Abstract
Canine transmissible venereal tumor (CTVT) is the oldest known somatic cell lineage. It is a transmissible cancer that propagates naturally in dogs. We sequenced the genomes of two CTVT tumors and found that CTVT has acquired 1.9 million somatic substitution mutations and bears evidence of exposure to ultraviolet light. CTVT is remarkably stable and lacks subclonal heterogeneity despite thousands of rearrangements, copy number changes and retrotransposon insertions. More than 10,000 genes carry non-synonymous variants and 646 genes have been lost. CTVT first arose in a dog with low genomic heterozygosity that may have lived approximately 11,000 years ago. The cancer spawned by this individual dispersed across continents approximately 500 years ago. Our results provide a genetic identikit of an ancient dog and demonstrate the robustness of mammalian somatic cells to survive for millennia despite a massive mutation burden.
Canine transmissible venereal tumor (CTVT) is a naturally occurring transmissible cancer. It is a clonal cell lineage that spreads within the domestic dog population by the allogeneic transfer of living cancer cells, usually during coitus. The disease manifests itself with the appearance of tumors most often associated with the external genitalia of male and female dogs. The first known report of CTVT was made in 1810 when it was described by a London veterinary practitioner as “an ulcerous state, accompanied with a fungous excrescence” which arises in “organs concerned in generation” (1). It has subsequently been reported in dog populations worldwide (2, 3) and is, to our knowledge, the oldest and most widely disseminated cancer in the natural world.
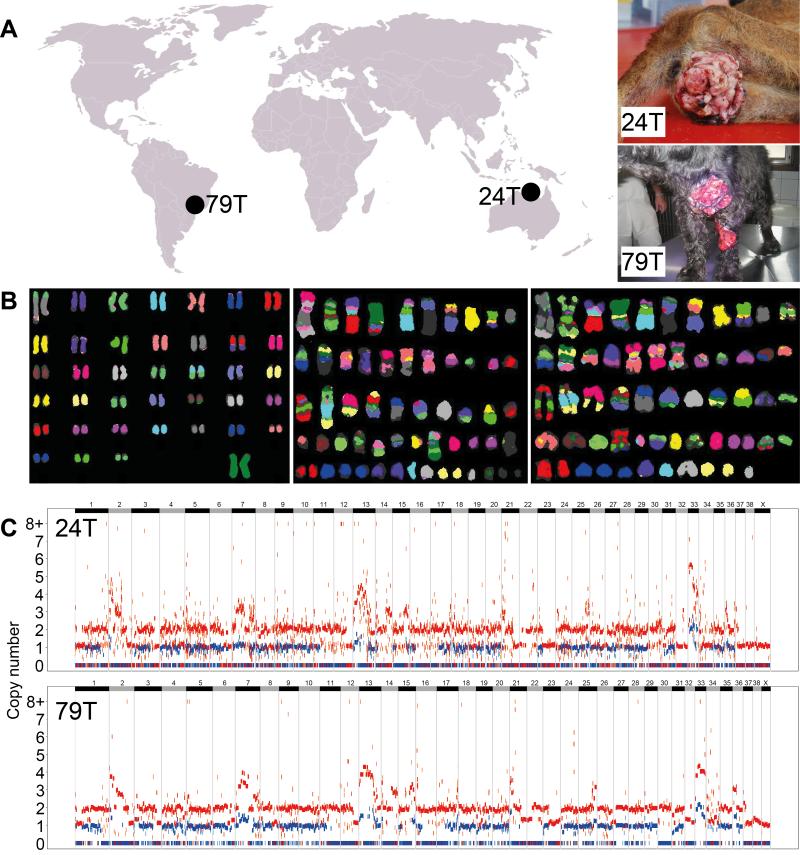
We sequenced the genomes of two CTVT tumors, collected in Maningrida, Australia (24T) and Franca, Brazil (79T), as well as the genomes of their respective hosts 24H, an Aboriginal Camp Dog, and 79H, an American Cocker Spaniel (Fig. 1A). We also prepared metaphases for cytogenetic analysis from two CTVTs collected in Cape Verde and Italy. Metaphase fluorescence in situ hybridization (FISH) using red fox chromosomes as probes revealed massive karyotypic rearrangement in the CTVT genome which was highly consistent between the two tumors analyzed (Fig. 1B). Despite the aneuploidy in CTVT detected using cytogenetics, copy number analysis revealed that the genome is largely diploid (including a large proportion of the genome that is diploid with loss of heterozygosity (LOH)) and that there has been minimal change in copy number status in either the 24T or 79T lineages since their divergence (Fig. 1C, Tables S1 and S2). In contrast to human tumors, most of which contain several detectable subclones (4), presumably due to positive selection for newly acquired mutations conferring selective advantage (5), we found no evidence for subclonality in CTVT metaphases or copy number plots. This suggests that CTVT is not undergoing positive selection at high frequency, possibly indicating that it is well adapted to its niche.
Figure 1. CTVT tumors, karyotypes and copy number.
(A) Samples sequenced in this study. Both tumor (24T, 79T) and host (24H, 79H) DNA was sequenced from the two individuals shown.
(B) Multiplex FISH using red fox probes to investigate karyotypes of a normal female dog (left) and CTVTs collected in Cape Verde (center) and Italy (right).
(C) CTVT genomic copy number for 24T (upper panel) and 79T (lower panel). Red and blue points represent total copy number and minor copy number (i.e. copy number of the allele present in fewer copies) respectively calculated using normalized read counts at each of 2,544,508 SNP loci. Chromosomes are represented by horizontal alternating black and gray bars.
CTVT’s massive burden of karyotypic abnormalities despite its largely diploid genomic copy number indicates that the genome has undergone large-scale copy neutral structural rearrangement. We found 2,118 candidate somatic structural variants that were shared between 24T and 79T, and 216 and 72 candidate somatic structural variants in 24T and 79T respectively that were unique to one tumor. We also searched for evidence of transposon mobilization in CTVT. We found 348 and 352 transposon insertions (involving both LINE and SINE elements) that were unique to 24T and 79T respectively, and are thus likely to represent somatic retrotransposition events that occurred after the divergence of 24T and 79T.
We identified 3.04 million substitution variants in 24T and 2.77 million in 79T after removing all single nucleotide polymorphisms (SNPs) known to segregate in the dog or wolf germline (including those identified in 24H or 79H). These variants will include somatic mutations as well as SNPs that have not been captured in previous canine sequencing efforts. We estimated the true number of somatic substitution mutations in CTVT by calculating the ratio of homozygous to heterozygous known SNPs in diploid regions that retain both parental chromosomes. Assuming that all homozygous variants in these regions are SNPs, we were able to estimate the number of unannotated heterozygous SNPs in each diploid segment using the homozygous to heterozygous ratio amongst annotated SNPs. This analysis indicated that at least 65% of unannotated variants in CTVT are likely to be somatic, corresponding to a total of ~1.9 million somatic mutations in CTVT. 103,667 and 109,119 variants were unique to 24T and 79T respectively, the majority of which probably arose as somatic mutations after the two tumors’ divergence as only 2,056 and 5,647 of these respectively occur in regions that have been lost in the other tumor. Although a range of total mutation counts are observed in human cancers, the majority have between 1,000 and 5,000 somatic single base substitution mutations (6). Thus CTVT has acquired several hundred times more somatic mutations than most human cancers.
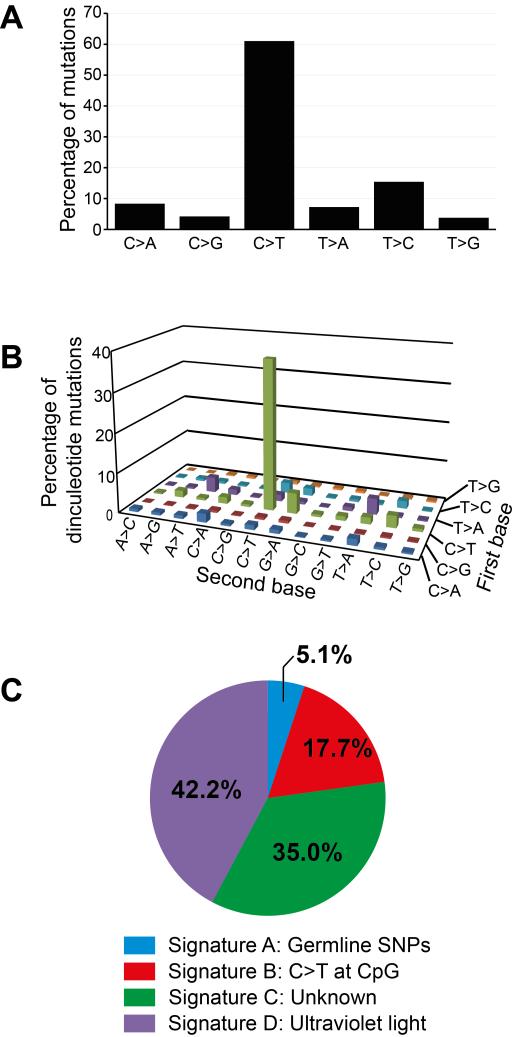
To ascertain the processes responsible for the mutations in CTVT, we characterized CTVT’s mutational spectrum and searched for known mutational signatures (6) in the CTVT genomes. The CTVT mutation spectrum was dominated by C>T (or G>A) mutations and CC>TT (or GG>AA) dinucleotide mutations (Fig. 2A and 2B). Four mutational signatures were identified in CTVT (labelled A to D, Fig. 2C), which were sufficient to explain 98% of the mutations in CTVT (0.96 Pearson correlation between the mutation set observed in CTVT and the mutation set reconstructed using four mutational signatures). The chemical events associated with some of these mutational signatures have been characterized. Signature A is associated with germline SNPs, and its contribution to CTVT variant sets probably reflects incomplete removal of germline inherited variants. Signature B defines the mutational signature characterized by C>T at CpG dinucleotides that is widely found in human cancers and is known to be correlated with patient age (6). Signature C (known as signature 5 in (6)) is also frequently present in a spectrum of human cancers, however its etiology is unknown (6). Signature D, which is characterized by C>T and CC>TT mutations, and in humans is predominantly observed in cancers of the skin and is known to be associated with exposure to ultraviolet light (6), explains 42% of the mutations in CTVT. These observations suggest that CTVT has been exposed at a low level to ultraviolet light during its evolution. Although CTVT tumors usually occur inside the genital orifice, they may be exposed to sunlight when they protrude from the vulva, ulcerate through preputial skin or occur on external surfaces such as the skin or conjunctiva (for example, see 24T and 79T, Fig. 1A). Furthermore, it is the very cells that are exposed on the surface of a tumor that are most likely to propagate the CTVT lineage by passage to new hosts.
Figure 2. CTVT mutations.
Analyses were performed on a set of 395,306 CTVT variants that were annotated as somatic due to their heterozygous status within genomic regions that have undergone both loss of heterozygosity (LOH) and duplication.
(A) Simple mutation spectrum in CTVT. Mutations are labelled in pyrimidine context.
(B) Dinucleotide mutation spectrum in CTVT. The “first base” was defined as the mutation with the lower chromosome coordinate and the “second base” is immediately adjacent to the first base on the same strand. The strand is displayed relative to the pyrimidine context of the first base. 3,518 dinucleotide mutations were included in the analysis.
(C) The proportion of mutations in CTVT explained by mutational signatures A to D.
More than 10,000 genes (10,955 in 24T and 10,546 in 79T) in CTVT carry at least one non-synonymous substitution variant that is not a known germline SNP. Table S3 lists those genes that we consider to be the highest confidence driver mutations in CTVT. These include a known rearrangement involving MYC (7), homozygous deletion of CDKN2A, a hemizygous nonsense mutation in SETD2 and a rearrangement involving ERG which creates a potential in-frame NEK1-ERG fusion gene. A census of genes that have been lost in CTVT by homozygous deletion or hemizygous nonsense mutation indicated that at least 646 genes, 2.8% of the 22,874 protein-coding genes annotated in the dog genome, are collectively dispensable for survival and proliferation of a somatic cell (Table S4).
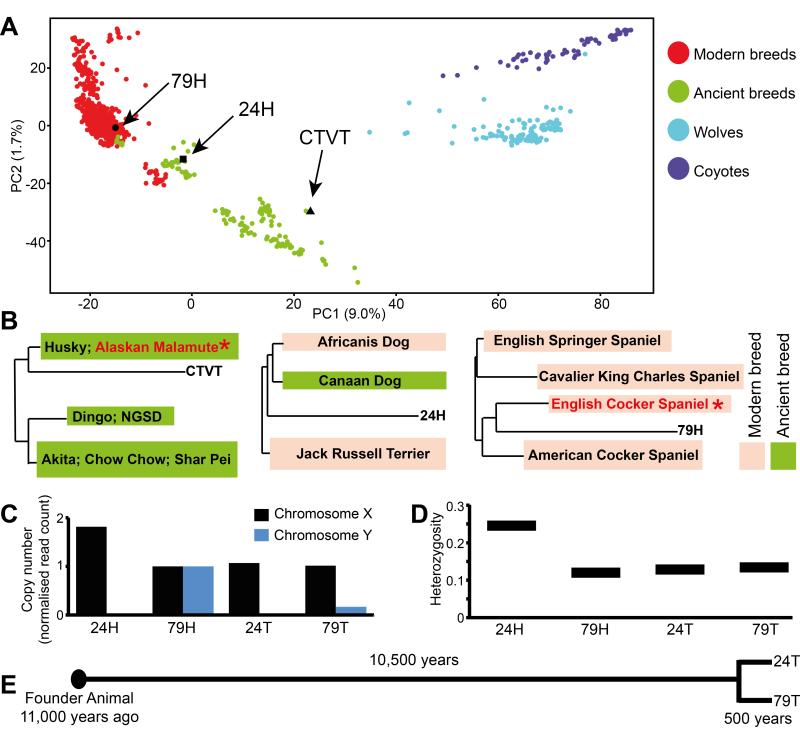
We next sought to reconstruct the phenotype of the CTVT Founder Animal and to estimate the age of the cancer that it spawned using the variants found in the CTVT genome. We compared the genotypes of 24T and 79T, as well as 24H and 79H, at 23,782 polymorphic SNP loci with those of 1,106 previously genotyped dogs, wolves and coyotes (8, 9). The result, displayed using principal components analysis (Fig. 3A), indicates that the CTVT Founder Animal was likely to have been a dog belonging to one of the “Ancient Breeds” (previous analyses were unable to distinguish between a wolf or an “Ancient Breed” dog origin (10)). Analysis of a pairwise distance tree indicated that, of the 86 breeds included in the analysis (9), the CTVT Founder Animal clusters most closely with Alaskan Malamutes and Huskies (11 Alaskan Malamutes have >95% probability after resampling genotypes of having one of the 16 genotypes closest to CTVT) (Fig. 3B and Table S5). As expected, 79H clustered most closely with Cocker Spaniels within the modern breeds (>95% probability after resampling that each of the six closest genotypes are English Cocker Spaniels) (Fig. 3A and 3B and Table S5). 24H, an Aboriginal Camp Dog, appears to have genetic contribution from both ancient and modern breeds (Fig. 3A and 3B and Table S5).
Figure 3. Tracing the CTVT Founder Animal.
(A) Principal component analysis of 1,106 wolves, dogs and coyotes using genotypes at 23,782 polymorphic SNP loci (8, 9). Each individual is represented by a single colored dot and positions of CTVT (inferred from the genotypes of 24T and 79T), 24H and 79H are indicated. Breeds were classified as Modern or Ancient according to (27).
(B) Positions of CTVT (left), 24H (center) and 79H (right) on pairwise distance tree comparing genotypes at 23,782 SNP loci with 1,106 other dogs, wolves and coyotes (8, 9). Only the closest breeds to CTVT, 24H and 79H are shown. Breeds containing members that most strongly clustered with CTVT and 79H after genotype resampling are marked with red text and * (see Table S5, 24H did not cluster strongly with individuals from any single breed). NGSD, New Guinea Singing Dog.
(C) Sex chromosome copy number of 24H (a female dog), 79H (a male dog), 24T and 79T determined by counting the number of reads aligning to X and Y chromosomes and normalized to 79H. Y chromosome reads in 79T are likely to be derived from contaminating host DNA.
(D) Proportion of annotated SNP loci in germline diploid regions that are heterozygous in 24H, 79H, 24T and 79T.
(E) Timeline for CTVT origin and divergence.
Recent studies have mapped several loci conferring canine phenotypic features, such as coat color, morphology and behavior (11-21). We examined the sequence of CTVT at a number of these loci to determine the likely phenotype of the Founder Animal (Table S6). Our analysis indicated that the Founder Animal was likely to have been of medium or large size with an agouti or solid black coat. It carried a mixture of “wolf-like” and “dog-like” alleles at loci that have been linked to dog domestication (21). 24T and 79T each carried a single X chromosome and had no evidence of a Y chromosome, as found in a previous analysis (22). This is consistent with either a male (after somatic Y chromosome loss) or a female (after somatic X chromosome loss) Founder Animal (Fig. 3C). Analysis of genome-wide heterozygosity indicated that the Founder Animal was relatively inbred (Fig. 3D).
Previous studies have estimated that CTVT is between 200 and 70,000 years old (10, 23). We sought to clarify the age of CTVT using the mutations associated with mutational signature B (Fig. 2C), which is correlated with patient age at diagnosis in many cancer types (6, 24). We estimated that 492,533 mutations in CTVT are likely to have been caused by this mutational process. Using the mutation rate of signature B in human medulloblastoma as a molecular clock (43.3 mutations of this signature genome-wide per year; we chose medulloblastoma because it is the human cancer with the closest correlation between number of mutations of this signature and patient age (6)), we estimate that CTVT may have first arisen approximately 11,368 years ago (lower and upper confidence intervals 10,179 and 12,873 years respectively). There is uncertainty in this estimate introduced by the possibilities that the accumulation of mutations of this signature is not clock-like in CTVT or that there are tissue or species specific differences in the rate of mutation accumulation between CTVT and human medulloblastoma. Applying this molecular clock to the mutations that occurred after divergence of the two tumors, we suggest that the most recent common ancestor of 24T and 79T may have existed approximately 460 years ago (458.2 years for 24T, 459.8 years for 79T) (Fig. 3E). It is interesting that the estimated timing of this divergence coincides with the era of rapid human global exploration.
The Founder Animal whose somatic cells first gave rise to CTVT was an “Ancient Breed” dog that may have lived about 11,000 years ago. The date of CTVT emergence together with the structure of its phylogenetic tree (23) and evidence for both wolf-like and dog-like alleles at loci associated with domestication is consistent with the possibility that CTVT may have first arisen within a genetically isolated population of early dogs whose limited genetic diversity facilitated the cancer’s escape from its hosts’ immune systems. Similarly, the Tasmanian devil facial tumor disease, the only other known naturally occurring clonally transmissible cancer, arose in an island population with low genetic diversity (25, 26). Populations with limited genetic diversity may be particularly susceptible to the emergence and spread of transmissible cancers.
The CTVT genome has illuminated the origins, history and evolution of the world’s oldest known cancer. It is remarkable that a somatic genome whose DNA would normally have survived for no more than 15 years during the life of one dog has continued to exist for several millennia as a parasitic life form. CTVT’s survival and global dominance is a testament to the ability of the mammalian somatic cell genome to adapt to and persist in a new ecological niche.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (grant reference 098051), the Kadoorie Charitable Foundation and a L’Oreal-UNESCO For Women in Science Fellowship (EPM). We are grateful to Andrew King, Susanna Cooke, Andrea Strakova, Maria Peleteiro, Cesaltina Semedo, Talita Mariana Morata Raposo, Rafael Ricardo Huppes, Cynthia Marchiori Bueno and the people of Maningrida. We thank members of the Wellcome Trust Sanger Institute Cancer Genome Project IT group and the Wellcome Trust Sanger Institute Core Sequencing and IT facilities. Additional sources of support included EMBO Long Term Fellowships (Lt-456-2010 (EPM) and ALTF-1287-2012 (IM)) and a Marie Curie IEF grant (JCMT). Genome sequence data reported in this study are available with accession number [to be finalized] in the European Nucleotide Archive.
Footnotes
Supplementary content includes Materials and Methods, Tables S1 to S8 and References (28 – 41).
REFERENCES AND NOTES
- 1.Blaine DP. A domestic treatise on the diseases of horses and dogs. T. Boosey; London: 1810. pp. 161–2. [Google Scholar]
- 2.Murchison EP. Oncogene. 2008;27(Suppl 2):S19–30. doi: 10.1038/onc.2009.350. [DOI] [PubMed] [Google Scholar]
- 3.Ganguly B, Das U, Das AK. Vet Comp Oncology. 2013 doi: 10.1111/vco.12060. published online EpubAug 25 (10.1111/vco.12060) [DOI] [PubMed] [Google Scholar]
- 4.Nik-Zainal S, et al. Cell. 2012 May 25;149:994. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, et al. New England J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov LB, et al. Nature. 2013 Aug 22;500:415. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzir N, et al. Proc Natl Acad Sci U S A. 1985 Feb;82:1054. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.vonHoldt BM, et al. Genome Res. 2011 Aug;21:1294. doi: 10.1101/gr.116301.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vonholdt BM, et al. Nature. 2010 Apr 8;464:898. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Cell. 2006 Aug 11;126:477. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadieu E, et al. Science. 2009 Oct 2;326:150. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candille SI, et al. Science. 2007 Nov 30;318:1418. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark LA, Wahl JM, Rees CA, Murphy KE. Proc Natl Acad Sci U S A. 2006 Jan 31;103:1376. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodman NH, et al. Mol Psychiatry. 2010 Jan;15:8. doi: 10.1038/mp.2009.111. [DOI] [PubMed] [Google Scholar]
- 15.Everts RE, Rothuizen J, van Oost BA. Anim Genet. 2000 Jun;31:194. doi: 10.1046/j.1365-2052.2000.00639.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoopes BC, Rimbault M, Liebers D, Ostrander EA, Sutter NB. Mamm Genome. 2012 Dec;23:780. doi: 10.1007/s00335-012-9417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmutz SM, Berryere TG. Anim Genet. 2007 Dec;38:539. doi: 10.1111/j.1365-2052.2007.01664.x. [DOI] [PubMed] [Google Scholar]
- 18.Sutter NB, et al. Science. 2007 Apr 6;316:112. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi Y, et al. Anim Genet. 2009 Oct;40:616. doi: 10.1111/j.1365-2052.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- 20.Vaysse A, et al. PLoS Genet. 2011 Oct;7:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Axelsson E, et al. Nature. 2013 Mar 21;495:360. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 22.Thomas R, Rebbeck C, Leroi AM, Burt A, Breen M. Chromosome Res. 2009;17:927. doi: 10.1007/s10577-009-9080-8. [DOI] [PubMed] [Google Scholar]
- 23.Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A. Evolution. 2009 Sep;63:2340. doi: 10.1111/j.1558-5646.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 24.Stephens PJ, et al. Nature. 2012 Jun 21;486:400. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murchison EP, et al. Cell. 2012 Feb 17;148:780. [Google Scholar]
- 26.Miller W, et al. Proc Natl Acad Sci of U S A. 2011;108:12348–12353. doi: 10.1073/pnas.1102838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larson G, et al. Proc Natl Acad Sci U S A. 2012 Jun 5;109:8878. doi: 10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindblad-Toh K, et al. Nature. 2005 Dec;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 29.Li G, et al. Genome Res. 2013 Sep 20;23:1486. doi: 10.1101/gr.154286.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Bioinformatics. 2009 Jul 15;25:1754. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F, et al. Genomics. 1999 Dec 1;62:189. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- 32.Jentsch I, Adler ID, Carter NP, Speicher MR. Chromosome Res. 2001;9:211. doi: 10.1023/a:1016696303479. [DOI] [PubMed] [Google Scholar]
- 33.Van Loo P, et al. Proc Natl Acad Sci U S A. 2010 Sep 28;107:16910. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsen G, et al. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nik-Zainal S, et al. Cell. 2012 May 25;149:979. [Google Scholar]
- 36.Flicek P, et al. Nucleic Acids Res. 2012 Jan;40:D84. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren W, et al. Bioinformatics. 2010 Aug 15;26:2069. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit AFA, Hubley R, Green P. 1996-2010.
- 39.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Cell Rep. 2013 Jan 31;3:246. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Gene Census [accessed Nov, 2013];COSMIC Database. http://cancer.sanger.ac.uk/cancergenome/projects/census/
- 41.Choi YK, Kim CJ. J Vet Sci. 2002 Dec;3:285. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.