Abstract
It is now clear that bacterial chromosomes rapidly separate in a manner independent of cell elongation, suggesting the existence of a mitotic apparatus in bacteria. Recent studies of bacterial cells reveal filamentous structures similar to the eukaryotic cytoskeleton, proteins that mediate polar chromosome anchoring during Bacillus subtilis sporulation, and SMC interacting proteins that are involved in chromosome condensation. A picture is thereby developing of how bacterial chromosomes are organized within the cell, how they are separated following duplication, and how these processes are coordinated with the cell cycle.
Introduction
Forty years ago, Jacob, Brenner and Cuzin presented a characteristically elegant model for bacterial chromosome segregation, in which the chromosomes, attached to the cell envelope near mid-cell, were separated by localized envelope growth between them [1]. Although this compelling model still appears in textbooks, recent cell biological experiments have revealed that bacterial chromosomes rapidly separate in a manner independent of cell elongation, rendering it unlikely that the model of Jacob et al. [2•-6•] is entirely correct. Central to the renewed debate has been the ability to precisely determine the subcellular distribution of proteins and DNA molecules within bacterial cells. In slowly growing Escherichia coli [7], Bacillus subtilis [8,9] and Caulobacter crescentus [10], origins and termini localize towards opposite poles of newborn cells (Figure 1). In B. subtilis [11,12] and E. coli [13,14], both regions move toward the cell center, where a stationary DNA replication factory catalyzes both leading and lagging strand synthesis [11]. Following origin duplication, and likely at a specific time in the cell cycle, the origins migrate towards opposite cell poles, while the termini separate close to or shortly after cell division, following chromosome decatenation [15••]. There are interesting variations on this emerging picture. For example, in C. crescentus, the DNA replication factory slowly marches from the cell pole to the mid-cell [16], while one replicated origin stays at the pole and the other migrates to the opposite pole [10]. When multiple chromosomes and plasmids co-exist in the same cell, similar replicons typically target to distinct sites within the same general region of the cell. For example, in Agrobacterium tumefaciens, which has one circular and one linear chromosome and two plasmids, each chromosomal origin and plasmid occupies distinct sites near the cell pole [17•].
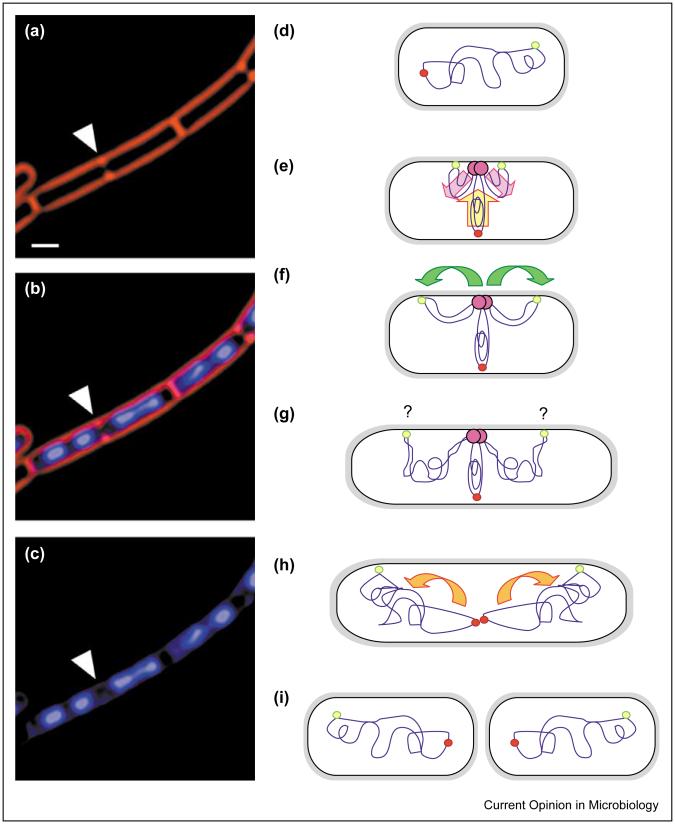
Figure 1.
Bacterial chromosome segregation. (a-c) Micrograph showing rapidly growing B. subtilis cells stained with FM 4-64 to visualize cell membranes (red) and DAPI to show chromosomal DNA (blue). During rapid growth, B. subtilis cells remain in septate chains because cell separation lags behind division, and multiple chromosomal replication cycles occur to enable rapid division. Thus, in cells that are about to divide (arrowhead), chromosome segregation often appears complete in the nascent daughter cells. In this case, the nascent daughter cell on the left has two apparently separate chromosomes, whereas that on the right has a bi-lobed nucleoid, which is likely to represent a partially replicated or segregated chromosome. Scale bar = 1 μm. (d-i) Model for chromosome segregation in slowly growing E. coli and B. subtilis. (d) In newborn cells, the origin (yellow-green circle) and terminus (red circle) appear at opposite cell poles. (e) At the initiation of chromosome replication, the chromosome rotates to bring the origin to the replisome (pink circle). The replicated DNA is extruded towards opposite cell poles (pink arrows), whereas the unreplicated DNA moves towards the replisome (yellow arrow). (f) The replicated origin DNA moves towards the cell poles (green arrows), (g) where it is anchored by an unidentified origin-binding protein (purple question mark). (h) The daughter chromosomes are separately condensed during replication by the concerted action of SMC and topoisomerases (orange arrows). (i) Finally, division results in two polarized daughter cells, in which the termini lie at each new pole, whereas the origins are at each older pole.
For simplicity here, we will consider bacterial chromosome segregation to consist of four steps, (i) the movement of newly replicated origins from mid-cell towards the cell poles, (ii) the anchoring of the origin to specific locations within the cell, (iii) the separate condensation of newly replicated chromosomes, and (iv) the resolution of dimeric and catenated chromosomes, although we recognize that these steps might not be completely distinct. These events are almost certainly coordinated with specific steps of cell division, and we will end this review by touching on this nearly uncharted territory. In this review, we discuss recent experiments that have shed light on each of these topics.
Origin movement
The rapid separation of origins through the viscous cytoplasm is likely to require a force-generating mechanism. Although the recently discovered cytoskeleton-like structures within bacterial cells (some of which are involved in plasmid segregation) raise the possibility that similar structures might be involved in chromosome segregation [18••-20••], the observation that DNA polymerase is a motor protein which resides in a stationary replisome, has led to the ‘extrusion-capture’ model for chromosome segregation [4•,11,21]. This model proposes that DNA replication itself pushes newly replicated DNA towards opposite sides of the cell, where the origins are captured by an as yet unidentified origin-binding protein. Vectoral origin movement has been proposed to be maintained by coupled transcription, translation and protein export, as well as factors involved in chromosome compaction, such as SMC (structural maintenance of chromosomes) [22,23]. Although the extrusion-capture model is appealing, certain results contradict with its most simple version. For example, in E. coli chromosomes engineered to initiate replication from integrated plasmid origins instead of the chromosomal origin, the chromosomal origin still localizes to the cell poles before origin-distal regions, emphasizing the importance of capturing chromosomal regions at specific locations within the cell [24].
Chromosome condensation
In growing bacteria, the replicating chromosome assumes a bi-lobed structure that occupies a small fraction of its volume in solution. This compaction crucially depends on negative superhelicity, which is maintained by topoisomerases, and also by the bacterial orthologs of the SMC proteins [25]. SMC proteins have a centrally located coiled-coil domain flanked by large globular domains that interact upon dimerization to form an ATPase. A flexible hinge region within the coiled-coil domain allows the protein to bend into V-shape molecules, perhaps regulating SMC dimerization and DNA binding activity [26,27,28••,29]. Eukaryotes typically have multiple forms of SMC that participate in many different aspects of chromosome dynamics, including sister chromosome cohesion, DNA condensation and DNA repair (reviewed in [30,31]), whereas bacteria usually have either a single SMC, or a functionally equivalent distant relative, MukB (as in the case of E. coli), that appear to act primarily in DNA condensation [32].In B. subtilis, C. crescentus and E. coli, smc or mukB mutants have decondensed chromosomes and chromosome segregation defects [10,33-35], but in B. subtilis, the newly replicated origins still move rapidly towards the cell poles [36]. Importantly, these defects can be suppressed by increasing the negative superhelicity of the chromosome, emphasizing the importance of supercoiling in chromosome compaction [21,25,37].
In many bacteria, as in eukaryotes, SMC interacts with accessory proteins such as E. coli MukE and MukF [38] and the recently identified and highly conserved B. subtilis ScpA and ScpB [39••,40••]. Mutants lacking either ScpA or ScpB have phenotypes identical to the smc mutant, and ScpA, ScpB, and SMC form a complex frequently located near mid-cell or quarter-cell positions [39••,41,42••]. The similarity in the position of this complex with the location of the DNA replication machinery suggested that SMC complexes might be closely associated and co-localized with the replisome, which may facilitate their ability to organize and condense newly replicated chromosomes. However, the picture that has emerged from localization studies is more complex. Although SMC proteins do form foci near replisomes in some cells, they are frequently located away from the replisome [38,42••,43-45]. Furthermore, chromatin immunoprecipitation experiments demonstrate that SMC binds many chromosomal regions, including sequences that have not yet been replicated in synchronized cell cultures [42••]. Thus, it is likely that SMC complexes both refold the chromosome after replication and maintain global chromosome organization.
Resolution of catenated chromosomes: the peculiarities of circularity
The final stages of chromosome segregation in bacteria with circular chromosomes (the majority of bacteria) include the resolution of chromosome dimers, which form when circular daughter chromosomes undergo an odd number of homologous recombination events [46], and the decatenation of topologically linked circular DNA molecules. Remarkably, in E. coli, both events are spatially and temporally regulated, occurring specifically at mid-cell during the final stages of cell division. This regulation is conferred by FtsK, an essential and conserved cell division protein consisting of two domains, an amino-terminal membrane domain involved in cell division, and a carboxy-terminal ATPase domain capable of moving along DNA in an ATP-dependent manner. This DNA tracking domain localizes the enzyme responsible for chromosome decatenation (TopoIV) to the division site, and also acts on XerC and XerD [47,48], the site-specific recombinase that mediates chromosome dimer resolution. It appears that FtsK aligns the dif sites at which recombination occurs [49], modifies the XerC/XerD/dif site interaction to promote XerC-mediated resolution of the holliday junction [50••], and finally exports one resolved chromosome into the appropriate daughter cell [15••]. FtsK is a key component of the division machinery, localizing to the septum before division and recruiting other division proteins to the septum [51]. The FtsK–XerCD–TopoIV interaction likely serves to ensure that these enzymes act only on the appropriate substrates (dimeric or catenated chromosomes), as only these will remain within the plane of the invaginating septum.
Plasmid and chromosomal Par proteins
The first partitioning determinants to be characterized were the par loci found on the low copy plasmids F and P1. These loci consist of a two-gene operon and a cis-acting centromere-like region (parAB and parS respectively, using the nomenclature of P1), and are sufficient to stablize plasmids and to mediate their appropriate localization. Most bacterial chromosomes also have par loci near their origins of replication, although E. coli and its close relatives apparently encode only the more distant ParA homolog MinD, which spatially regulates cell division. The ParA proteins are ATPases that interact with ParB, which binds to the parS site adjacent to the operon [52-54]. The mechanisms by which the ParAB proteins contribute to DNA segregation remains unclear, however, it has recently been demonstrated that the analogous but unrelated ATPase of R1 plasmid (ParM) forms extended, actin-like helical filaments within cells [20••,55,56,57••]. These dynamic structures could generate the force needed for movement of plasmids to opposite cell poles, if polymerization occurred between plasmid molecules, or they could provide a linear scaffold along which the plasmids could move. It is possible that the function of ParA proteins is mechanistically similar to ParM, as the E. coli ParA homolog, MinD, also forms extended filaments in E. coli that are likely to mediate the pole to pole oscillation of MinC [19••,58]. Other oscillating ParA homologs include the E. coli plasmid pB171 [59] and the B. subtilis chromosomal ParA protein Soj, the oscillation of which also depends on MinD [60•].
The chromosomally encoded Par proteins are in some ways similar to the plasmid proteins, as the chromosomal par loci are sufficient to stabilize plasmids and direct their correct positioning within the cell [61,62]. In addition, mutations in the chromosomal par genes increases the proportion of anucleate cells, albeit often only during specific phases of growth [63-66]. However, there are striking differences between plasmid and chromosomal Par proteins. First, in B. subtilis, localization of the ParB homolog Spo0J depends on the chromosomal position of its binding site; moving this site away from the origin is not sufficient to target this new region to the location normally occupied by the origin, indicating the existence of a residual origin-targeting mechanism [67••]. Second, B. subtilis mutants lacking Spo0J/Soj have an increased chromosomal copy number, most likely owing to increased and asynchronous chromosome replication, suggesting that these proteins regulate chromosome replication [67••,68]. The relatively benign effects of mutations on chromosome partitioning during growth suggests the existence of additional mechanisms by which chromosomes are positioned within the cell, as is supported by recent studies of chromosome positioning during B. subtilis sporulation.
B. subtilis sporulation and the identification of origin anchoring determinants
The asymmetric cell division event that marks the onset of sporulation in B. subtilis has provided an ideal system for studying chromosome dynamics, and has allowed the identification of proteins required for polar chromosome anchoring (Figure 2). At the onset of B. subtilis sporulation, the chromosome is reorganized from the bi-lobed structure typical of growing cells to an elongated axial filament [69] in which the origin-proximal 30% of the future forespore chromosome is condensed near one cell pole [70,71]. Polar septation thereby traps the forespore chromosome in the septum, and the remaining portion is translocated across the septum by the SpoIIIE DNA translocase, an FtsK homolog that exports DNA from the cell in which it is synthesized [72,73]. The orientation of the chromosome and targeting of the chromosomal origin region to the extreme cell poles depends on specific regions within the chromosome and on specific DNA binding proteins. A large chromosomal region adjacent to, but not including the replicative origin, was shown to be necessary for polar origin positioning during sporulation [74] and two DNA sequences within and downstream of dnaA (located within the origin region) are required for chromosome positioning during growth [75].
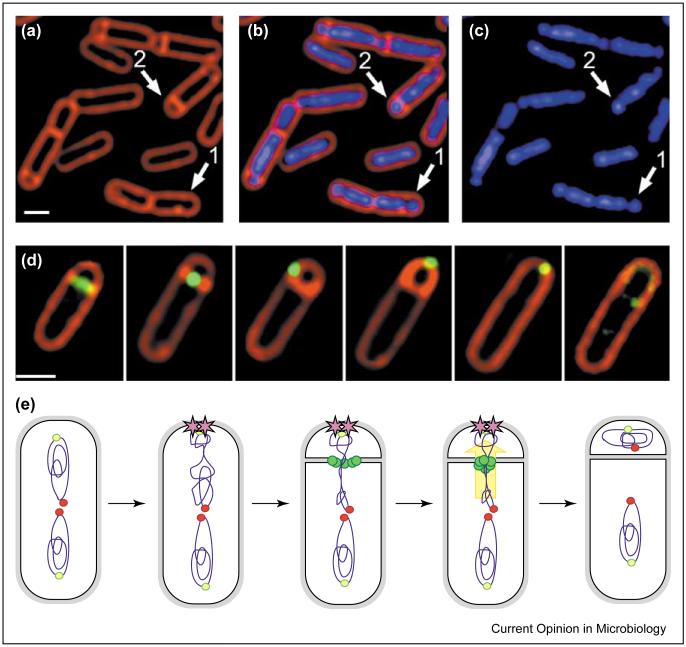
Figure 2.
Chromosome segregation during B. subtilis sporulation. (a-c) Micrograph showing B. subtilis cells early in sporulation, following staining with FM 4-64 to visualize cell membranes (red) and DAPI to show DNA (blue). Prior to polar septation, the origin-proximal 30% of the future forespore chromosome condenses near one cell pole (arrow 1); the asymmetrically-positioned sporulation septum is synthesized between these two chromosome domains (arrow 2). (d) Localization of the SpoIIIE DNA translocase (green) together with cell membranes (red). The protein first forms a ring at the site of cell division, then assembles a focus at the septal midpoint, and finally it relocalizes to the cell pole, where it participates in the final stages of the phagocytosis-like process of engulfment [82]. Scale bars = 1 μm. (e) Model for chromosome segregation during sporulation. At the onset of sporulation, the origins (yellow-green circles) are brought to the cell poles by an interaction with RacA (purple stars), while the temini remain near mid-cell (red circles). During septation, the SpoIIIE DNA translocase (green circles) assembles at the septal midpoint, and translocates the forespore chromosome across the septum (yellow arrow).
Recent studies have demonstrated that the sporulation-specific anchoring of the origin to the extreme cell pole depends on RacA, a polarly localized DNA binding protein [76••,77••]. RacA is transiently expressed early in sporulation, and in racA mutants, the sporulation septum often fails to trap DNA, producing anucleate forespores. A mutant lacking both RacA and the Spo0JSoj (Par) system has unexpectedly severe effects on chromosome orientation during sporulation, suggesting that the RacA protein acts together with the chromosomal par systems to mediate polar chromosome positioning during sporulation [77••]. However, because RacA is not produced during growth, it is likely that an as yet unidentified protein contributes to origin positioning during growth.
Morphological checkpoints
Clearly, in bacteria, as in eukaryotes, morphological checkpoints must serve to coordinate cell division with chromosome replication and segregation. One such checkpoint is likely to be conferred by B. subtilis Soj, which in addition to contributing to chromosome orientation and replication, acts as a transcription factor governing production of proteins required for synthesis of the sporulation septum [78]. Similarly, in C. crescentus, the CtrA transcription factor directly regulates chromosome replication and the production of cell division proteins (as well as proteins involved in morphogenesis), whereas the ParAB proteins couple chromosome replication and cell division [79].In B. subtilis additional links between chromosome segregation and cell division include the DivIVA cell division protein, which is necessary for the polar localization of both RacA and MinCD; indeed, divIVA mutants produce a high frequency of anucleate forespores [80]. The polar localization of MinD is in turn required for both the assembly of Soj onto the chromosome [60•] and to regulate the polarity of SpoIIIE DNA translocase assembly [81•]. No doubt the future will see the identification of additional proteins involved in both chromosome dynamics and septation, as well as the molecular elucidation of additional regulatory checkpoints governing the bacterial cell cycle.
Conclusions
The past two years have seen tremendous advances in our understanding of how chromosomal organization is achieved inside of bacterial cells. Proteins specifically involved in anchoring chromosomal origins near the cell pole have been identified. The bacterial SMC chromosome condensation apparatus has been more fully described, opening the way to address how it functions mechanistically. Some of the questions that remain to be addressed include: how do chromosomal origins find their proper positions during exponential growth? Which proteins are responsible for the rapid and directed movement of chromosomal DNA? And how is the timing of chromosome segregation regulated and coordinated with chromosome replication and cell division?
Acknowledgements
The author’s research is supported by grants from the National Institutes of Health (KP, GM57045) and from the National Science Foundation (KP, NSF 0135955; JP, NSF 0215752). EB is supported by a National Institutes of Health postdoctoral fellowship (GM65692).
Abbreviations
- SMC
structural maintenance of chromosomes
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
••of outstanding interest
- 1.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol. 1963;28:329–348. [Google Scholar]
- 2•.Jensen RB, Wang SC, Shapiro L. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat Rev Mol Cell Biol. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- See annotation for [3•]
- 3•.Jenal U, Stephens C. The Caulobacter cell cycle: timing, spatial organization and checkpoints. Curr Opin Microbiol. 2002;5:558–563. doi: 10.1016/s1369-5274(02)00378-8. [DOI] [PubMed] [Google Scholar]
- The Caulobacter cell cycle is reviewed [2•,3•]. Caulobacter provides an ideal bacterium for chromosome segregation studies, as synchronous cultures can readily be obtained.
- 4•.Draper GC, Gober JW. Bacterial chromosome segregation. Annu Rev Microbiol. 2002;56:567–597. doi: 10.1146/annurev.micro.56.012302.160729. [DOI] [PubMed] [Google Scholar]
- See annotation [6•]
- 5•.Ryan KR, Shapiro L. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu Rev Biochem. 2003;72:367–394. doi: 10.1146/annurev.biochem.72.121801.161824. [DOI] [PubMed] [Google Scholar]
- See annotation [6•]
- 6•.Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- References [4•-•6] provide excellent comprehensive reviews on bacterial chromosome segregation.
- 7.Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharpe ME, Errington J. A fixed distance for separation of newly replicated copies of oriC in Bacillus subtilis: implications for co-ordination of chromosome segregation and cell division. Mol Microbiol. 1998;28:981–990. doi: 10.1046/j.1365-2958.1998.00857.x. [DOI] [PubMed] [Google Scholar]
- 9.Webb CD, Graumann PL, Kahana JA, Teleman AA, Silver PA, Losick R. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 10.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 12.Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 13.Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- 14.Koppes LJ, Woldringh CL, Nanninga N. Escherichia coli contains a DNA replication compartment in the cell center. Biochimie. 1999;81:803–810. doi: 10.1016/s0300-9084(99)00217-5. [DOI] [PubMed] [Google Scholar]
- 15••.Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- This comprehensive paper reports the co-localization of distinct chromosomal regions, as well as the co-localization of these regions with the cell division protein FtsZ, and the RarA protein, which is likely to be a component of the replisome. Surprisingly, separation of the termini frequently occurs after septation, implying that FtsK must serve to translocate one terminus into the nascent daughter cell. This asymmetric DNA translocase activity is strikingly similar to that of B. subtilis SpoIIIE, which acts in the mother cell to translocate DNA into the forespore.
- 16.Jensen RB, Wang SC, Shapiro L. A moving DNA replication factory in Caulobacter crescentus. Embo J. 2001;20:4952–4963. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Kahng LS, Shapiro L. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J Bacteriol. 2003;185:3384–3391. doi: 10.1128/JB.185.11.3384-3391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not all bacteria contain a single circular chromosome, and the mechanism by which a bacterium coordinates the replication and segregation of multiple chromosomes remains unclear. Here the authors report the first study of chromosome segregation in a bacterium with two chromosomes, one of which is linear. The results indicate that independent episomes are anchored to unique positions within the cell.
- 18••.Carballido-Lopez R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- Beautiful cell biological investigations of the dynamics of the Mbl protein, a widely conserved bacterial protein which is structurally similar to actin, and which assembles dynamic helical filaments. Mbl plays an essential role in bacterial cell shape determination; in its absence normally rod-shaped bacteria become lemon-shaped.
- 19••.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This paper demonstrates that Min proteins produce coiled structures extending from one cell pole to another, providing new insight into the mechanism by which Min proteins function to measure the length of the bacterial cell.
- 20••.Moller-Jensen J, Jensen RB, Lowe J, Gerdes K. Prokaryotic DNA segregation by an actin-like filament. Embo J. 2002;21:3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ParM partitioning protein of plasmid R1 is shown to form cytoskeletal actin-like filaments that participate in plasmid movement.
- 21.Sawitzke J, Austin S. An analysis of the factory model for chromosome replication and segregation in bacteria. Mol Microbiol. 2001;40:786–794. doi: 10.1046/j.1365-2958.2001.02350.x. [DOI] [PubMed] [Google Scholar]
- 22.Dworkin J, Losick R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA. 2002;99:14089–14094. doi: 10.1073/pnas.182539899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- 24.Gordon GS, Shivers RP, Wright A. Polar localization of the Escherichia coli oriC region is independent of the site of replication initiation. Mol Microbiol. 2002;44:501–507. doi: 10.1046/j.1365-2958.2002.02901.x. [DOI] [PubMed] [Google Scholar]
- 25.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 28••.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. Embo J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutations in the hinge domain of Bacillus subtilis SMC are shown to disrupt both dimerization and DNA binding, suggesting that the hinge region might play an important role in modulating SMC–DNA interactions.
- 29.Lowe J, Cordell SC, van den Ent F. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J Mol Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- 30.Cobbe N, Heck MM. Review: SMCs in the world of chromosome biology-from prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- 31.Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 32.Graumann PL. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie. 2001;83:53–59. doi: 10.1016/s0300-9084(00)01218-9. [DOI] [PubMed] [Google Scholar]
- 33.Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriya S, Tsujikawa E, Hassan AK, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 35.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partitioning of E. coli. Embo J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graumann PL. Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions. J Bacteriol. 2000;182:6463–6471. doi: 10.1128/jb.182.22.6463-6471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitao T, Nordstrom K, Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- 38.Ohsumi K, Yamazoe M, Hiraga S. Different localization of SeqAbound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol Microbiol. 2001;40:835–845. doi: 10.1046/j.1365-2958.2001.02447.x. [DOI] [PubMed] [Google Scholar]
- 39••.Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. Embo J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See annotation for [40••]
- 40••.Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- The identification and characterization of a pair of conserved proteins (ScpA and ScpB) that interact with Bacillus subtilis SMC and function in chromosome condensation are described [39••,40••]
- 41.Volkov A, Mascarenhas J, Andrei-Selmer C, Ulrich HD, Graumann PL. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol Cell Biol. 2003;23:5638–5650. doi: 10.1128/MCB.23.16.5638-5650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Lindow JC, Kuwano M, Moriya S, Grossman AD. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol Microbiol. 2002;46:997–1009. doi: 10.1046/j.1365-2958.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- This paper shows that Bacillus subtilis SMC localizes in foci at positions similar to the that of the cellular replisome and that localization depends on ScpA and ScpB but not on ongoing replication. SMC was also shown to frequently, but not always, co-localize with the replisome.
- 43.Graumann PL, Losick R, Strunnikov AV. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J Bacteriol. 1998;180:5749–5755. doi: 10.1128/jb.180.21.5749-5755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.den Blaauwen T, Lindqvist A, Lowe J, Nanninga N. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol Microbiol. 2001;42:1179–1188. doi: 10.1046/j.1365-2958.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- 45.Jensen RB, Shapiro L. Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein. J Bacteriol. 2003;185:3068–3075. doi: 10.1128/JB.185.10.3068-3075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendricks EC, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000;36:973–981. doi: 10.1046/j.1365-2958.2000.01920.x. [DOI] [PubMed] [Google Scholar]
- 47.Perals K, Capiaux H, Vincourt JB, Louarn JM, Sherratt DJ, Cornet F. Interplay between recombination, cell division and chromosome structure during chromosome dimer resolution in Escherichia coli. Mol Microbiol. 2001;39:904–913. doi: 10.1046/j.1365-2958.2001.02277.x. [DOI] [PubMed] [Google Scholar]
- 48.Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corre J, Louarn JM. Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J Bacteriol. 2002;184:3801–3807. doi: 10.1128/JB.184.14.3801-3807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- A beautiful study which demonstrates that like SpoIIIE, FtsK is capable of translocating along DNA in an ATP-dependent manner, and which also elucidates the biochemical mechanism by which FtsK participates in the resolution of chromosome dimers.
- 51.Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- 52.Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 53.Hiraga S. Dynamic localization of bacterial and plasmid chromosomes. Annu Rev Genet. 2000;34:21–59. doi: 10.1146/annurev.genet.34.1.21. [DOI] [PubMed] [Google Scholar]
- 54.Gordon GS, Wright A. DNA segregation in bacteria. Annu Rev Microbiol. 2000;54:681–708. doi: 10.1146/annurev.micro.54.1.681. [DOI] [PubMed] [Google Scholar]
- 55.Jensen RB, Gerdes K. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. Embo J. 1999;18:4076–4084. doi: 10.1093/emboj/18.14.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen RB, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR–ParC complex. J Mol Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- 57••.van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. Embo J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This paper reports the crystal structure of ParM and describes the striking structural similarities between ParM and actin.
- 58.Rothfield L. New insights into the developmental history of the bacterial cell division site. J Bacteriol. 2003;185:1125–1127. doi: 10.1128/JB.185.4.1125-1127.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebersbach G, Gerdes K. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc Natl Acad Sci USA. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.Autret S, Errington J. A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis. Mol Microbiol. 2003;47:159–169. doi: 10.1046/j.1365-2958.2003.03264.x. [DOI] [PubMed] [Google Scholar]
- Here the authors clarify the controversial localization pattern of Soj (chromosomal versus polar). They also identify a novel role for MinD, a protein that spatially regulates cell division, in regulating the dynamic localization of Soj, demonstrating an intriguing link between the cell division and chromosome segregation machineries.
- 61.Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 63.Godfrin-Estevenon AM, Pasta F, Lane D. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol Microbiol. 2002;43:39–49. doi: 10.1046/j.1365-2958.2002.02735.x. [DOI] [PubMed] [Google Scholar]
- 64.Ireton K, Gunther NW, 4th, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J Bacteriol. 2000;182:1313–1320. doi: 10.1128/jb.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiol. 2002;148:537–548. doi: 10.1099/00221287-148-2-537. [DOI] [PubMed] [Google Scholar]
- 67••.Lee PS, Lin DC, Moriya S, Grossman AD. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J Bacteriol. 2003;185:1326–1337. doi: 10.1128/JB.185.4.1326-1337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This paper suggests a role for Spo0J in the regulation of chromosome replication, and also demonstrates that the Spo0J binding site is not sufficient to convey positional information on new regions of the chromosome.
- 68.Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 69.Ryter A, Jacob F. Segregation of the nuclei during growth and germination of B. subtilis. C R Acad Sci Hebd Seances Acad Sci D. 1967;264:2254–2256. [PubMed] [Google Scholar]
- 70.Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- 71.Pogliano J, Sharp MD, Pogliano K. Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J Bacteriol. 2002;184:1743–1749. doi: 10.1128/JB.184.4.1743-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 73.Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002;295:137–139. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu LJ, Errington J. A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. Embo J. 2002;21:4001–4011. doi: 10.1093/emboj/cdf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kadoya R, Hassan AK, Kasahara Y, Ogasawara N, Moriya S. Two separate DNA sequences within oriC participate in accurate chromosome segregation in Bacillus subtilis. Mol Microbiol. 2002;45:73–87. doi: 10.1046/j.1365-2958.2002.03016.x. [DOI] [PubMed] [Google Scholar]
- 76••.Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- See annotation for [77]
- 77••.Wu LJ, Errington J. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49:1463–1475. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
- Papers [76,77] report the identification of a sporulation-specific protein necessary for the dramatic change in chromosome architecture which occurs at the initiation of sporulation, when the chromosome becomes anchored to both cell poles, forming an elongated structure known as the axial filament. Importantly, localization of this protein (RacA) requires DivIVA, which could explain why the latter protein has two separable roles in cell division and in chromosome segregation [80].In addition, evidence is provided [77] that the DivIVA/RacA and Soj–Spo0J systems act in a concerted fashion to target the chromosomal origin to the cell pole.
- 78.Cervin MA, Spiegelman GB, Raether B, Ohlsen K, Perego M, Hoch JA. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol. 1998;29:85–95. doi: 10.1046/j.1365-2958.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 79.Figge RM, Easter J, Gober JW. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol Microbiol. 2003;47:1225–1237. doi: 10.1046/j.1365-2958.2003.03367.x. [DOI] [PubMed] [Google Scholar]
- 80.Thomaides HB, Freeman M, El Karoui M, Errington J. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 2001;15:1662–1673. doi: 10.1101/gad.197501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81•.Sharp MD, Pogliano K. MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. Embo J. 2002;21:6267–6274. doi: 10.1093/emboj/cdf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This paper reveals an additional role for the MinCD heterodimer, which spatially regulates cell division during vegetative growth, in determining the polarity of chromosome segregation during sporulation.
- 82.Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]