counting of the penetrating cells under a microscope at 200× magnification on 10 random fields in each well. Each experiment was performed in triplicate.
In vivo metastasis assays
Mice were handled performed with best humane practices and were cared for in accordance with NIH Animal Care and Use Committee guidelines. Cells were harvested from tissue culture flasks performed with trypsin and washed three times with PBS. Mice were injected with 1×106 cells in 0.1 ml PBS through tail vein. The mice were then monitored overall health, and total body weight. After 4 weeks of injection, the mice were sacrificed. The liver tissues were observed with naked eyes and the number of visible tumours in liver surface was counted. The liver tissues were made serial sections before being haematoxylin and eosin dyed and observed under a light microscope. Each experimental group contained 6–10 mice.
Luciferase reporter assay
HEK 293T cells were grown in 10% FBS in DMEM to 80–90% confluence in 24-well plates. Cells were co-transfected with 100 ng firefly luciferase reporter vector containing the DICER1 3’-UTR (named pGL3-DOCER1-3’-UTR) or its 3’-UTR mutant (named pGL3-DOCER1-3’-UTRmut) and 8 ng of the control vector containing Renilla luciferase, pRL-TK (Promega), in a final volume of 0.5 ml performed with lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured consecutively performed with dualluciferase assays (Promega) 48 hrs after transfection.
Western blot analysis
Cells were washed twice with Hanks’s balanced salt solution and lysed directly in RIPA buffer (50 mM Tris-HCl (pH 7.4), 1% (v/v) Triton X-100, 1 mM EDTA, 1 mM leupeptin, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 1 mM Na3VO4). The lysates were centrifuged at 14,000 rpm for 30 min. at 4°C and the supernatants were collected. Cell lysate (60 μg) was respectively separated by SDS-polyacrylamide gel electrophoresis, blotted onto nitrocellulose membrane, and incubated with a primary antibody: anti-DICER1 (Sigma-Aldrich, Shanghai, China) diluted 1:1000, or anti-β-actin (Sigma-Aldrich) diluted 1:5000. After repeated washing, the membranes were incubated with horseradish-peroxidase-conjugated anti-mouse secondary antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) diluted 1:2000. The bands were visualized using the enhanced chemiluminescence (ECL) system (Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA).
Statistical analysis
Each experiment was repeated at least three times. Student’s t-test was used to investigate the significance of the difference between the covariates. The Kaplan–Meier method was performed to calculate the patients’ survival durations. Survival durations in the patient groups were calculated with the log-rank test. All statistical analyses were conducted using SPSS version 11.05 software (SPSS, Chicago, IL, USA). P < 0.05 was considered as statistically significant and error bars represent S.E.M.
Results
MiR-107 is up-regulated in gastric cancer and associated with tumour metastasis and worse prognosis
In previous studies [5–6], miR-107 was reported to be significantly overexpressed in gastric cancer tissues compared with the matched normal tissues. In an attempt to explore the expression and significance of miR-107 in gastric carcinogenesis, we first confirmed the miR-107 expression levels in 50 cases of gastric cancer tissues and the matched normal tissues by quantitative reverse-transcription PCR. Our results showed that the expression of miR-107 is significantly up-regulated in gastric cancer tissues when compared with matched normal tissues (Fig. 1A, P < 0.0001). When these 50 tumours were stratified based on clinical progression, we found that miR-107 expression was remarkably up-regulated in primary tumours that subsequently metastasized, when compared to those primary tumours that did not recur (Fig. 1A). Moreover, high miR-107 levels correlated strongly with increased distant disease-free survival relative to tumours with low miR-107 (5-year metastasis-free survival of 24% versus 76%, median survival of 20 months versus not reached, P = 0.0071; Fig. 1B). These results suggested that overexpression of miR-107 is closely related to the increase of gastric cancer metastasis, and may play an important role in the pathological process.
Fig 1.
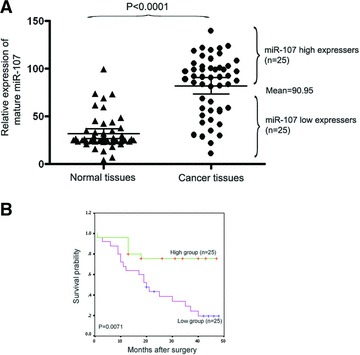
MiR-107 is up-regulated in gastric cancer and associated with tumour metastasis and worse prognosis. (A) The expression level of mature miR-107 in gastric cancer samples or normal control tissues performed with quantitative PCR. A significant difference was detected in the mean value of miR-107 expression between these two groups (P < 0.0001). The same gastric cancer samples were divided into two groups, according to the mean expression of miR-107 (mean = 90.95). Cases with levels of miR-107 below the mean were miR-107 low expressers (n = 25), and those with levels of miR-107 above the mean were miR-107 high expressers (n = 25). (B) Kaplan–Meier survival curve and log-rank test for gastric cancer patients between high and low miR-218 expressers. MiR-107 expression demonstrated a significant relationship with patient survival (log-rank, P = 0.0071).
MiR-107 promotes gastric cancer cell invasion and metastasis in vitro and in vivo
To determine whether miR-107 has a causal function on gastric cancer metastasis, we first examined the expression of mature miR-107 in various gastric originated cell lines and found that miR-107 was significantly up-regulated in gastric cancer cell lines (Fig. 2A), especially in SGC-7901 and MKN-45, which were demonstrated to have invasive ability to penetrate Matrigel-coated transwell in vitro invasion assay and metastatic ability to metastasize mainly to liver instead of lung and other organs in vivo tail vein metastatic assay [11]. We silenced the expression of miR-107 with retroviral miRNA sponge [9] in SGC-7901 and MKN-45 cells. We found that miR-107 sponge specifically inhibited miR-107 function by nearly >3.0-fold, when compared with the control sponge in SGC-7901 and MKN-45 cells (Fig. 2B).
Fig 2.
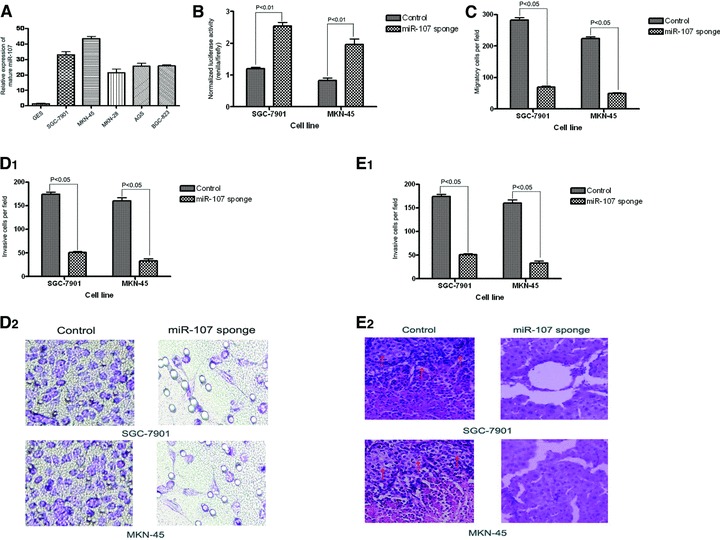
MiR-107 promotes gastric cancer cell invasion and metastasis in vitro and in vivo. (A) The expression level of mature miR-107 in various gastric cancer cell lines was determined using quantitative PCR. (B) MiR-107 sponge inhibition function was assayed in SGC-7901 or MKN-45 cells. Luciferase reporter assays were used to examine the sponge inhibition efficiency. (C, D) Transwell migration assay (C) and Matrigel invasion assay (D) of SGC-7901 or MKN-45 cells transfected with miR-107 sponge or its control retrovirus. Magnification in D2, 200×. (E) Mice were injected with 1×106 cells through tail vein. Four weeks later, the mice were sacrificed. The liver tissues were observed with naked eyes and the number of visible tumours in liver surface was counted (E1). The liver tissues were made serial sections before being haematoxylin and eosin dyed and observed under a light microscope (E2). N = 3; error bars represent S.E.M. P-values obtained using a one-sided Student’s t-test. P < 0.05 versus cells transfected with control retrovirus.
Given that expression of miR-107 is highly associated with the metastasis and prognosis of gastric cancer, we wondered whether miR-107 could play an important part in gastric cancer cell invasion and metastasis. Migration and invasion assays showed that down-regulation of miR-107 could significantly inhibit migration and invasion of SGC-7901 and MKN-45 cells, when compared with their control groups (Fig. 2C, D). Tail vein metastatic assay in nude mice was further adopted to examine the in vivo metastatic ability of SGC-7901-miR-107-sponge and MKN-45-miR-107-sponge. Compared with control cells transfected with control sponge, i.v. inoculation of SGC-7901-miR-107-sponge and MKN-45-miR-107-sponge cells led to significantly less visible tumours in liver surface (Fig. 2E, both P < 0.05). Taken together, these observations suggest that miR-107 is a positive metastatic regulator for gastric cancer.
MiR-107 post-transcriptionally down-regulates DICER1 expression by directly targeting its 3′-UTR
To understand the mechanisms by which miR-107 mediate tumour invasion and metastasis, the target prediction program TargetScan [12], PicTar [13] or miRanda (microrna.org and miRBase) was used to search for predicted direct target genes of miR-107. Among the hundreds targets of miR-107, three genes—CHRD, NF2 and DICER1—were previously implicated as tumour suppressor genes [14–20]. DICER1 was of particular interest, because its expression has been found to be progressively lost in gastric cancers showing increasing degrees of malignancy [20]. TargetScan and PicTar bioinformatics analysis of the 3′-UTRs of DICER1 revealed one putative binding site for miR-107, which is highly conserved across various species (Fig. 3A).
Fig 3.

MiR-107 post-transcriptionally down-regulates DICER1 expression by directly targeting its 3′-UTR. (A) The target site of miR-107 in DICER1 3′-UTR is conserved among mammalian species (shown in white). (B) Predicted duplex formation between miR-107 and the targeted DICER1 3′-UTR. The DICER1 3′-UTR mutant is identical to the wild-type, except that it had four point substitutions (red) disrupting pairing to miR-107 seed. (C) pGL3-DICER1 3′-UTR reporter plasmid in which the luciferase coding sequence had been fused to the 3′-UTR of DICER1 was co-transfected into HERK 293T cells with pcDNA3.1 or pcDNA3.1–miR-107. Luciferase activity was normalized relative to a simultaneously transfected Renilla expression plasmid. The 3′-UTR-Mut indicates the introduction of alterations into the seed complementary sites shown in Figure 2B. N = 3; error bars represent S.E.M. P-values obtained using a one-sided Student’s t-test. P < 0.05 versus cells transfected with control. (D) Western blots of DICER1 in SGC-7901 or MKN-45 cells after miR-107 sponge or control retrovirus infection. β-Actin antibody was used as a internal control. The level of DICER1 protein expression level was indicated by the ratio of DICER1/actin (bottom). Representative of three experiments with similar results.
To investigate the potential interaction experimentally, the human DICER1 3′-UTR was subcloned after the firefly luciferase open reading frame (ORF) and co-transfected into HEK 293T cells with the miR-107-expressing vector (pcDNA-3.1-miR-107). An analogous reporter with point substitutions disrupting the target sites (Fig. 3B) was also co-transfected with pcDNA-3.1-miR107. Our results showed that pcDNA-3.1-miR-107 produced a 60% decrease in relative luciferase activity compared with pcDNA3.1-transfected cells, when the full-length wild-type 3′-UTR of DICER1 was present (Fig. 3C). However, the relative luciferase activity did not drop as sharply in UTRs that contained mutant binding sites as in those that contained wild-type binding sites, which indicates that this reduction was sequence-specific (Fig. 3C).
To demonstrate that inhibited expression of miR-107 has a direct effect on DICER1 target. We assessed DICER1 protein expression levels after miR-107 inhibition by Western blot. We found that inhibition of miR-107 expression in SGC-7901 and MKN-45 cells increased the protein level of DICER1 (Fig. 3D). Those results really demonstrated that the endogenous DICER1 is direct target of miR-107.
To exclude the possibility that DICER1 is regulated by other predicted miRNAs in parallel with miR-107, we used TargetScan, PicTar and miRanda (miRBase) to search all miRNAs predicted to target DICER1. Besides miR-107, six candidate mature miRNAs (miR-103, miR-29a, miR-29b, miR-29c, miR-196a, miR-196b) were commonly predicted to target DICER1 by all of the three algorithms. We then test the expression of six candidate miRNAs (miR-103, miR-29a, miR-29b, miR-29c, miR-196a, miR-196b) in gastric cancer tissues and their paired adjacent non-tumour gastric cancer tissues, our results showed that miR-29b, and miR-29c were down-regulated in gastric cancer tissues compared with their paired adjacent non-tumour gastric cancer tissues. However, we did not find any expression differences of miR-103, miR-29a, miR-196a and miR-196b in gastric cancer tissues compared with their paired adjacent non-tumour gastric cancer tissues (Fig. S1). In conclusion, our results indicated that endogenous DICER1 is direct target of miR-107 in gastric carcinoma.
DICER1 inhibits gastric cancer cell migration and invasion in vitro and in vivo
Previous reports indicated that the expression of DICER1 is decreased during the progression of gastric cancer, especially in progressive gastric cancers, which indicating DICER1 may play an important role in the development of cancer and the epigenetical regulation involved [20]. However, the biopathological significance of DICER1 in human malignancies, especially in gastric cancer, is still unknown. To better understand the potential role of DICER1 in miR-107-mediated tumour invasion and metastasis, SGC-7901 cells were stable infected with pcDNA-3.1-DICER1 or pcDNA-3.1 vector alone (Fig. 4A). Remarkably, overexpression of DICER1 can strongly inhibit gastric cancer cell migration and invasion in vitro (Fig. 4B, C). Tail vein metastatic assay in nude mice also showed that ectopic expression of DICER1 in SGC-7901 cells led to significantly less visible tumours in liver surface (Fig. 4D, both P < 0.05), compared with control cells. This phenotype was similar to the one which was induced by the inhibition of miR-107.
Fig 4.
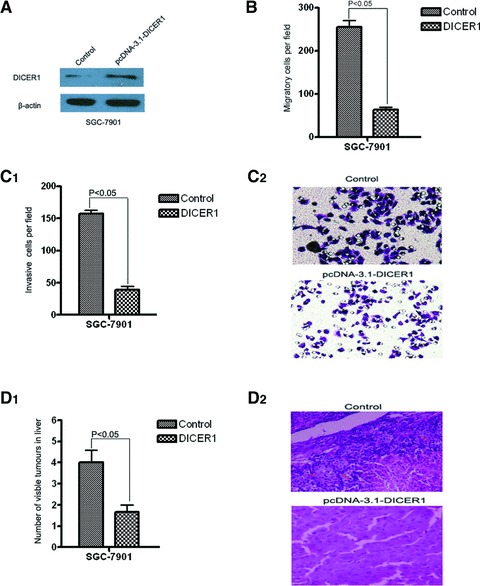
DICER1 inhibits gastric cancer cell migration and invasion in vitro and in vivo. (A) Western blots of DICER1 in SGC-7901 cells after pcDNA3.1-DICER1 or control transfection. β-Actin antibody was used as an internal control. (B, C) Transwell migration assay (B) and Matrigel invasion assay (C) of SGC-7901 cells transfected with miR-107 sponge or its control retrovirus. Magnification in C2, 200×. (D) Mice were injected with 1×106 cells through tail vein. Four weeks later, the mice were sacrificed. The liver tissues were observed with naked eyes and the number of visible tumours in liver surface was counted (C1). The liver tissues were made serial sections before being haematoxylin and eosin dyed and observed under a light microscope (C2). N = 3; error bars represent S.E.M. P-values obtained using a one-sided Student’s t-test. P < 0.05 versus cells transfected with control.
Ablation of dicer restores the miR-107 KO phenotype on gastric cancer cell migration and invasion in vitro and in vivo
Considered DICER1 is frequently down-regulated in gastric cancer and inhibits gastric cancer cell migration and invasion, and considered miR-107 can post-transcriptionally regulate the expression of DICER1 at the mRNA and protein levels by directly binding to its 3′-UTR, we hypothesized that down-regulation of DICER1 could directly reverse miR-107 KO phenotype on tumour invasion and metastasis. We stable transfected DICER1-specific siRNA and control vector into miR-107-knockdown SGC-7901 cell line and investigated the relevant metastasis trail in vitro and in vivo. The inhibition of DICER1 in this stable cell line was confirmed through Western blot analysis (Fig. 5A). Importantly, transwell assays indicated that the knockdown of DICER1 significantly prompted miR-107-knockdown SGC-7901 cells migration and invasion in vitro (Fig. 5B, C) and in vivo (Fig. 5D), which were inhibited by miR-107 deletion, suggesting that DICER1 is a direct and functional target for miR-107.
Fig 5.
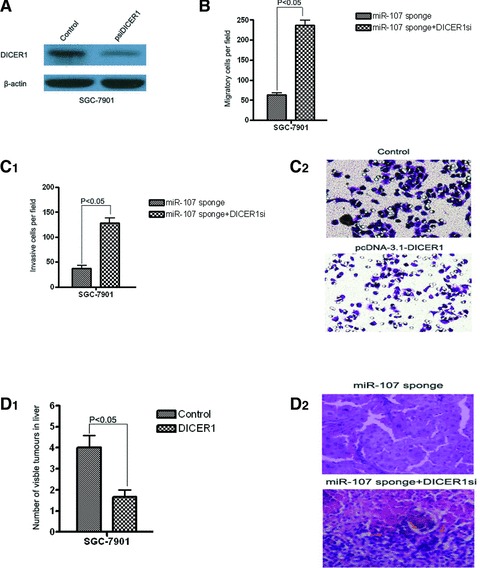
Inhibition of DICER1 promotes miR-107 deletion-mediated gastric cancer cell migration and invasion in vitro and in vivo. (A) Western blots of DICER1 in SGC-7901-miR-107 sponge cells after psiDICER1 or control transfection. β-Actin antibody was used as an internal control. (B, C) Transwell migration assay (B) and Matrigel invasion assay (C) of SGC-7901-miR-107 sponge cells transfected with psiDICER1 or control. Magnification in C2, 200×. (D) Mice were injected with 1×106 cells through tail vein. Four weeks later, the mice were sacrificed. The liver tissues were observed with naked eyes and the number of visible tumours in liver surface was counted (C1). The liver tissues were made serial sections before being haematoxylin and eosin dyed and observed under a light microscope (C2). N = 3; error bars represent S.E.M. P-values obtained using a one-sided Student’s t-test. P < 0.05 versus cells transfected with control.
Discussion
Metastasis, the spread and growth of tumour cells to distant organs, represents the most devastating attribute of cancer. A notable feature of this process is the variation in metastatic tissue tropism displayed by different types of cancer [21]. In the case of gastric cancer, metastasis is the most frequent pattern of recurrence in patients with gastric carcinoma [22]. About half of patients with serosa-invasive gastric carcinoma develop recurrence and die of this disease during the first 2 years, even if curative resection is performed [23]. The 5-year survival rate of patients with metastasis and recurrence is only 2% [24]. An incomplete understanding of the molecular and cellular mechanisms underlying metastasis hinders the development of effective therapies that would eliminate or ameliorate this condition.
Recently, miRNAs have been shown to be potential tools for cancer progression, diagnosis and prognosis [8]. With the advent of miRNA expression profiles, significant efforts have been made to correlate miRNA expressions with tumour development [25–27]. Because single miRNA can regulate hundreds of downstream genes, the information gained from miRNAs may be complementary to that from the expression profiling of protein-coding genes. Thus, identification of the actions of miRNAs could add new layers of complexity to our understanding of cancer dynamic.
In this study, we demonstrated that miR-107 is commonly up-regulated in gastric cancers and can promote gastric cancer cell migration, invasion and metastasis both in vitro and in vivo. A direct and functional target of miR-107 was also identified. The target gene, DICER1, is frequently repressed and functions as a metastasis suppressor in gastric cancer. Taken together, these findings indicate that miR-107 plays a fundamental role in gastric carcinogenesis, especially in the process of gastric cancer metastasis.
Recent findings have shown that miR-107 is involved in various biological and pathological processes. For example, miR-107 expression is reduced in Alzheimer’s disease brain neocortex and tends to be significantly relative to AD progresses [28]. Nelson et al. also reported that the expression of miR-107 was decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1 [29]. Yamakuchi et al. reported that miR-107 was involved in regulate hypoxic signalling and tumour suppressor gene TP53 could transcriptional activate miR-107 then inhibit hypoxia inducible factor-1β (HIF-1β) and colon cancer angiogenesis [30]. Tang et al. reported that miR-107 was involved in human non-small cell lung cancer cell cycle regulation and introduction of synthetic miR-107 could suppress growth of the human non-small cell lung cancer cell lines [31]. Interestingly, Lee et al. reported that miR-107 was overexpressed in pancreatic cancer and their cell lines performed with Northern blot assays. Further experiments showed that enforced expression of miR-107 in MiaPACA-2 and PANC-1 cells down-regulated in vitro growth, through repression of the putative miR-107 target, cyclin-dependent kinase 6 [32]. However, little is known about the role and underlying molecular mechanism of miR-107 in human cancer, especially in gastric cancer metastasis. In this report, we have, for the first time, established the role of miR-107 in tumour invasion and metastasis. Moreover, we identified DICER1, which functions as a ribonuclease, as a direct and functional target of miR-107. Although the expression of miR-107 is often up-regulated in gastric cancer and pancreatic cancer, it has been reported to be down-regulated in human non-small cell lung cancer cell lines, which seems to conflict with our observations in gastric cancer and Lee’s results in pancreatic cancer. We postulate that that this might be a consequence of different cancer heterogeneity, different tumour types have its own specific genetic background. Another reason is that Tang et al. only detected that the miR-107 expression level in human small cell lung cancer cell lines, which could not on behalf of its expression in tumour tissues.
DICER1 encodes a protein possessing an RNA helicase motif containing a DEXH box in its amino terminus and an RNA motif in the carboxy terminus. It is encoded protein functions as a ribonuclease and is required by the RNA interference and small temporal RNA (stRNA) pathways to produce the active small RNA component that represses gene expression. Previous reports showed that DICER1 expression is decreased during the progression of gastric cancer, especially in progressive gastric cancers, which indicating DICER1 may play an important role in the development of cancer and the epigenetical regulation involved [19]. We report for the first time that the enhanced expression of DICER1 could inhibit gastric cancer cell migration and invasion. Moreover, we also demonstrated that miR-107 can bind to one site of the 3′-UTR of DICER1 and dramatically decrease the level of DICER1 protein expression, which provides the first line of demonstrated in support of a mechanism for DICER1 regulation at the post-transcriptional level.
In summary, our results showed that miR-107, acts as an oncogene miRNA, can significantly promote gastric cancer cell invasion and metastasis through up-regulation its functional target of DICER1, which is down-regulated in gastric cancer and can inhibit gastric cancer cell migration and invasion. The newly identified miR-107/DICER1 axis provides a new insight into the pathogenesis of gastric cancer, especially with respect to invasion and metastasis, and represents a new, prognosis marker and potential therapeutic target for gastric cancer.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting Information
The expression level of mature miR-103 (A),miR-29a (B), miR-29b (C), miR-29c (D),miR-196a (E), and miR-196b (F) were detected ingastric cancer samples and normal control tissues performed withquantitative PCR.
References
- 1.Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Zhang Y, Ding J, et al. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–85. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 6.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 10.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–70. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 11.Pan Y, Zhao L, Liang J, et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886–8. doi: 10.1096/fj.06-6138fje. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 13.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 14.Moll F, Millet C, Noel D, et al. Chordin is underexpressed in ovarian tumors and reduces tumor cell motility. FASEB J. 2006;20:240–50. doi: 10.1096/fj.05-4126com. [DOI] [PubMed] [Google Scholar]
- 15.Patrick DL, Ramsey SD, Spencer AC, et al. Economic evaluation of aquatic exercise for persons with osteoarthritis. Med Care. 2001;39:413–24. doi: 10.1097/00005650-200105000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Bensenor LB, Barlan K, Rice SE, et al. Microtubule-mediated transport of the tumor-suppressor protein Merlin and its mutants. Proc Natl Acad Sci USA. 2010;107:7311–6. doi: 10.1073/pnas.0907389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, You L, Cooper J, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–90. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morales FC, Molina JR, Hayashi Y, et al. Overexpression of ezrin inactivates NF2 tumor suppressor in glioblastoma. Neuro Oncol. 2010;12:528–39. doi: 10.1093/neuonc/nop060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng ZH, Sun XJ, Fu WN, et al. Decreased expression of DICER1 in gastric cancer. Chin Med J (Engl) 2007;120:2099–104. [PubMed] [Google Scholar]
- 21.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 22.Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12:89–96. doi: 10.1006/scbi.2001.0416. [DOI] [PubMed] [Google Scholar]
- 23.Hiratsuka M, Iwanaga T, Furukawa H, et al. [Important prognostic factors in surgically treated gastric cancer patients] Gan To Kagaku Ryoho. 1995;22:703–8. [PubMed] [Google Scholar]
- 24.Yoo CH, Noh SH, Shin DW, et al. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87:236–42. doi: 10.1046/j.1365-2168.2000.01360.x. [DOI] [PubMed] [Google Scholar]
- 25.Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–7. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 27.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–12. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 28.Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer’s disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–9. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang WX, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–23. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamakuchi M, Lotterman CD, Bao C, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci USA. 2010;107:6334–9. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y, Forrest AR, Maeno E, et al. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KH, Lotterman C, Karikari C, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression level of mature miR-103 (A),miR-29a (B), miR-29b (C), miR-29c (D),miR-196a (E), and miR-196b (F) were detected ingastric cancer samples and normal control tissues performed withquantitative PCR.


