Abstract
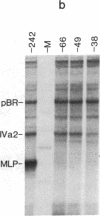
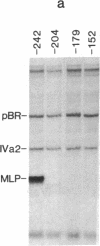
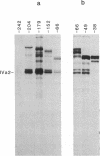
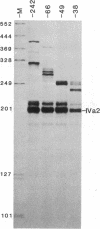
The adenovirus IVa2 gene, which is expressed at an intermediate time in the viral infectious cycle, is separated from the adenovirus major late promoter (MLP) 5' start site by 210 base pairs and is transcribed from the opposite strand. In contrast to the MLP, the IVa2 gene does not contain a "TATA" box upstream from its 5' start sites. By using a series of deletion mutants, two upstream control regions that are rich in cytidine residues, one proximal to the cap site at nucleotide positions -39 to -48 and a distal domain between nucleotide positions -152 and -242 have been identified as essential for IVa2 transcription (IVa2 cap site is nucleotide position + 1). Transcription efficiency is decreased by 70-90% after the deletion of a proximal C-rich domain when either linear or supercoiled DNAs were used as template. However, distal sequences functioned as transcriptional control domains only with covalently closed DNA templates. The deletion of both the proximal and distal regions from covalently closed DNA templates reduces the levels of IVa2 transcription by a factor of 100-150. When the plasmid pAd242 that contains the 5' start sites of adenovirus MLP and IVa2 is transcribed, there is essentially a complete suppression of transcription of the adenovirus IVa2 gene. The transcription efficiency of IVa2 is increased 10-fold after deletion of the MLP cap site. A model based on a shared entry site for RNA polymerase II and competition between major late and IVa2 promoters is proposed to explain the in vitro transcriptional results.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker C. C., Ziff E. B. Promoters and heterogeneous 5' termini of the messenger RNAs of adenovirus serotype 2. J Mol Biol. 1981 Jun 25;149(2):189–221. doi: 10.1016/0022-2836(81)90298-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brady J., Radonovich M., Vodkin M., Natarajan V., Thoren M., Das G., Janik J., Salzman N. P. Site-specific base substitution and deletion mutations that enhance or suppress transcription of the SV40 major late RNA. Cell. 1982 Dec;31(3 Pt 2):625–633. doi: 10.1016/0092-8674(82)90318-x. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Hernandez N., Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983 Sep 22;305(5932):336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- Concino M., Goldman R. A., Caruthers M. H., Weinmann R. Point mutations of the adenovirus major late promoter with different transcriptional efficiencies in vitro. J Biol Chem. 1983 Jul 10;258(13):8493–8496. [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Fire A., Baker C. C., Manley J. L., Ziff E. B., Sharp P. A. In vitro transcription of adenovirus. J Virol. 1981 Dec;40(3):703–719. doi: 10.1128/jvi.40.3.703-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman S. A., Van Beveren C., Verma I. M. Identification of a RNA polymerase II initiation site in the long terminal repeat of Moloney murine leukemia viral DNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5411–5415. doi: 10.1073/pnas.78.9.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding C. R., Russell W. C. S1 sensitive sites in adenovirus DNA. Nucleic Acids Res. 1983 Jan 11;11(1):21–36. doi: 10.1093/nar/11.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Delimitation of far upstream sequences required for maximal in vitro transcription of an H2A histone gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):297–301. doi: 10.1073/pnas.79.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., de Boer E., Shewmaker C. K., Flavell R. A. DNA sequences necessary for transcription of the rabbit beta-globin gene in vivo. Nature. 1982 Jan 14;295(5845):120–126. doi: 10.1038/295120a0. [DOI] [PubMed] [Google Scholar]
- Hen R., Sassone-Corsi P., Corden J., Gaub M. P., Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S., Takeuchi K., Suzuki Y. In vitro characterization of the fibroin gene promoter by the use of single-base substitution mutants. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7258–7262. doi: 10.1073/pnas.79.23.7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jat P., Roberts J. W., Cowie A., Kamen R. Comparison of the polyoma virus early and late promoters by transcription in vitro. Nucleic Acids Res. 1982 Feb 11;10(3):871–887. doi: 10.1093/nar/10.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Natarajan V., Madden M. J., Salzman N. P. Preferential stimulation of transcription from simian virus 40 late and adeno IVa2 promoters in a HeLa cell extract. J Biol Chem. 1983 Dec 10;258(23):14652–14655. [PubMed] [Google Scholar]
- Orkin S. H., Sexton J. P., Cheng T. C., Goff S. C., Giardina P. J., Lee J. I., Kazazian H. H., Jr ATA box transcription mutation in beta-thalassemia. Nucleic Acids Res. 1983 Jul 25;11(14):4727–4734. doi: 10.1093/nar/11.14.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne T. F., Gaynor R. B., Berk A. J. The TATA homology and the mRNA 5' untranslated sequence are not required for expression of essential adenovirus E1A functions. Cell. 1982 May;29(1):139–148. doi: 10.1016/0092-8674(82)90098-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schreier P. H., Cortese R. A fast and simple method for sequencing DNA cloned in the single-stranded bacteriophage M13. J Mol Biol. 1979 Mar 25;129(1):169–172. doi: 10.1016/0022-2836(79)90068-8. [DOI] [PubMed] [Google Scholar]
- Zarucki-Schulz T., Tsai S. Y., Itakura K., Soberon X., Wallace R. B., Tsai M. J., Woo S. L., O'Malley B. W. Point mutagenesis of the ovalbumin gene promoter sequence and its effect on in vitro transcription. J Biol Chem. 1982 Sep 25;257(18):11070–11077. [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]