Summary
Xenopus is ideal for systematic decoding of cis-regulatory networks because its evolutionary position among vertebrates allows one to combine comparative genomics with efficient transgenic technology in one system. Here we have identified and analyzed the major enhancer of FoxE3/Lens1, a gene essential for lens formation that is activated in the presumptive lens ectoderm (PLE) when commitment to the lens fate occurs. Deletion and mutation analyses of the enhancer based on comparison of Xenopus-mammalian sequences and in vitro and in vivo binding assays identified two essential transcriptional regulators; Otx2, a homeodomain protein expressed broadly in head ectoderm including the PLE, and Su(H), a nuclear signal transducer of Notch signaling. A Notch ligand, Delta2, is expressed in the optic vesicle adjacent to the PLE, and inhibition of its activity led to loss or severe reduction of FoxE3 expression followed by failure of placode formation. Ectopic activation of Notch signaling induced FoxE3 expression within head ectoderm expressing Otx2, and additional misexpression of Otx2 in trunk ectoderm extended the Notch-induced FoxE3 expression posteriorly. These data provide the first direct evidence of involvementof Notch signaling in lens induction. The obligate integration of inputs of a field-selector (Otx2) and localized signaling (Notch) within target cis-regulatory elements may be a general mechanism of organ-field specification in vertebrates (as in Drosophila). This concept is also consistent with classical embryological studies of many organ systems involving a “multiple-step induction”.
Keywords: competence, genomics, induction, lens, Notch, Xenopus
Introduction
While many signaling factors and transcriptional regulators are essential for organ formation in vertebrates, little is known about how these multiple inputs are integrated to generate the specific locations and identities of particular organs. This is mainly because of the laborious nature of analysis of cis-regulatory elements on which transcriptional and signaling inputs directly converge; enhancer searches by traditional genome walking is a slow and painstaking process. However, recent remarkable advances in genome analytic tools have revolutionized the process (Wasserman and Sandelin, 2004). Putative enhancers are predicted in silico within entire genomes as clusters of transcription factor-binding motifs and/or intergenic sequences conserved among related species. Especially in vertebrates, enhancer activities are often mapped to conserved non-coding elements associated with developmental regulatory genes (Woolfe et al., 2005).
From this perspective, we have investigated mechanisms of lens induction in Xenopus, where essential tools for genomics-based cis-regulatory analysis, i.e. the whole genome sequence (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html) and a highly efficienttransgenesis technique, have become recently available (Offield et al., 2000; Smith, 2005). Comparison of Xenopus tropicalis and mammalian genome sequences is expected to be very useful to yield biologically meaningful information owing to their evolutionary distance (350 million years) (Muller et al., 2002).
Embryological studies have shown that lens induction is a stepwise process that begins when a broad domain of the animal cap ectoderm acquires a lens-forming competence at mid-gastrula stages (Grainger, 1992). This lens-competent field is subsequently narrowed down to the non-neural ectoderm surrounding the anterior margin of the newly formed neural plate. Accompanying this field-restriction process, the lens-forming ability of the competent region is enhanced by planar signals provided by the adjacent anterior neural plate. This “lens-biased” region corresponds to (or includes) the part of non-neural ectoderm termed pre-placodal ectoderm, which will give rise to the lens, otic, olfactory, and adenohypophyseal placodes at later stages. After neural tube formation, only the lateral part of the lens-biased ectoderm makes contact with the developing optic vesicle and at this stage determination occurs, followed several hours later by differentiation into lens tissue(lens placode and subsequently, the lens vesicle).
This stepwise commitment process is likely to be mediated, at least in part, by several transcription factor genes that exhibit distinct but overlapping expression during the course of lens-field specification (Ogino and Yasuda, 2000). Otx2, the earliest of these genes, exhibits expression from the end of gastrulation in the pre-placodal ectoderm and the adjacent anterior neural plate (Blitz and Cho, 1995; Zygar et al., 1998). Pax6 and Six3 show more restricted expression in the lens-field within the pre-placodal ectoderm (Zhou et al., 2000; Zygar et al., 1998). After neural tube formation, their expression is followed by activation of lens-specific transcription factor genes such as mafB, L-maf, and Pitx3, exclusively in the presumptive lens ectoderm (PLE) overlying the optic vesicle (Ishibashi and Yasuda, 2001; Pommereit et al., 2001).
The expression of Otx2, Pax6, and Six3 implies their involvement in the establishment of lens competence/bias, and recent significant progress made by mouse genetic studies supports this view (Lang, 2004). However, little is still known about how their activities are sequentially integrated to narrow down the lens-field, and how signaling from the opticvesicle is involved in this process. To address this question, we chose to study regulation of a forkhead family gene, Lens1, which is the earliest of the genes that are expressed primarily in the PLE overlying the optic vesicle (Kenyon et al., 1999), and which is activated at the time when commitment to the lens fate occurs (Grainger, 1992). Synteny analysis using Metazome 1.1 (http://www.metazome.net/) showed that Xenopus tropicalis Lens1 locus is orthologous to mouse Foxe3 and therefore we refer to it as Xenopus FoxE3 in this study. Analyses of mouse mutants have shown that Foxe3 is essential for lens epithelial proliferation and lens vesicle closure (Blixt et al., 2000; Brownell et al., 2000).
Using in vivo and in silico approaches, we identify and characterize an enhancer of X. tropicalis FoxE3 responsible for its PLE-specific expression, and demonstrate that Xenopus-mammalian genome comparison is a powerful strategy for prediction and further detailed analysis of vertebrate cis-regulatory elements. A “co-transgenesis” assay where separate enhancer and reporter element constructs were co-injected was also developed to facilitate the rapid survey of possible enhancer activities of the predicted cis-regulatory elements. Our analysis has led to the first recognition that Notch signaling (Lai, 2004) is alens-inducing signal, and revealed a role for Otx2 acting in concert with Notch signaling to specify the lens-field. The data presented here reveal one of the first molecular mechanisms found to underlie stepwise determination of the lens, as well as suggesting a general mechanism for how organ progenitor cells are segregated within a broader “zone of competence” during vertebrate development.
Materials and methods
Plasmid constructs
pBSSK+EGFP was generated by introducing an EGFP-poly(A) cassette excised from pEGFP-1 (Clontech) into pBluescript SK+ (Stratagene). −10.6kGFP was generated by introducing the FoxE3 promoter region (−10632 to +118) isolated from a X. tropicalis genomic DNA library (a gift from Dr. Richard Harland) into pBSSK+EGFP. This promoter sequence is followed by an open reading frame whose nucleotide sequence has 93% identity with that of Xenopus laevis Lens1/FoxE3 (Kenyon et al., 1999) in X. tropicalis genome assembly 4.1 (scaffold 1: 2902082-2903173, http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). We assigned the transcription start site (+1) to the 5' end of the putative 5' untranslated region (UTR) (scaffold 1: 2901906-2902081) that was predicted according to its homology to the 5' UTR of X. laevis FoxE3 (94% identity) to number the flanking sequence. βGFP was generated by introducing chicken β-actin basal promoter (−55 to +53) excised from pβLuc (Ogino and Yasuda, 1998) into pBSSK+EGFP. A series of base-substitution mutant constructs (mt1–mt9) were generated from Xt462-βGFP using the Quikchange Site-Directed Mutagenesis Kit (Stratagene) with the following primers and their complements (mutated sequences are underlined):
mt1, 5'-CAAGGAGAGTGAAATGAGAGGTACCATGTTTTCATCATCCG-3';
mt2, 5'-GAGTGAAATGAGATAATCCATCGATTCATCATCCGTAGGCC-3';
mt3, 5'-CTCTTTTCACAAGCCATGGTACGTACTTTATTAGGCTGAGC-3';
mt4, 5'-CATGGGCCGTACTTTATTAGGTACCGCAGTTCTGGGCCTGTAAG-3';
mt5, 5'-ATGCAGAATGGCAGAAACCGGTAGGCCCAGTACATTTTCC-3';
mt6, 5'-CAACATCAGATTTTCCTACATCTAGAGTGCAGAAATCCCACAC-3';
mt7, 5'-CTACAGATAGAGTGCAGAAATCTAGAACATGTCCAAATCTGTTAACATC-3';
mt8, 5'-GCAGAAATCCCACACATGTGGCCATCTGTTAACATCTGACATG-3';
mt9, 5'-CCACACATGTCCAAATCGATTAACATCTGACATGAAGTC-3'.
Otx-Su(H)-βGFP, mtOtx-Su(H)-βGFP, and Otx-mtSu(H)-βGFP were generated by introducing the following double-stranded oligonucleotides into βGFP (Otx- and Su(H)-binding sequences are underlined):
Otx-Su(H), 5'-ctagaGGGATTAGAGTTCCCACACGGGATTAAATTTCCCACGGAGGATTAGGGTTCCCACAAGGGATTAGATTTCCCACg-3';
mtOtx-Su(H), 5'-ctagaGGCGGTAGAGTTCCCACACGGCGGTAAATTTCCCACGGAGCGGTAGGGTTCCCACAAGGCGGTAGATTTCCCACg-3';
Otx-mtSu(H), 5'-ctagaGGGATTAGAGTTCGGTCACGGGATTAAATTTCGGTCGGAGGATTAGGGTTCGGTCAAGGGATTAGATTTCGGTCg-3'.
Details of other reporter constructs are described in the Results. pGEM-XFoxE3 was generated by cloning of the coding sequence of X. laevis FoxE3 into pGEM-T Easy vector (Promega). pCS2+GR-Otx2-En was generated by introducing the coding sequence of a GR ligand-binding domain isolated from pCS2+GR-Su(H)VP16 (Rones et al., 2000) into the 5' end of the Otx2 coding sequence of pCS2+Otx2-En (Gammill and Sive, 2001).
Xenopus transgenesis
Transgenic embryos were generated by a sperm nuclear transplantation method (Kroll and Amaya, 1996), and their GFP expression was detected by in situ hybridization (Sive et al., 2000) for maximum sensitivity. The fraction of embryos that developed normally until scoring stages (stages 22–24 (Nieuwkoop and Faber, 1967)) varied between 10–20% of total injected embryos depending on egg quality. However, the frequency of reporter gene expression within the group of normal embryos was quite constant. In the case of three independent assays using Xt462-βGFP, the average fraction of embryos with PLE-specific GFP expression in a normal group was 21 ±1.7 %. This construct was injected in parallel with a series of its mutant constructs (mt1–mt9) as a control to monitor transgenesis efficiency. For co-transgenesis, the 462 bp enhancer fragment of Xenopus FoxE3, amplified by PCR, was mixed with a β-actin promoter-GFP cassette excised from βGFP in a molar ratio of 4:1 and directly used for transgenesis.
In situ hybridization and RNA injections
In situ hybridization analyses of Delta1, Delta2, Otx2, γ1-crystallin, and Rx were performed as described (Blitz and Cho, 1995; Chitnis et al., 1995; Jen et al., 1997; Mathers et al., 1997; Offield et al., 2000). The antisense probes for GFP, FoxE3, Serrate1, and Notch2 were generated using pBSSK+EGFP, pGEM-XFoxE3, pBS-XSerrate1 (a gift from Dr. C. Kintner), and an EST clone (NCBI accession number: BX855333), respectively.
Capped mRNAs for microinjections were transcribed from pCS2+GR-Su(H)VP16, pCS2+GR-Su(H)DBM (Rones et al., 2000), pCS2+NICD (Chitnis et al., 1995), pCS2+XOtx2-GR (Gammill and Sive, 1997), pCS2+GR-Otx2-En, pCS2+XOtx2 (Blitz and Cho, 1995), pCS2+XDelta1, pCS2+XDelta1Stu (Chitnis et al., 1995), pCS2+XDelta2, pCS2+XDelta2Tr (Jen et al., 1997), pCS2+EGFP, and pCS2+nlacZ. For lacZ staining, magenta-gal was used as the substrate (Rones et al., 2000).
Results
Identification of the FoxE3 cis-regulatory element by both classic promoter deletion assays and in silico analysis
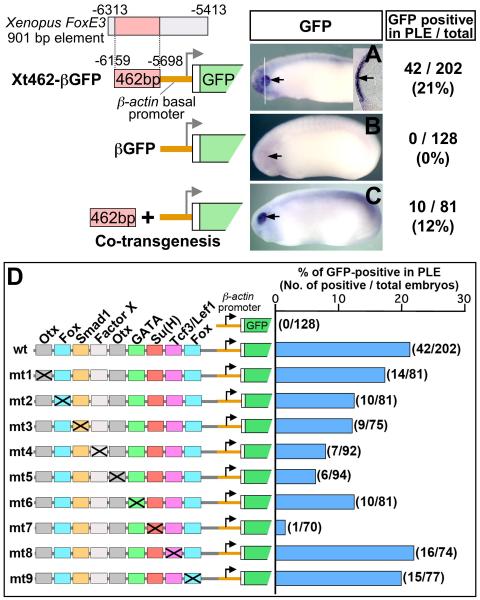
PLE-specific expression of FoxE3 in early tailbud embryos of X. tropicalis (Fig. 1A) was indistinguishable from the previously reported FoxE3 expression in X. laevis (Kenyon et al., 1999). This expression was closely recapitulated by GFP expression in transgenic X. laevis embryos generated with a reporter construct carrying a 10.6 kbp upstream sequence of X. tropicalis FoxE3 (Fig. 1B, −10.6kGFP, see Fig. S1 in the Supplementary Material for details). Sequences responsible for PLE-specific expression were narrowed down by a series of promoter deletion assays (Fig. 1C–G), which identified a 901 bp element (−6313 to −5413) as the one that essentially recapitulates the activity of the −10.6 kbp promoter when linked to the basal promoter region (−640 to +118) (Fig. 1G).
Fig. 1.
In vivo deletion analysis identifies a 901 bp enhancer that directs PLE-specific expression of FoxE3. (A) FoxE3 expression in X. tropicalis embryos (stage 23) detected by in situ hybridization. (B–G) GFP expression detected by in situ hybridization in representative transgenic embryos (stages 22–24) generated with reporter constructs shown to the left. White and black arrows in (A–G) indicate the PLE. An arrowhead in (B) indicates ectopic GFP expression in the presumptive oral ectoderm. Numbers of embryos with GFP expression in the PLE and the total number of normally (or near normally) developing embryos injected with each construct are indicated to the right with the percentage of GFP-positive cases. The 901 bp element necessary for PLE-specific expression is boxed with a red broken line. *The expression in (D) was positive in the PLE but very spotty and broad as shown in the left panel.
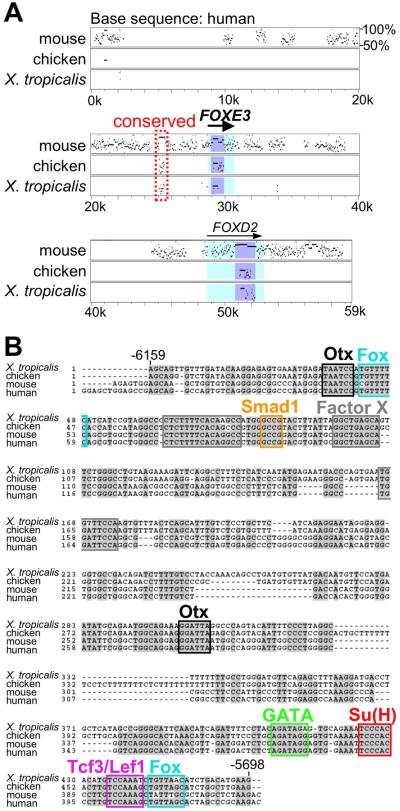
We also approached the same question from a bioinformatic perspective. To examine a possible relationship between conserved non-coding elements distributed around vertebrate FoxE3 loci and the functionally identified 901 bp enhancer, we aligned a nearly 60 kb sequence of the human FOXE3 locus with orthologous mouse, chicken, and X. tropicalis sequences using a genome alignment program, MultiPipmaker (http://pipmaker.bx.psu.edu/pipmaker/) (Fig. 2A). In this alignment, the percent identity plot (pip) shows both the position in the human sequence and the degree of similarity for each aligning segment between the human and other sequences. The pip in the human-mouse alignment (Fig. 2A, top row) indicates extensive sequence conservation between these two species. However, the pip in the human-chicken and the human-Xenopus alignments (Fig. 2A, second and third rows) indicates that only one region is conserved in all four species besides the coding region (Fig. 2A, red box). This conserved region corresponds to the 462 bp sequence between positions −6159 and −5698 in X. tropicalis FoxE3 locus, and is included in the 901 bp element (−6313 to −5413) described above.
Fig. 2.
In silico analysis of FoxE3 cis-regulatory element. (A) Genomic sequence of the human FOXE3 locus (−29.0 kb to +30.4 kb) is aligned with its orthologous mouse, chicken, and X. tropicalis sequences using MultiPipMaker. The aligned sequences were downloaded from the VISTA Browser (http://genome.lbl.gov/vista/index.shtml) or from the Xenopus tropicalis v4.1 genome site (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). A non-coding region conserved from human to Xenopus is boxed with a red broken line. Black arrows indicate exons of human FOXE3 and its neighboring gene, FOXD2, with their orientations. Coding and untranslated sequences are shaded with light blue and light cyan, respectively. The scale at the bottom of the alignment indicates relative positions in the human FOXE3 locus. (B) The conserved non-coding element of Xenopus FoxE3 (−6159 to −5698) identified by MultiPipMaker is aligned with its orthologous chicken, mouse and human sequences using ClustalW (http://www.ebi.ac.uk/clustalw/). Sequences conserved in at least three species are shaded with gray. Putative transcription factor binding motifs mapped in conserved sequences are boxed with different colors. Three gray boxes indicate conserved sequences that do not match to any known binding motifs.
Details of sequence conservation in the 462 bp element were further analyzed by phylogenetic footprinting (Fig. 2B). The evolutionary distance from Xenopus to human resolves the 462 bp element into discontinuous stretches of conserved sequences of 6–11 bp in length, each of which may predict transcription factor binding motifs. Eight of these stretches are identical or similar to known transcription factor-binding motifs and include target sequences of three signaling pathways: a Smad1-binding motif for BMP signaling (Kusanagi et al., 2000); a Su(H) (also known as CBF1/RBP-Jκ/Lag-1)-binding motif for Notch signaling (Tun et al., 1994); and a Tcf3/Lef1-binding motif for canonical Wnt signaling (Eastman and Grosschedl, 1999). Other predicted motifs are two Otx binding motifs (Gan et al., 1995), two Fox motifs (Kaufmann et al., 1995), and a GATA motif (Ko and Engel, 1993).
Identification of transcription factor-binding motifs essential for PLE-specific expression
The 901 bp enhancer identified by the deletion analysis contains not only the conserved 462 bp but also surrounding nonconserved sequences. Transgenic embryos generated with a construct where the conserved 462 bp sequence alone was linked to a heterologous basal promoter (chicken β-actin basal promoter) exhibited GFP expression in the PLE similar to the embryos carrying the 901 bp enhancer construct (Xt462-βGFP in Fig. 3A, compare with Fig. 1G), whereas the construct with the β-actin promoter alone (βGFP) did not drive any detectable expression in any embryos (Fig. 3B). Interestingly, PLE-specific expression was also detected when the 462 bp element amplified by PCR was co-injected with the βGFP cassette (Fig. 3C), an approach taken because this transgenic method is known to produce a concatemer of transgenic inserts. This novel, cloning-less “co-transgenesis” strategy is a powerful tool for the quick survey of enhancer activities of conserved non-coding elements that are widely distributed in vertebrate genomes (see Discussion).
Fig. 3.
Mapping of regulatory motifs essential for PLE-specific activity of the FoxE3 enhancer. (A–C) Representative transgenic embryos (stages 22–24) generated with GFP reporter constructs shown on the left. Black arrows indicate the PLE. Numbers of embryos with GFP expression in the PLE and the total number of normally (or near normally) developing embryos injected with each construct are indicated on the right side with percentages of the GFP-positive cases. A white line in (A) indicates the plane of a transverse section shown as an inset. A black arrow in the inset indicates GFP expression in the PLE overlying the optic vesicle. The embryo shown in (C) was generated by co transgenesis, i.e. co-injection of the 462 bp enhancer of Xenopus FoxE3 amplified by PCR along with the βGFP cassette. (D) Identification of transcription factor-binding motifs essential for PLE-specific expression by mutation analysis. wt is the construct used in Fig. 3A (Xt462-βGFP). mt1-mt9 were generated from wt/Xt462-βGFP by introducing a base-substitution mutation (marked with a cross) into each of the conserved transcription factor-binding motifs. The right graph shows percentages of embryos with GFP expression in the PLE in total developed embryos injected with the constructs shown in the left side. Actual numbers of GFP-positive cases and that of total embryos are indicated in parenthesis.
Transgenic embryos generated with a construct where the 462 bp Xenopus element was replaced with the orthologous 423 bp element of mouse Foxe3 (−3529 to −3107) used for the phylogenetic footprinting (Fig. 2B) exhibited GFP expression that was indistinguishable from that driven by the Xenopus element (compare Fig. S2A and B in the Supplementary Material), suggesting that the sum of the discontinuous stretches of short conserved sequences identified by the phylogenetic footprinting is sufficient to account for the expression. To evaluate the role of each short conserved sequence, we introduced base-substitution mutations individually into all of the eight putative transcription factor-binding motifs mapped there and into one of the conserved motifs with no similarity to known transcription factor-binding motifs (indicated as Factor X motif in Fig. 2B). The mutant constructs were generated from Xt462-βGFP, which drove PLE-specific expression in 21% of the generated embryos in transgenic assays as described (Fig. 3A and “wt” in Fig. 3D). None of the mutations led to additional ectopic expression, but the percentage of embryos with PLE-specific expression was decreased to different extents (Fig. 3D). The most striking result was obtained with the construct carrying a mutation in the Su(H) motif (Fig. 3D, mt7), which completely abolished the expression in all cases except one (n = 70). Even in this one positive case, the expression was very faint (not shown). The mutation of the 3'-most Otx motif and the mutation of the unknown Factor X motif decreased the positive cases to 6% and 8%, respectively (Fig. 3D, mt5 and mt4). The mutation of the 5'-most Fox motif, Smad1 motif, or GATA motif all somewhat decreased the positive cases, to about 12% (Fig. 3D; mt2, mt3, and mt6). The mutation of the 5' Otx motif, Tcf3/Lef1 motif, and 3' Fox motif did not significantly reduce the percentage of positive cases (Fig. 3D; mt1, mt8, and mt9). Chi square test (http://www.graphpad.com/quickcalcs/chisquared1.cfm) shows that the differences of the percentage of positive cases between the wild type and the Su(H), 3'-most Otx, or unknown Factor X mutant constructs are statistically significant (P < 0.0001, P = 0.0006, and P = 0.0018, respectively), whereas the differences observed in other cases are not (P > 0.05). These results show that the Su(H), 3'-most Otx, and unknown Factor X motifs are essential for the enhancer activity, and other motifs might be for boosting its level and/or involved in the regulation at different developmental stages.
Regulation of FoxE3 expression and lens placode formation by Notch signaling
Gel retardation assays showed direct binding of Xenopus Su(H) protein to the putative Su(H) motif identified in the FoxE3 enhancer in vitro, and chromatin immunoprecipitation experiments confirmed in vivo binding of the Su(H) protein to the enhancer (Fig. S3A and S4A in the Supplementary Material). Su(H) is ubiquitously expressed and activates transcription only when it forms a nuclear complex with the intracellular domain of Notch receptor (NICD) that is translocated from the cytoplasm upon activation by a ligand (Fig. 7A; (Lai, 2004)). We found that a Xenopus homologue of mammalian Notch2, which was newly identified in this study, is expressed during the course of lens formation in the pre placodal ectoderm, PLE, and developing lens vesicle (Fig. 4A–B', and Fig. S5 in the Supplementary Material).
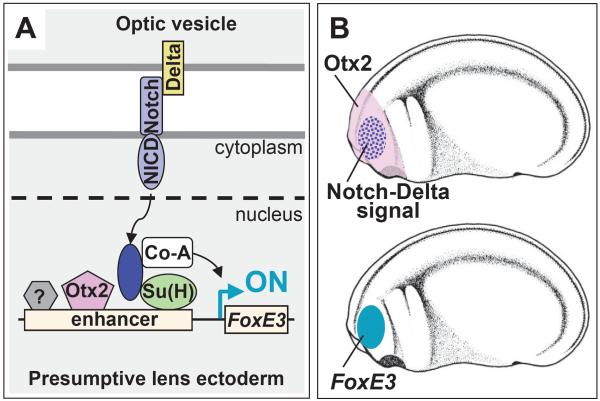
Fig. 7.
A model of FoxE3 activation. (A) Integration of Otx2 and Notch inputs on FoxE3 enhancer. Otx2 and Su(H) are proposed to bind to FoxE3 enhancer prior to receiving Notch signaling, but remain in a quiescent status. In response to stimuli of Delta ligands, Notch intracellular domain (NICD) is cleaved from its extracellular domain and translocated into the nucleus. While this model shows a direct interaction between membrane bound Notch and Delta, we do not know the exact mechanism by which this signaling may occur in this system (see Discussion). In the nucleus, NICD forms a complex with Su(H) and activates transcription by recruiting a co-activator (Co-A). Otx2 synergistically stimulates this transcription. An unidentified factor(s) (indicated with a question mark) may contribute to boosting and refining the PLE-specific expression. (B) Otx2 is broadly expressed in the head ectoderm that includes PLE, while localized Notch signaling is provided to the lens field from the underlying optic vesicle (upper panel). FoxE3 is expressed in the PLE, which is the region where there is both Otx2 expression and Notch signaling (lower panel).
Fig. 4.
Comparative expression analysis of Notch signaling components and FoxE3 in X. laevis embryos, showing that Notch2 and FoxE3 are expressed in PLE while Delta1 and Delta2 are expressed in presumptive retina. Expression of Notch2 (A–B'), Delta1 (C–E'), Delta2 (F–H'), and FoxE3 (I–K') was detected by in situ hybridization from neural plate stages to early tailbud stages. Regions circled with black broken lines in (C) and (F) are the approximate presumptive retina fields, where neither Delta1 nor Delta2 is expressed. Arrows in (B, B'), (D–E'), (G–H'), and (J–K') indicate expression of Notch2, Delta1, Delta2, and FoxE3, respectively. A black triangle in (C) indicates Delta1 expression in the anterior neural ridge, and white triangles in (A) and (I) indicate Notch2 and FoxE3 expression in the pre-placodal ectoderm, respectively. White lines in (E), (H) and (K) indicates the planes of transverse eye sections shown in (E'), (H') and (K').
Regarding Notch ligand genes, Delta1 (Chitnis et al., 1995), Delta2 (Jen et al., 1997), and Serrate1 (Kiyota et al., 2001), have been identified in Xenopus. To assess involvement of Notch ligands in FoxE3 regulation, their expression was compared with FoxE3 expression during the course of lens-field formation. In neural plate-stage embryos, neither Delta1 nor Delta2 is expressed in the presumptive retina fields that will give rise to optic vesicles (Fig. 4C and F; (Chitnis et al., 1995)). Interestingly, Delta1 expression in the anterior neural ridge is adjacent to FoxE3 expression in the pre-placodal ectoderm at this stage ((Bourguignon et al., 1998; Kenyon et al., 1999), compare Fig. 4C and I) where it may act as an early signal involved in FoxE3 expression. Accompanying neural tube formation, both Delta genes exhibit strong up-regulation in the developing optic vesicle (Fig. 4D–E' and G–H'), which is followed by FoxE3 expression in the PLE (Fig. 4J–K'; (Kenyon et al., 1999)). The cells expressing Delta genes are located in the most outer region of the optic vesicle and make contact with the overlying PLE cells expressing FoxE3 (Fig. 4E', H' and K'). Serrate1 is not expressed in these tissues at the neurula or early tailbud stages, but later is expressed in the developing lens placode as reported (not shown; (Kiyota et al., 2001)).
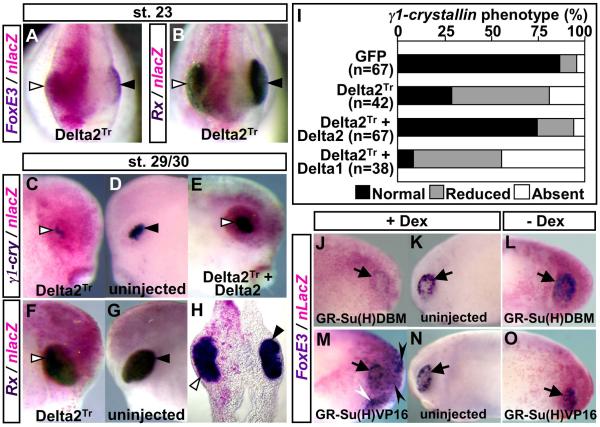
To examine possible roles of the Delta genes in FoxE3 regulation, we blocked Delta1 and Delta2 activities using their dominant negative forms, Delta1Stu and Delta2Tr, respectively (Chitnis et al., 1995; Jen et al., 1997). mRNA encoding either Delta1Stu, Delta2Tr, or GFP was injected along with a lineage tracer, nlacZ mRNA (nuclear lacZ, 50 pg), into one dorsal blastomere of eight-cell stage X. laevis embryos. The injected embryos were fixed at the early tailbud stages (stages 22–24), and stained for lacZ to trace distribution of the injected mRNAs. Only the embryos that showed lacZ staining in the optic vesicle were subjected to in situ hybridization with FoxE3 probe. Control injections using GFP mRNA did not have any significant effects on FoxE3 expression (n = 55). Embryos injected with Delta1Stu mRNA exhibited loss or severe reduction of FoxE3 expression on the injected side (100%, n = 22), but this phenotype appeared to be associated with head abnormalities caused by the expression of this construct (not shown). While the effect seen here is consistent with a role for Delta1 in lens formation, we did not study Delta1 further because of the complexity in interpreting the cause of lens defects in light of the head abnormalities seen in these experiments. In contrast, injection of Delta2Tr mRNA led to a very specific phenotype; FoxE3 expression was lost or severely reduced (75%, n = 32, Fig. 5A) in the PLE overlying the optic vesicle stained with lacZ, but the optic vesicle itself, which was marked by expression of a retina-specific homeobox gene Rx (Mathers et al., 1997), did not show any detectable abnormalities (100%, n = 37, Fig. 5B).
Fig. 5.
Effects of manipulation of Notch signaling on FoxE3 expression and subsequent lens placode formation. (A, B) Frontal view of embryos injected with mRNA encoding Delta2Tr (500 pg), fixed at stage 23, and subjected to lacZ staining (magenta) and in situ hybridization with FoxE3 or Rx probe (purple or deep purple staining). White and black triangles in (A–H) indicate in situ hybridization signals on injected and uninjected sides of embryos, respectively. (C–D, F–G) The injected and uninjected sides of embryos injected with Delta2Tr mRNA, fixed at stages 29/30, and hybridized with γ1-crystallin or Rx probe. (H) A transverse head section of the embryo shown in (F, G). (E) The injected side of an embryo injected with both Delta2Tr mRNA (500 pg) and wild-type Delta2 mRNA (500 pg), fixed at stage 29, and hybridized with γ1-crystallin probe. (I) Summary of Delta2Tr mRNA injection experiments. GFP mRNA (1000 pg) was injected as a control. (J, K) The injected and uninjected sides, respectively, of an embryo injected with GR-Su(H)DBM mRNA (1000 pg) and induced with Dex. Arrows in (J–O) indicate endogenous FoxE3 expression in the PLE. (L) The injected side of an embryo injected with GR-Su(H)DBM but not induced with Dex. (M, N) The injected and uninjected sides, respectively, of an embryo injected with GR-Su(H)VP16 mRNA (1000 pg) and induced with Dex. Black and white arrowheads in (M) indicate ectopic FoxE3 expression in the ectoderm overlying the anterior brain and that surrounding the cement gland, respectively. (O) The injected side of an embryo injected with GR-Su(H)VP16 but not induced with Dex.
In addition to FoxE3, we examined expression of a lens differentiation marker, γ1-crystallin (Offield et al., 2000), by in situ hybridization to investigate late phenotypes of embryos expressing Delta2Tr. At late tailbud stages (stages 29/30), the lens placode of the uninjected sides showed clear γ1-crystallin expression (Fig. 5D), but the γ1-crystallin-positive cells on the injected sides formed a tiny cell mass or were absent (71%, n = 42, Fig. 5C and I). Expression analysis of Rx showed that the optic vesicle of the injected sides still had no significant defects, at least through these stages (100%, n = 47, Fig. 5F–H). The down-regulation of γ1-crystallin by Delta2Tr was rescued by co-injection of mRNA encoding wild-type Delta2 (75%, n = 67, Fig. 5E and I) but not by Delta1 (n = 38, Fig. 5I), indicating the specific activity of Delta2 for lens induction. These results show that Delta2 activity in the optic vesicle is necessary for FoxE3 expression in the PLE and subsequent lens placode formation.
To examine how the responses to Notch signaling may directly impinge on FoxE3 expression in lens cells, we used a construct for an inducible dominant negative form of Su(H), GR-Su(H)DBM (Rones et al., 2000). This construct was generated by fusing the human glucocorticoid receptor ligand binding domain (GR) to a modified version of Xenopus Su(H), which contains a point mutation in its DNA-binding domain. This GR-Su(H)DBM protein, which inhibits Notch signaling in response to dexamethasone (Dex) by sequestering NICD from endogenous Su(H), allowed us to circumvent possible head defects that could be caused by constitutive inhibition of Notch signaling. mRNA encoding GR-Su(H)DBM was injected along with nlacZ mRNA into one dorsal blastomere of four-cell stage X. laevis embryos. The injected embryos were cultured in the absence of Dex until stages 15–16, and then maintained with Dex (10 μM) either present (induced) or absent (uninduced) until fixation at early tailbud stages (stages 22–24). This time period was chosen to yield functional GR-Su(H)DBM protein at the time when endogenous FoxE3 expression is up-regulated in the PLE following neural tube closure. The fixed embryos were subjected to lacZ staining to select embryos where expression was targeted to the anterior ectoderm including the PLE. As observed in embryos injected with Delta2Tr mRNA, FoxE3 expression was lost or severely reduced on injected sides of the Dex-treated embryos (55%, n = 33, Fig. 5J). Down-regulation of FoxE3 was not observed on either uninjected sides of any of these embryos (Fig. 5K) or injected sides of any sibling embryos untreated with Dex (n = 65, Fig. 5L), indicating that Dex itself has no effect on FoxE3 expression and the FoxE3 down-regulation in the Dex-treated embryos depended on the activation of GR-Su(H)DBM by hormone treatment.
Effects of ectopic activation of Notch signaling on FoxE3 expression were examined using an inducible active form of Su(H), GR-Su(H)VP16 (Xenopus Su(H) fused to GR and to VP16 activation domain), which mimics Notch pathway activation in response to Dex (Rones et al., 2000). Similar to GR-Su(H)DBM, GR-Su(H)VP16 was expressed by injecting its mRNA, and the resulting embryos were cultured with or without Dex until fixation at stages 22–24. Ninety-five percent of the injected embryos treated with Dex exhibited ectopic FoxE3 expression in the anterior region of the injected sides stained with lacZ (n = 55, Fig. 5M). In contrast to the extended lacZ staining, the ectopic FoxE3 expression was spotty and localized in a domain of the ectoderm overlying the anterior brain and that surrounding the cement gland. Ectopic FoxE3 expression was not observed on either the uninjected sides of any of these embryos (Fig. 5N) or the injected sides of any sibling embryo untreated with Dex (n = 69, Fig. 5O).
The activation and inhibition of Notch signaling using the active and dominant negative forms of Su(H), respectively, induced up-regulation and down-regulation of FoxE3. These results show the essential role of Notch signaling in PLE-specific FoxE3 expression, and that Notch signaling in the PLE is likely to be activated by Delta2 expressed in the adjacent optic vesicle. Interestingly, the ectopic FoxE3 expression induced by the active form of Su(H) was regionally restricted to part of the anterior ectoderm, which suggests pre-localization of a factor providing competence to respond to Notch signaling there. In addition, this restricted ectopic expression is consistent with a role for Notch signaling as a cue to turn on FoxE3 at the right place within this competent domain.
Otx2 confers competence to activate FoxE3 in response to Notch signaling
As a candidate factor responsible for the regional competence to respond to Notch signaling, we examined Otx2, for two reasons. First, we identified a putative Otx motif as among the most essential of the transcription factor-binding motifs in the FoxE3 enhancer (Fig. 3D, mt5). Second, during the time window chosen for the activation of GR-Su(H)VP16 (from the neural plate to early tailbud stages), Otx2 shows diffuse expression in the head ectoderm including not only the PLE but also the surrounding region where the ectopic FoxE3 expression was observed (Fig. 6A–C”; (Zygar et al., 1998), compare Fig. 5M and 6C).
Fig. 6.
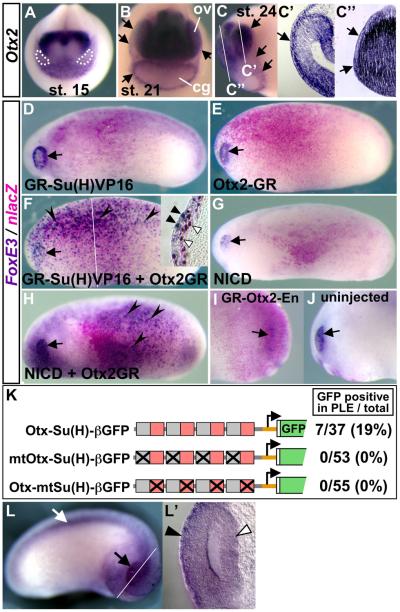
Otx2 confers competence to activate FoxE3 in response to Notch signaling. (A–C") Expression of Otx2 in the anterior ectoderm detected by in situ hybridization. At the neural plate stages, Otx2 is expressed in the anterior ectoderm including the presumptive lens fields, which are circled with white broken lines in (A). Arrows in (B) indicate broad expression in the ectoderm that overlies the optic vesicles (ov) and surrounds the cement gland primordium (cg). Arrows in (C) indicate the border of ectodermal Otx2 expression. White lines in (C) indicate the planes of transverse sections shown in (C') and (C”). Arrows in (C') and (C”) respectively indicate expression in the PLE overlying the optic vesicle and in the ectoderm overlying the forebrain. (D–H) Notch-Otx2 combination activates FoxE3 in the trunk ectoderm. Embryos injected with mRNAs indicated in each panel were induced with Dex, and then subjected to lacZ staining and in situ hybridization with FoxE3 probe. Ectopic FoxE3 expression was not detected in embryos injected with mRNA encoding GR-Su(H)VP16 (1000 pg) (D), Otx2-GR (250 pg) (E), or NICD (1000 pg) (G), whereas it was detected in embryos injected with both GR-Su(H)VP16 (750 pg) and Otx2-GR (250 pg) (F), or both NICD (750 pg) and Otx2-GR (250 pg) (H). Arrowheads in (F) and (H) indicate ectopic FoxE3 expression. Arrows in (D–H) indicate endogenous FoxE3 expression in the PLE. A white line in (F) indicates the plane of a transverse section shown as an inset. Black triangles in the inset indicate overlaps of FoxE3 expression and nuclear lacZ staining in the ectodermal cells, and white triangles indicate cells in the underlying mesoderm layer showing nuclear lacZ staining but no FoxE3 expression. (I, J) The injected and uninjected sides, respectively, of an embryo injected with mRNA encoding GR-Otx2-En (250 pg), induced with Dex from stage 18, and then subjected to lacZ staining and in situ hybridization with FoxE3 probe at stage 22. Arrows indicate the PLE. (K) Transgenic experiments using Otx-Su(H) reporter constructs. Numbers of embryos with GFP expression in the PLE and the total number of normally (or near normally) developing embryos injected with the constructs shown on the left are indicated on the right side with percentages of the GFP-positive cases. Gray and red boxes respectively indicate Otx- and Su(H)-binding motifs in the constructs, and crosses indicate base-substitution mutations introduced there. (L, L') A representative transgenic embryo generated with Otx-Su(H)-βGFP. Black and white arrows indicate GFP expression in the eye and spinal cord, respectively. A white line indicates the plane of a transverse eye section shown in (L'). Black and white triangles indicate GFP expression in the PLE and optic vesicle, respectively.
Gel retardation assays showed direct binding of Xenopus Otx2 protein to the putative Otx motif identified in the FoxE3 enhancer in vitro, and chromatin immunoprecipitation experiments confirmed in vivo binding of endogenous Otx2 protein to the enhancer (Fig. S3B and S4B in the Supplementary Material). To test whether Otx2 misexpression confers competence to respond to Notch signaling in vivo, we used an inducible form of Xenopus Otx2, GR-fused Otx2 (Otx2-GR), whose activity can be controlled by Dex treatment (Gammill and Sive, 1997). This construct allows us to circumvent severe gastrulation defects and spina bifida caused by misexpression of wild type Otx2 in Xenopus embryos (Blitz and Cho, 1995). mRNAs encoding GR-Su(H)VP16 and Otx2-GR were injected separately or together into a ventral blastomere of four-cell stage embryos to target expression in the trunk region instead of the head ectoderm expressing endogenous Otx2. All injected embryos were treated with Dex at stages 15–16, fixed at stages 22–24, and subjected to lacZ staining and in situ hybridization with the FoxE3 probe.
None of embryos misexpressing either GR-Su(H)VP16 or Otx2-GR exhibited ectopic FoxE3 expression in the trunk region where the lacZ staining indicated broad distribution of the injected mRNAs (n = 66 and n = 52, respectively, Fig. 6D and E). This indicates that neither Notch signaling nor Otx2 activity is sufficient for FoxE3 activation. However, when embryos were co-injected with GR-Su(H)VP16 and Otx2-GR mRNAs, 91% of them (n = 54) exhibited striking FoxE3 expression in the trunk region throughout the lacZ-stained region (Fig. 6F, black arrowheads). This trunk expression was mostly restricted to the ectoderm layer, and not in the underlying neural or mesodermal tissues (Fig. 6F, inset). Ectopic FoxE3 expression was also detected in embryos that were induced with Dex in the presence of a protein synthesis inhibitor, cycloheximide, verifying a direct effect of GR-Su(H)VP16 and Otx2-GR on the FoxE3 promoter (Fig. S6 in the Supplementary Material).
The trunk expression was not likely to result from an increase in the total amount of misexpressed transcription factors, since the total amount of mRNAs co-injected in this experiment (750 pg of GR-Su(H)VP16 and 250 pg for Otx2-GR) was kept the same as that for the misexpression of GR-Su(H)VP16 alone (1000 pg). The trunk expression was also observed when wild-type Xenopus Otx2 was misexpressed instead of Otx2-GR in combination with GR-Su(H)VP16 (95%, n = 22, not shown). This indicates that the ectopic expression was not associated with the GR ligand-binding domain fused to Otx2, though many of the Otx2-injected embryos exhibited a spina bifida phenotype that is characteristic of misexpression of wild-type Otx2.
The Otx2-dependent activation of FoxE3 was also observed when NICD was misexpressed instead of GR-Su(H)VP16 to activate Notch signaling: misexpression of NICD alone in the trunk ectoderm did not induce ectopic FoxE3 expression in any injected embryos (n = 33, Fig. 6G), but combination of NICD and Otx2-GR did (49%, n = 45, Fig. 6H, black arrowheads). This showed that, endogenous Su(H) activates FoxE3, as well as the artificial construct GR-Su(H)VP16, if Notch is activated.
In addition to these gain of function experiments, we designed loss of function experiments for Otx2 to verify that its activity is required for FoxE3 expression in the PLE. Since constitutive loss of Otx2 activity impairs anterior neural fate determination (Gammill and Sive, 2001), we chose to inject mRNA encoding an inducible dominant negative form of Otx2, GR-Otx2-En. We generated this construct by fusing a coding sequence of GR to a dominant negative form of Otx2, Otx2-En (Xenopus Otx2 fused to the minimal repressor domain of Engrailed), which has been shown previously to specifically block Otx2 function (Gammill and Sive, 2001). The injected embryos induced with Dex from stage 18 onward did not exhibit any detectable head abnormalities (Fig. 6I and J), suggesting that anterior neural defects were circumvented by the use of this inducible construct. Loss or significant reduction of FoxE3 expression in the PLE was detected on their injected sides by in situ hybridization (58%, n = 33, Fig. 6I), but not in either their uninjected sides (Fig. 6J) or injected sides of any sibling embryos untreated with Dex (n = 25, not shown).
These experiments demonstrate that Otx2 provides the competence to activate FoxE3 in response to Notch signaling. To test whether Otx2 and Notch inputs are sufficient to direct PLE- specific expression, we generated a reporter construct carrying four copies of a pair of consensus Otx- and Su(H)-binding motifs in front of the β-actin promoter-GFP cassette (Otx-Su(H)-βGFP). Transgenic embryos injected with this Otx-Su(H) reporter exhibited GFP expression not only in the PLE but also in the optic vesicle and spinal cord (Fig. 6K–L'), though the expression in the PLE did not appear so strong as that driven by FoxE3 enhancer. The eye-specific expression was not detected when transgenic embryos were generated with reporter constructs where either Otx or Su(H) motifs were mutated (mtOtx-Su(H)-βGFP and Otx-mtSu(H)-βGFP, respectively; Fig. 6K) or when a reporter construct that carried eight copies of Su(H) motifs and no copy of the Otx motif was used (not shown). These reporter assays suggest that both Otx2 and Notch inputs are necessary and sufficient to drive expression in PLE, but additional inputs are required for boosting expression level in the PLE and repressing expression in neural tissues to direct the more defined expression of the FoxE3 enhancer.
Discussion
A model of FoxE3 activation
We have presented gain of function and loss of function strategies demonstrating that Otx2 activity and Notch signaling directly converge on FoxE3 enhancer to direct PLE-specific expression (Fig. 7A). These are the first data directly showing involvement of Notch signaling in lens induction. Based on these results, we propose a stepwise model of FoxE3 activation (Fig. 7B): (1) Otx2 regionally provides the competence to respond to Notch signaling within head ectoderm including PLE; (2) Delta2 signaling from the optic vesicle locally activates Notch signaling in the overlying PLE to turn on FoxE3 expression within this broader competent region. The results of the transgenic assay using the Otx-Su(H) reporter suggest that other transcription factors may contribute to boosting and refining the PLE-specific expression in combination with Otx2 and Su(H). An unknown factor that binds to the Factor X motif (Fig. 2B and 3D), and FGF, BMP, and SIP1 signaling, whose inhibition causes reduction of FoxE3 expression in mouse embryos, may be involved in this process (Faber et al., 2001; Yoshimoto et al., 2005).
Because the dominant-negative form of Delta1 induced head defects in embryos, we could not examine possible roles for Delta1 in FoxE3 regulation and subsequent lens formation. However, neural tube formation is accompanied by a dynamic change in Delta1 expression from the anterior neural ridge to the optic vesicle (Fig. 4C–E'), which may be responsible for the shift of FoxE3 expression from the pre-placodal ectoderm to the PLE (Fig. 4I–K'). FoxE3 expression in the PLE of embryos expressing the dominant negative form of Delta2 was severely reduced but remained in some cases, which may be due to some contributions of Delta1 to FoxE3 regulation.
As in Xenopus, Otx2 is expressed in the PLE of mouse embryos, and the lens placode of Otx2 heterozygous mutant mice fail to form a normal lens vesicle on an Otx1 homozygous mutant background (Martinez-Morales et al., 2001). deltaC, a zebrafish Notch ligand gene that has the highest sequence similarity to Xenopus Delta2, is expressed in the developing optic vesicle as Xenopus Delta2 (Smithers et al., 2000), whereas a mammalian homologue of Delta2/deltaC has not been identified yet. Interestingly, Jagged1, a mammalian homologue of Xenopus Serrate1, is expressed in the optic vesicle of rat embryos (Bao and Cepko, 1997), and deleted in the mouse mutant Coloboma (Cm), whose lens fails to detach from the ectoderm as in the FoxE3 mutant mouse, dysgenetic lens (dyl) (Blixt et al., 2000; Brownell et al., 2000; Theiler and Varnum, 1981; Xue et al., 1999). Hence the role of Delta2/deltaC in the lens induction of lower vertebrates might be taken over by Jagged1 in mammals. Regarding Notch receptors, mammalian Notch2 and Notch3 are expressed in the developing lens (Lindsell et al., 1996), but their expression in earlier stages has not been characterized in detail. In mammalian embryos, cells in the optic vesicle and lens ectoderm are separated by a space but connected by cytoplasmic extensions (McAvoy, 1980), which may permit direct contact for Notch signaling. It is also possible that Notch ligands may have secreted forms that are involved in Notch signaling (Qi et al., 1999).
Otx2-Notch interactions in lens determination programs, and their analogy to selector-signaling system in Drosophila
The data presented here have significant implications for molecular mechanisms underlying the stepwise determination of the lens. Otx2 expression in head ectoderm may constitute a part of the lens-forming competence and/or bias that were suggested by embryological studies, and Notch signaling is likely one of the inducing signals provided from the optic vesicle to turn on the lens specification programs in this competent/biased ectoderm. Unlike FoxE3, expression of a lens differentiation marker, γ1-crystallin, was not induced in the trunk ectoderm by misexpression of Otx2-GR and GR-Su(H)VP16 (not shown), suggesting that the Otx2-Notch combination is not sufficient to activate whole lens differentiation programs. However, severe reduction or loss of γ1-crystallin-positive lens placode cells in embryos expressing the dominant negative Delta2 suggests a crucial role for Notch signaling in lens specification. The lens differentiation programs are presumably turned on when a set of all terminal regulators, such as FoxE3 and L-maf, is activated in the PLE by different but possibly overlapping mechanisms.
Genetic studies in mouse have shown that Pax6 lies upstream of Mab21l1, and Mab21l1 lies upstream of FoxE3 (Yamada et al., 2003). We found that the combination of Otx2 and Notch signaling induced ectopic FoxE3 expression in the trunk ectoderm without activating Pax6 (not shown). These findings imply that the Pax6-Mab21l1 pathway controls FoxE3 expression indirectly through regulation of Otx2 and/or Notch signaling. Notch2 may be a downstream target of Pax6, since the broad expression of Notch2 in the head ectoderm is, as development proceeds, gradually localized to the lens and olfactory fields expressing Pax6 (Fig. 4A–B' and Fig. S5 in the Supplementary Material).
Interestingly, the combinatorial mechanism of FoxE3 regulation is quite similar to the selector-signaling system in Drosophila, in which selective gene activation by signals for cell fate specification is achieved by obligate integration of both inputs of field-specific transcription factors (selectors) and signal-activated transcription factors at the level of their target cis-regulatory elements (Guss et al., 2001). Although this system has not been previously examined in vertebrate development, our study suggests that the same mechanism underlies the “competence-dependent induction” of the lens. Since classic embryological studies demonstrate a competence-dependent induction in many vertebrate organ systems (Gurdon, 1992), a selector-signaling system may be broadly used in vertebrates for specifying a variety of organ and tissue identities by reiteratively using a limited number of signaling pathways.
Xenomics (Xenopus genomics) for analysis of genomic regulatory networks for development
The results of our classic-style deletion analysis are in close agreement with those of the comparative analysis of human to Xenopus genomes, which demonstrate the advantage of the use of the Xenopus genome for in silico prediction of conserved regulatory elements in vertebrates. The conserved enhancer of mouse Foxe3 identified in our study (−3529 to −3107) is included in the lens element that was independently identified by Kondoh's group by deletion analysis in transgenic mice (−4.40k to −2.63k; (Yoshimoto et al., 2005)), showing that Xenopus and mouse assays give consistent results.
An important challenge in the post-genomic era is clearly to untangle the complex wiring of gene regulatory networks (GRNs) controlling development, growth and differentiation. As shown in the pioneering study of the GRN for sea urchin endomesoderm specification (Davidson et al., 2002), this type of study requires a high-throughput assay system for comprehensive analysis of cis-regulatory elements. The mammalian-Xenopus genome comparison and an approach developed in the course of this study--co-transgenesis--which quickly tests enhancer activities by co-injection of PCR products along with the basal promoter-GFP cassette, will allow Xenopus to serve as a vertebrate model system that fulfills this requirement.
Supplementary Material
Acknowledgements
We acknowledge Ms Renee Aloise and Dr. Lyle Zimmermann for their early work attempting to clone the Xenopus FoxE3 promoter. We wish to thank Drs. Milan Jamrich, Mark Mercola, Chris Kintner, Ira L. Blitz, Ken W. Y. Cho, Hazel Sive, and David Turner for kindly providing us their plasmids. We also especially thank Mr. William B. McConnell, and Ms. Hong Jin for their expert technical assistance in this study, and other members of the Grainger lab for ongoing helpful discussions. This work was supported in part by a postdoctoral fellowship for research abroad from JSPS (Japan Society for the Promotion of Science) to H.O and by grants RR13221, EY06675, EY10283 and EY17400 to R.M.G from NIH.
References
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–34. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Cho KW. Anterior neurectoderm is progressively induced during gastrulation: the role of the Xenopus homeobox gene orthodenticle. Development. 1995;121:993–1004. doi: 10.1242/dev.121.4.993. [DOI] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–54. [PMC free article] [PubMed] [Google Scholar]
- Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125:4889–900. doi: 10.1242/dev.125.24.4889. [DOI] [PubMed] [Google Scholar]
- Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–6. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–78. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol. 1999;11:233–40. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–38. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Sive H. Identification of otx2 target genes and restrictions in ectodermal competence during Xenopus cement gland formation. Development. 1997;124:471–81. doi: 10.1242/dev.124.2.471. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Sive H. otx2 expression in the ectoderm activates anterior neural determination and is required for Xenopus cement gland formation. Dev Biol. 2001;240:223–36. doi: 10.1006/dbio.2001.0470. [DOI] [PubMed] [Google Scholar]
- Gan L, Mao CA, Wikramanayake A, Angerer LM, Angerer RC, Klein WH. An orthodenticle-related protein from Strongylocentrotus purpuratus. Dev Biol. 1995;167:517–28. doi: 10.1006/dbio.1995.1046. [DOI] [PubMed] [Google Scholar]
- Grainger RM. Embryonic lens induction: shedding light on vertebrate tissue determination. Trends Genet. 1992;8:349–55. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The generation of diversity and pattern in animal development. Cell. 1992;68:185–99. doi: 10.1016/0092-8674(92)90465-o. [DOI] [PubMed] [Google Scholar]
- Guss KA, Nelson CE, Hudson A, Kraus ME, Carroll SB. Control of a genetic regulatory network by a selector gene. Science. 2001;292:1164–7. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Yasuda K. Distinct roles of maf genes during Xenopus lens development. Mech Dev. 2001;101:155–66. doi: 10.1016/s0925-4773(00)00585-2. [DOI] [PubMed] [Google Scholar]
- Jen WC, Wettstein D, Turner D, Chitnis A, Kintner C. The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development. 1997;124:1169–78. doi: 10.1242/dev.124.6.1169. [DOI] [PubMed] [Google Scholar]
- Kaufmann E, Muller D, Knochel W. DNA recognition site analysis of Xenopus winged helix proteins. J Mol Biol. 1995;248:239–54. doi: 10.1016/s0022-2836(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Moody SA, Jamrich M. A novel fork head gene mediates early steps during Xenopus lens formation. Development. 1999;126:5107–16. doi: 10.1242/dev.126.22.5107. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Jono H, Kuriyama S, Hasegawa K, Miyatani S, Kinoshita T. X-Serrate-1 is involved in primary neurogenesis in Xenopus laevis in a complementary manner with X-Delta-1. Dev Genes Evol. 2001;211:367–76. doi: 10.1007/s004270100165. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–22. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–83. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Kusanagi K, Inoue H, Ishidou Y, Mishima HK, Kawabata M, Miyazono K. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol Biol Cell. 2000;11:555–65. doi: 10.1091/mbc.11.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lang RA. Pathways regulating lens induction in the mouse. Int J Dev Biol. 2004;48:783–91. doi: 10.1387/ijdb.041903rl. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales JR, Signore M, Acampora D, Simeone A, Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development. 2001;128:2019–30. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–7. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- McAvoy JW. Cytoplasmic processes interconnect lens placode and optic vesicle during eye morphogenesis. Exp Eye Res. 1980;31:527–34. doi: 10.1016/s0014-4835(80)80011-x. [DOI] [PubMed] [Google Scholar]
- Muller F, Blader P, Strahle U. Search for enhancers: teleost models in comparative genomic and transgenic analysis of cis regulatory elements. Bioessays. 2002;24:564–72. doi: 10.1002/bies.10096. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis. North-Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- Offield MF, Hirsch N, Grainger RM. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–97. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–8. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Sequential activation of transcription factors in lens induction. Dev Growth Differ. 2000;42:437–48. doi: 10.1046/j.1440-169x.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- Pommereit D, Pieler T, Hollemann T. Xpitx3: a member of the Rieg/Pitx gene family expressed during pituitary and lens formation in Xenopus laevis. Mech Dev. 2001;102:255–7. doi: 10.1016/s0925-4773(01)00305-7. [DOI] [PubMed] [Google Scholar]
- Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–4. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–76. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- Sive H, Grainger R, Harland R. Early Development of Xenopus laevis - A LABORATORY MANUAL. Cold Spring Harbor Laboratory Press; New York: 2000. [Google Scholar]
- Smith JC. Xenopus genetics and genomics. Mech Dev. 2005;122:259–62. doi: 10.1016/j.mod.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Smithers L, Haddon C, Jiang YJ, Lewis J. Sequence and embryonic expression of deltaC in the zebrafish. Mech Dev. 2000;90:119–23. doi: 10.1016/s0925-4773(99)00231-2. [DOI] [PubMed] [Google Scholar]
- Theiler K, Varnum DS. Development of coloboma (Cm/+), a mutation with anterior lens adhesion. Anat Embryol (Berl) 1981;162:121–6. doi: 10.1007/BF00318098. [DOI] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-Jκ. Nucleic Acids Res. 1994;22:965–71. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–87. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–30. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- Yamada R, Mizutani-Koseki Y, Hasegawa T, Osumi N, Koseki H, Takahashi N. Cell-autonomous involvement of Mab21l1 is essential for lens placode development. Development. 2003;130:1759–70. doi: 10.1242/dev.00399. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A, Saigou Y, Higashi Y, Kondoh H. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132:4437–48. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]
- Zhou X, Hollemann T, Pieler T, Gruss P. Cloning and expression of xSix3, the Xenopus homologue of murine Six3. Mech Dev. 2000;91:327–30. doi: 10.1016/s0925-4773(99)00270-1. [DOI] [PubMed] [Google Scholar]
- Zygar CA, Cook TL, Grainger RM., Jr. Gene activation during early stages of lens induction in Xenopus. Development. 1998;125:3509–19. doi: 10.1242/dev.125.17.3509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.