Abstract
Heparin (HP), an important anticoagulant polysaccharide, is produced in a complex biosynthetic pathway in connective tissue-type mast cells. Both the structure and size of HP are critical factors determining the anticoagulation activity. A murine mastocytoma (MST) cell line was used as a model system to gain insight into this pathway. As reported, MST cells produce a highly sulfated HP-like polysaccharide that lacks anticoagulant activity (Montgomery RI, Lidholt K, Flay NW, Liang J, Vertel B, Lindahl U, Esko JD. 1992. Stable heparin-producing cell lines derived from the Furth murine mastocytoma. Proc Natl Acad Sci USA 89:11327–11331). Here, we show that transfection of MST cells with a retroviral vector containing heparan sulfate 3-O-sulfotransferase-1 (Hs3st1) restores anticoagulant activity. The MST lines express N-acetylglucosamine N-deacetylase/N-sulfotransferase-1, uronosyl 2-O-sulfotransferase and glucosaminyl 6-O-sulfotransferase-1, which are sufficient to make the highly sulfated HP. Overexpression of Hs3st1 in MST-10H cells resulted in a change in the composition of heparan sulfate (HS)/HP and CS/dermatan sulfate (DS) glycosaminoglycans. The cell-associated HS/HP closely resembles HP with 3-O-sulfo group-containing glucosamine residues and shows anticoagulant activity. This study contributes toward a better understanding of the HP biosynthetic pathway with the goal of providing tools to better control the biosynthesis of HP chains with different structures and activities.
Keywords: heparan sulfate, heparin, mastocytoma, proteoglycan, sulfotransferase
Introduction
Heparin (HP) is a widely used therapeutic anticoagulant (Linhardt 2003). Consequently, it represents an important biosynthetic target. HP is a highly sulfated form of the ubiquitous glycosaminoglycan (GAG), heparan sulfate (HS), and is biosynthesized in the Golgi by a common pathway (Esko et al. 2009). Biosynthesis begins in the endoplasmic reticulum where xylose is covalently attached to the serine residue of a core protein through the action of a xylosyltransferase. Next, two galactose residues are added by galactosyltransferases to the xylose, followed by glucuronic acid (GlcA) addition by glucuronosyltransferase I. ExtL3 and possibly ExtL2 add the first N-acetylglucosamine residue, which is subsequently extended by a copolymerase complex of Ext-1 and Ext-2 (Kobayashi et al. 2000; Presto et al. 2008). The growing chain is further modified through the introduction of sulfo groups in various positions and epimerization of d-GlcA into l-iduronic acid (IdoA). These processes are catalyzed by sulfotransferases (N-acetylglucosamine N-deacetylase-N-sulfotransferase(s) (Ndst), uronyl 2-O-sulfotransferase (Hs2st), glucosaminyl 6-O-sulfotransferase(s) (Hs6st) and glucosaminyl 3-O-sulfotransferase(s) (Hs3st)) and the glucuronyl C5-epimerase (Glce), which converts GlcA into IdoA. The HS chain is highly heterogeneous, due to incomplete modifications, resulting in highly modified domains that alternate with less highly modified domains. HP has a somewhat simpler structure than HS as it is highly modified and contains a large percentage of trisulfated disaccharide (TriS, →α-l-IdoA2S→α-D-GlcNS6S→) repeating units (Esko et al. 2009). Although the enzymes and their position within the HS biosynthetic pathway are well known, the biosynthetic production of a clinically acceptable, pharmaceutical HP represents a major metabolic engineering challenge.
While HS is largely a resident of the extracellular matrix or external cell membrane (Hassell et al. 1980; Bernfield et al. 1999), connective tissue mast cells naturally produce HP and store it intracellularly in granules (Katz et al. 1986; Wang and Kovanen 1999). Thus, mast cells are useful in understanding HP biosynthesis and the biological role of HP. However, isolation, propagation and maintenance of primary connective tissue mast cells are extremely challenging. These issues have made research into the role of mast cells in HP production problematic. Primary mast cells have mainly been used to study various aspects of inflammatory processes (Young et al. 1987; Theoharides and Cochrane 2004). Mice deficient in the biosynthetic enzyme N-deacetylase-N-sulfotransferase isoform 2 (Ndst2 isoform) show no obvious pathological phenotype but produce mast cells with an undersulfated HP, having a structure more closely resembling that of HS (Forsberg et al. 1999). Mast cells deficient in Ndst1 produce higher quantities of HP with higher level of sulfation (Dagalv et al. 2011). A study using mutant mouse mast cells deficient in the biosynthetic enzyme Glce highlighted its importance for the subsequent introduction of O-sulfo groups into HP (Feyerabend et al. 2006). An immortalized mast cell line, C57.1, expresses the transcript for Hs3st (Shworak et al. 1997), but little else has been established regarding the HP biosynthetic pathway in this cell line.
The Furth murine mastocytoma (MST) was one of the earliest models used to study HP biosynthesis (Furth et al. 1957; Hurst et al. 1978; Uhlin-Hansen et al. 1997). Cells derived from the tumor express the proteoglycan (PG), serglycin, which carries either multiple HP chains or, to a lesser degree, chondroitin sulfate (CS)-E GAG chains (Lidholt et al. 1995). In 1992, stable clonal cell lines (MST) were isolated from a tumor cell suspension (Montgomery et al. 1992). The HP chain produced by MST cells has a structure similar to that of pharmaceutical HP except that it lacks 3,6-O-sulfoglucosamine residues required for anticoagulant activity, which is essential for the pharmacological function of HP (Montgomery et al. 1992). It has also been shown that MST cell GAGs are associated with cytoplasmic granules (Montgomery et al. 1992).
MST cells and their various subclones are important models for elucidation of the HP biosynthetic pathway. In the current study, MST cells and MST-10H cells (an MST clone into which heparan sulfate 3-O-sulfotransferase-1 (Hs3st1) had been stably transfected) were used to gain insight into various aspects of HP biosynthesis with the ultimate aim of producing a pharmaceutical anticoagulant HP from cultured mammalian cells. The transcript levels of the GAG chain polymerization and modification enzymes were examined. The effect that modification of the HP biosynthetic pathway in MST-10H cells produced on the structure and activity of the newly biosynthesized HP chains was also studied.
Results and discussion
HP is a widely used and important therapeutic anticoagulant (Linhardt 2003). Applications include its use during heart surgery in cardiopulmonary bypass, in hemodialysis and treatment of atrial fibrillation and as a general anticoagulant during surgery (Doty et al. 1979; Langenecker et al. 1994; Billett et al. 2010). HP is biosynthesized in the same pathway as HS (Esko and Selleck 2002). Although the individual steps in the pathway are well established, it has been difficult to understand the best way to modify this pathway to produce HP from non-animal sources with the structure and activity of pharmaceutical HP. This limitation was clearly demonstrated using a system engineered to produce HP in Chinese hamster ovary (CHO) cells expressing two crucial enzymes of the HP biosynthetic pathway, Ndst2 and Hs3st1. The GAG chains expressed by these cells displayed both a low level of TriS disaccharide and low anticoagulant activity (Baik et al. 2012). To gain further insight into the regulation of HP production, we examined the HP biosynthetic pathway in mast cells, the natural producers of HP, with the aim of applying this knowledge to HP production through metabolic engineering. The use of primary mast cells in this endeavor is complicated by difficulties associated with their isolation and propagation. Therefore, we used MST cells, which produce HP that is comparable in structure to therapeutic HP (Montgomery et al. 1992), and do not have the same drawbacks as primary mast cells. The MST-10H cell line was engineered by transfecting the Hs3st1 gene into MST cells.
MST cells express sulfated GAGs
MST and MST-10H cells grow as aggregates in static suspension conditions (Supplementary data, Figure S1A) (Montgomery et al. 1992). Staining with the positively charged stain ruthenium red was performed to show that MST and MST-10H cells expressed negatively charged, sulfated GAGs (Supplementary data, Figure S1B). Staining confirmed that both cell lines contain sulfated GAGs.
Expression profile of HP biosynthetic enzymes in MST and MST-10H cells
Since MST cells do not produce anticoagulant HP, we used reverse transcription–polymerase chain reaction (RT–PCR) to determine whether MST cells express Hs3st1 (the critical enzyme required for anticoagulant HP production) at transcript level. Although MST cells expressed Hs3st1 mRNA, the protein product was not detected by western blotting. MST-10H cells expressed not only Hs3st1 mRNA, as expected, but also the Hs3st1 protein as detected by western blotting (Figure 1). The mRNA levels of other HP biosynthetic enzymes in MST and MST-10H cells were also examined by RT–PCR (Table I and Supplementary data, Figure S2). Transcripts for all of the biosynthetic enzymes required for HP synthesis were detected in MST and MST-10H cells.
Fig. 1.
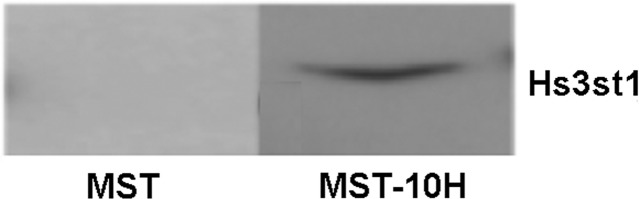
Western blot analysis of MST and MST-10H cells for the presence of 3-O-sulfotransferase 1. Expression of Hs3st1 protein is not detectable in parent MST cells, whereas it is detectable in MST-10H cells, which have been transfected with Hs3st1 gene. Note that this figure is a composite of two gels used in order to examine a much higher concentration of extract from MST cells to ensure that Hs3st1 protein was below the limit of detection by western blotting.
Table I.
Expression of HS/HP biosynthetic enzymes in MST and MST-10H cells
| Cell type | Ext1 | Ext2 | Ndst1 | Ndst2 | Glce | Hs2st | Hs6st1 | Hs6st2 | Hs6st3 | Hs3st1 | Hs3st5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT–PCR | MST | + | + | + | + | + | + | + | + | + | + | ?a |
| MST-10H | + | + | + | + | + | + | + | + | + | + | ? |
Transcripts for HS/HP biosynthetic enzymes were detected in both MST and MST-10H cells.
aRT–PCR of Hs3st5 from MST and MST-10H cells yielded a band of incorrect size.
Intracellular localization of HP biosynthetic enzymes
Cellular localization of the key HP biosynthetic enzymes, Ndst2, Hs3st1, Hs6st1 and Hs2st1, was analyzed using immunofluorescence (Supplementary data, Figure S3). The 58 kDa Golgi protein was used as a marker for Golgi membranes in the cells. The marker protein was localized in intact MST and MST-10H cells juxtaposed with the nucleus, thereby confirming its Golgi association. Ndst2, Hs6st1 and Hs2st1 were co-localized with the Golgi marker, suggesting their Golgi association in MST cells. Hs3st1 protein was detected at a basal level in MST cells with a substantially increased level in MST-10H cells. In MST-10H cells, Ndst2, Hs3st1, Hs6st1 and Hs2st1 enzymes were co-localized with 58 kDa Golgi protein.
In summary, in the parental MST cell line, Hs3st1 was undetectable using western blotting. However, in the MST-10H cell line prepared by transfection with the Hs3st1 gene, this protein was clearly detected by western blotting. Using immunofluorescence measurements, the signal for the Hs3st1 protein was significantly increased in MST-10H cells.
Expression of HP core proteins in MST and MST-10H cells
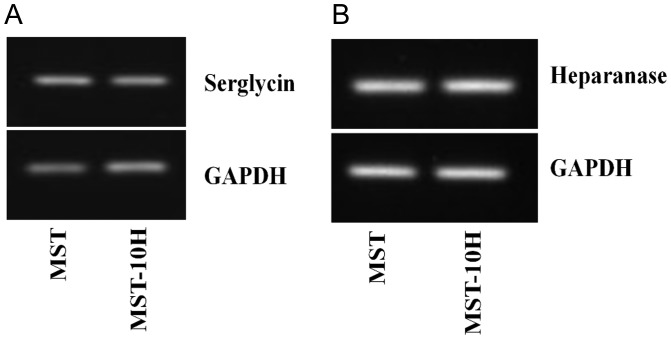
Serglycin is an HP core protein that has been shown to be expressed in mouse mastocytoma cells (Lidholt et al. 1995). Its expression in MST and MST-10H was confirmed by RT–PCR (Figure 2A).
Fig. 2.
Detection of serglycin core protein (A) and heparanase (B) in MST and MST-10H cells by RT–PCR. Transcripts for serglycin and heparanase were detected in both MST and MST-10H cells.
HP processing by heparanase in MST and MST-10H cells
The action of heparanase, a lysosomal endo-β-glucuronidase, upon HP chains is an important step in HP bioprocessing (Ogren and Lindahl 1971). The biosynthesis of GAG chains takes place in the Golgi on the serglycin core protein before the HP PG is stored in cytoplasmic granules (Kolset and Pejler 2011). When mast cells degranulate or when pharmaceutical HP is purified from mast cell-rich tissues (i.e. porcine intestines and bovine lungs), the resulting HP GAG chains have a considerably reduced molecular weight (MW ∼ 15,000) (Linhardt and Gunay 1999). The details of the final processing of HP, such as whether heparanase acts on the initial packaging of HP in the mast cell granules or during degranulation, are not known. We confirmed the expression of heparanase in both MST and MST-10H cells by RT–PCR (Figure 2B). The average MW of HP isolated from MST and MST-10H cells was ∼10–12 kDa, consistent with the idea that the chains were processed by endogenous heparanase.
HS/HP disaccharide structure in MST and MST-10H cells
GAG chains were isolated, digested with HP and chondroitin lyases and the resultant disaccharides were collected and analyzed by liquid chromatography-mass spectrometry (LC-MS) to establish the structure of the HS/HP chains synthesized by MST and MST-10H cells (Table II). HP is comprised of eight disaccharides that can be generated through exhaustive treatment with HP lyases I, II and III and detected by LC-MS. HS/HP chains with sites having 3-O-sulfo groups are resistant at these sites toward HP lyases, and do not afford disaccharide products (Xiao et al. 2011). The levels of HS/HP from the cell pellet fraction and from cell culture media were assessed. Bovine lung HP was used as a positive control. The total quantity of total GAG isolated from the MST cell pellet was 1.9 µg/107 cells, and MST culture media was 8.1 µg/107 cells. Most of the GAG coming from the MST cell pellet was HP/HS, while most of that coming from the cell media was CS/DS. We observed 1.8 μg HS/HP/107 cells with 86% of the HP/HS found in the cell pellet, comparable with the reported values of ∼0.4 μg HS/HP/106 cells with ∼85% of the HP/HS GAG in the intracellular granules (Montgomery et al. 1992).
Table II.
Composition of HS/HP isolated from MST and MST-10H cells
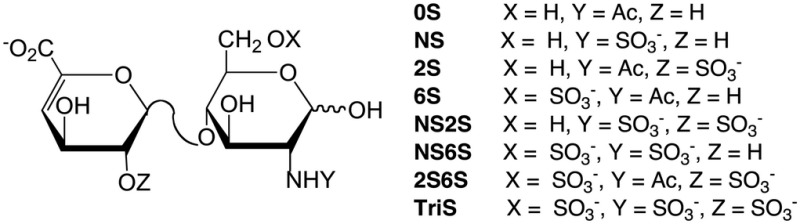 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Total amount of GAG (µg/107 cells) | HS/HP disaccharides (wt. %)a |
Ratio of HS/HP to CS/DS | |||||||
| 0S | NS | 2S | 6S | NS2S | NS6S | 2S6S | TriS | |||
| MST cell pellet | 1.9 | 6.5 | 14.2 | 1.2 | 8.9 | 9.1 | 18.3 | n/db | 41.8 | 14:1 |
| MST-10H cell pellet | 6.1 | 1.8 | 7.2 | 0.7 | 2.1 | 4.3 | 20.6 | n/d | 63.3 | 4:1 |
| MST cell media | 8.1 | 6.9 | 10.0 | 20.3 | 3.9 | 10.5 | 17.5 | n/d | 30.8 | 1:27 |
| MST-10H cell media | 49.5 | 12.3 | 11.7 | 10.5 | 4.5 | 11.4 | 18.8 | n/d | 30.8 | 1:32 |
| HP (bovine lung) | – | 0.1 | 0.3 | 0.2 | 0.1 | 10.6 | 6.7 | n/d | 81.9 | – |
aData an average of two replicate analyses.
bThe 2S6S disaccharide structure was not detected in any of the samples.
The major disaccharide comprising HP from bovine lungs is TriS, measured at 81.9% (see Table II for structure of HP/HS disaccharides). In the MST cell pellet, all disaccharides were detected, except for 2S6S, with TriS being the major disaccharide (41.8%). Other disaccharides include NS6S 18.3%, NS2S 9.1%, NS 14.2%, 6S 8.9%, 2S 1.2% and 0S 6.5%. The HS/HP isolated from culture media of MST cells was composed of TriS 30.8%, 2S 20.3%, NS6S 17.5%, NS2S 10.5%, NS 10%, 6S 3.9%, 0S 6.9%. Trypan blue staining confirmed that the cells used in this sample were intact; hence, the GAGs in the media were secreted or shed from the cell surface. While much less HS/HP was found in the medium sample, the reduced level of TriS in the MST medium, compared with the cell pellet, suggests that the medium contained more HS and the cell pellet sample contained more HP. In the MST-10H cells, the amount of total GAG found in the culture medium was much greater (49.5 µg/107 cells) than in cells (6.1 µg/107 cells). Analysis of HS/HP, the major GAG component of the MST-10H cell pellet, showed a high level of TriS content (63.3%). Other disaccharides include NS6S 20.6%, NS2S 4.3%, NS 7.2%, 6S 2.1%, 2S 0.7% and 0S 1.8%. Again, while HS/HP represents a small portion of the GAG found in the culture medium of MST-10H cells, it consisted of TriS 30.8%, NS6S 18.8%, NS2S 11.4%, NS 11.7%, 6S 4.5%, 2S 10.5% and 0S 12.3%.
HP chains isolated from both MST and MST-10H cell lines more closely resemble pharmaceutical HP than HS (Linhardt and Gunay 1999) in that they all contain high levels of TriS disaccharide. MST-10H cells have a higher amount of TriS disaccharide, comparable with that of many commercial porcine mucosal HPs (Fu et al. 2013).
Since CS/DS and HS/HP share early stages of their biosynthetic pathways, we also determined the ratio between these classes of GAGs in MST and MST-10H cells. The quantity of HS/HP was higher than that of CS/DS in the cell pellet fraction from MST and MST-10H (14:1 and 4:1, respectively). The higher level of HS/HP in the cell pellet is consistent with the intracellular storage of HP in mast cells. In contrast, the CS/DS quantity was greater than HS/HP in culture media from both MST and MST-10H cells with the [HS/HP]/[CS/DS] ratio being 1:27 and 1:32, respectively. Questions regarding why the increase in the expression of HS/HP-modifying enzymes affects both the overall biosynthesis and the ratio between HS/HP and CS/DS production remain to be answered. Perhaps, there is a mechanism that allows the cells to sense the increase in the availability of the modifying enzymes and cells respond to that by increasing the bioproduction of GAGs. One could hypothesize that some form of feedback inhibition exists that is relieved by changing the sulfation state of the HS/HP chain. But since there is no increase in other enzymes except Hs3st1, cells might utilize the CS/DS pathway. As MST cells are normally HS/HP-producing cells, they could have a pool of CS/DS enzymes that are not being actively used. MST-10H cells could take advantage of that pool and use it to produce more CS/DS. This and other hypotheses need to be tested in the future in order to have a deeper comprehension of the GAG biosynthetic pathway in MST cells. Interestingly, we observed a similar result in CHO cells, where addition of two modification enzymes (NDST2 and Hs3st1), resulted in a significant increase in GAG production (Baik et al. 2012).
HP extracted with other GAGs from animal tissues, such as porcine intestines and bovine lungs, typically undergoes fractionation steps in which highly sulfated pharmaceutical HP chains are collected (Linhardt and Gunay 1999). Thus, we used similar steps during GAG purification where three different fractions of HS/HP were eluted with various salt concentrations and collected. Fraction I was eluted with 0.5 M NaCl and primarily contained nonsulfated precursor chains (heparosan) and under sulfated HS chains. Fraction II was eluted with 1 M NaCl and contained HS chains and HP chains. Fraction III was eluted with 2 M NaCl and contains primarily highly sulfated HP chains. The ratio between Fractions I–III was determined to be 1:9.5:42 for MST and 1:99:400 for MST-10H. Thus, the largest fraction of HS/HP GAG isolated from MST and MST-10H cells is the most highly sulfated HP fraction.
HP isolated from MST-10H cells contains tetrasaccharide structures comparable with those found within the AT-binding sites of pharmaceutical HP
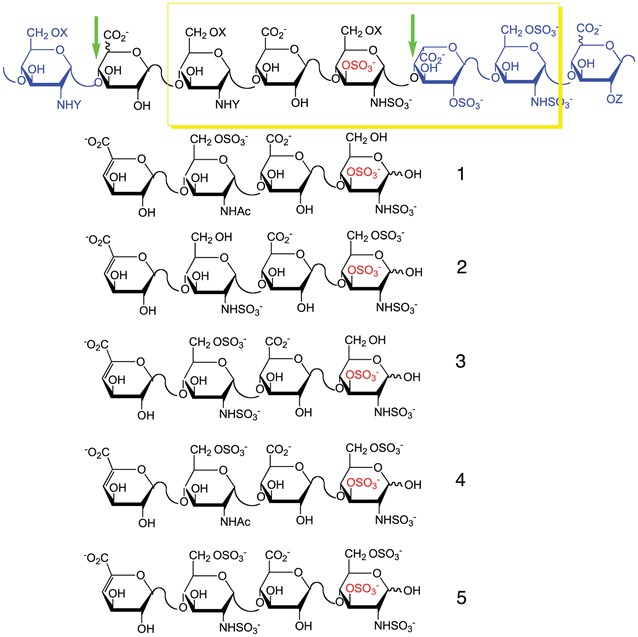
The anticoagulant activity of HP is determined by the presence of the AT-binding sites, which are pentasaccharides of defined sequence, containing a 3-O-sulfo group. This AT-binding site domain in HP (including the pentasaccharide (bold) and its surrounding residues, where S is sulfo and Ac is acetyl) consists of α-D-GlcN(Ac/S)(6S α/β1-4d-GlcA/l-IdoA→α-d-GlcN(Ac/S)(6S)→β-d-GlcA→α-d-GlcNS3S(6S)→α-l-IdoA2S→α-d-GlcN6S→α/β-d-GlcA/l-IdoA(2S (Shworak et al. 1997). The HP lyases used in our analyses, however, are able to cleave the linkages between GlcN(Ac/S)(6S)→GlcA/IdoA and GlcA/IdoA(2S) (Xiao et al. 2011) leading to tetrasaccharide fragments containing three of the five residues present in the AT-binding site (shown in italics). HP from MST and MST-10H cells was analyzed for the presence of pentasaccharide-binding sites by treatment with HP lyase II followed by tetrasaccharide analysis. This technique established the presence of five variations of the AT-binding sequence (Table III). Bovine lung HP, used as a positive control, also contains these 5 variations: 1 ΔUA-GlcNAc6S-GlcA-GlcNS3S (where ΔUA is 4-deoxy-α-L-threo-hex-4-eno-pyranosyluronic acid); 2 ΔUA-GlcNS-GlcA-GlcNS3S6S; 3 ΔUA-GlcNS6S-GlcA-GlcNS3S; 4 ΔUA-GlcNAc6S-GlcA-GlcNS3S6S and 5 ΔUA-GlcNS6S-GlcA-GlcNS3S6S. HP isolated from MST-10H cells expressed all of the above-mentioned sequences in ratios that were similar to those of bovine lung HP (Table III and Supplementary data, Figure S5). The most common tetrasaccharide structure was structure 2 (3.6 and 6.7% for bovine lung HP and HP from MST-10H cells, respectively), followed by 3 (3.1 and 2.1% for bovine lung HP and HP from MST-10H cells, respectively), 4 (1.4 and 3.4% for bovine lung HP and HP from MST-10H cells, respectively), 5 (1.2 and 0.4% for bovine lung HP and HP from MST-10H cells, respectively) and 1 (0.4 and 0.3% for bovine lung HP and HP from MST-10H cells, respectively) for both bovine lung HP and HP from MST-10H cells. None of these tetrasaccharides was detected in MST cells indicating the absence of AT-binding site pentasaccharides and the absence of 3-O-sulfo groups needed for AT-mediated anticoagulant activity. HP chains isolated from MST-10H cells have the same 3-O-sulfo group-containing tetrasaccharides, which are found in anticoagulant HP, suggesting that HP from MST-10H is functionally active as an anticoagulant.
Table III.
Distribution of 3-O-sulfo group containing tetrasaccharide structures in bovine lung HP, MST and MST-10H cells
 | ||||||
|---|---|---|---|---|---|---|
| Samples | 1a (%) | 2 (%) | 3 (%) | 4 (%) | 5 (%) | Total (%) |
| Bovine lung HP | 0.4c | 3.6 | 3.1 | 1.4 | 1.2 | 9.7 |
| MST-10H cells | 0.3 | 6.7 | 2.1 | 3.4 | 0.4 | 12.9 |
| MST cells | n/db | n/d | n/d | n/d | n/d | n/d |
The box surrounds the AT-binding site structural variants (where X and Z can be H or SO3− and Y can be Ac or SO3−). The portion of the structure containing 3-O-sulfo group (residues 2-5 counting from the left) corresponds to the tetrasaccharide structures identified and described in the text. The remainder of the structure corresponds to residues flanking the excised tetrasaccharide within the HP chain. The arrows show the HP lyase II cleavage sites.
aStructures of the sequences are presented above.
bn/d, not detected.
cAmount of each tetrasaccharide structure is given as a weight percent from total amount of both disaccharide and tetrasaccharides detected by LC-MS in a given sample.
Anticoagulant activity of HP chains isolated from MST and MST-10H cells
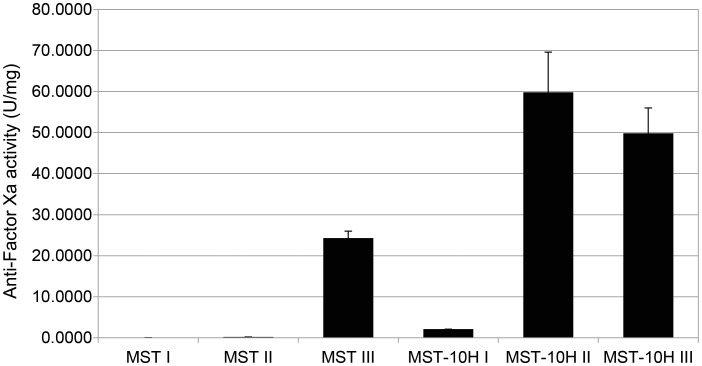
Anti-Factor Xa assay was used to determine the anticoagulant activity of HP chains isolated from MST and MST-10H cells. Preliminary evaluation of the cell pellet associated HS/HP GAGs from MST and MST-10H showed ∼2 and ∼40 U/mg of anti-factor Xa activity, respectively. Since commercial anticoagulant HP is generally purified through a fractionation step to enrich its anticoagulant HP chains from its low anticoagulant activity, less sulfated HS chains, we examined Fractions I–III obtained from each cell line for anti-Factor Xa activity. The results of these assays, shown in Figure 3, clearly demonstrate that the activity of the MST cell pellet GAGs resides in Fraction III, while that of the MST-10H cell pellet resides in both Fractions II and III. Neither MST nor MST-10H cell pellets showed much activity in the low salt fraction I. In contrast, MST-10H HP could be found in both Fraction II (1 M NaCl elution) and Fraction III at specific activities corresponding to 30–40% of those of pharmaceutical HP. These results are consistent with the substantial levels of 3-O-sulfo group-containing tetrasaccharides found in MST-10H HP.
Fig. 3.
Anti-factor Xa activity of MST and MST-10H HP fractions. Anticoagulant activity was detected only in Fraction III (highly sulfated fraction) of HS/HP isolated from MST cells. Increased anticoagulant activity was observed in Fraction II (moderately sulfated) and Fraction III (highly sulfated) of HS/HP isolated from MST-10H cells.
Conclusion
Together, these findings suggest that the HP biosynthetic pathway is a complex yet elegant system with interplay among a large array of enzymes. The expression of Ext1, Ndst2, Hs2st1 and Hs6st1 was sufficient to produce highly sulfated HP chains in MST and MST-10H cells. Overexpression of Hs3st1 was sufficient to result in anticoagulant activity for HP chains isolated from MST-10H cells. Yet in CHO-S cells transfected with Hs3st1, only a modest increase was detected in the anticoagulant activity of HP chains (Baik et al. 2012). This research provides insight into the HP biosynthetic pathway that should assist in designing a successful strategy to metabolically engineer the production of a pharmaceutical HP molecule.
Materials and methods
MST and MST-10H cell culture
Mastocytoma cell line MST has been described (Montgomery et al. 1992). The mastocytoma cell line MST-10H was generated by transfection of MST cells with murine Hs3st1 using the retroviral vector pLXRN pseudotyped with VSV-G (Clonetech, Mountain View, CA). Cells were grown as aggregates in static suspension conditions in complete media containing Dulbecco's modified Eagle medium/F12 (Life Technologies, Grand Island, NY) and 15% fetal bovine serum (Life Technologies). In addition, culture media for MST-10H cells contained Geneticin® at a final concentration of 400 µg/mL. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Ruthenium red staining
Cells were smeared onto a microscope slide, air-dried and stained with 0.005% Ruthenium red (Sigma-Aldrich, St. Louis, MO) for 30 min at room temperature and examined using brightfield microscopy (Gustafson and Pihl 1967).
Total RNA isolation and qRT–PCR reactions
Total RNA was extracted from the MST and MST-10H cells using RNeasy mini kits (Qiagen, Germantown, MD) according to the manufacturer's instructions. The amount of RNA was assessed via ultraviolet (UV) on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). 60–90 ng of total RNA per sample was used in reverse transcription and polymerase chain reactions using a SuperScript III Platinum One-Step RT–PCR kit (Life Technologies). A list of primers is supplied (Supplementary data, Table SI). RT–PCR products were separated by agarose gel electrophoresis and visualized using ethidium bromide.
Protein isolation, quantification and immunoblotting
For total protein extraction, exponentially growing cells were lysed in Nonidet-P40 lysis buffer (Boston Bioproducts, Ashland, MA) on ice for 30 min in the presence of a cocktail of protease and phosphatase inhibitors (Thermo Fisher Scientific), which comprise AEBSF, aprotinin, bestatin, E-64, leupeptin and pepstatin A. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). Forty micrograms of total protein from each sample was loaded and separated on a 4–20% polyacrylamide gel (Thermo Fisher Scientific) at 150 V, with Tris-hydroxyethyl piperazineethanesulfonic acid-sodium dodecyl sulphate buffer used as the running buffer. Proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA), probed with relevant primary antibodies, and subsequently detected using the appropriate horseradish peroxidase (HPR)-conjugated secondary antibody with chemiluminescent (Super Signal West Pico enhanced chemiluminescence (ECL) substrate, Thermo Fisher Scientific) exposure on high-performance chemiluminescence film (Amersham Hyperfilm ECL, GE Healthcare). The following were the primary and secondary antibodies used: Anti-γ-tubulin (T3320, Sigma-Aldrich); anti-Ext1 (sc-11039, Santa Cruz Biotechnology, Santa Cruz, CA), anti-Ext2 (sc-11045, Santa Cruz Biotechnology), anti-Ndst2 (sc-16764, Santa Cruz Biotechnology), anti-Hs3st1 (sc-104313, Santa Cruz Biotechnology), anti-Hs6st1 (sc-109943, Santa Cruz Biotechnology), anti-Hs6st3 (sc-84308, Santa Cruz Biotechnology), anti-Glce (H00026035-B01P, Abnova, Taipei City, Taiwan); anti-58K Golgi protein (ab27043, Abcam, Boston, MA), goat anti-rabbit HRP-conjugated (31 460), goat anti-mouse HRP-conjugated (31430, Thermo Fisher Scientific); donkey anti-goat HRP-conjugated (sc-2020, Santa Cruz Biotechnology).
Immunocytochemistry
Cell suspensions were smeared on microscope slides (poly-L-lysine treated) and air-dried for 30 min. Slides were fixed with 4% paraformaldehyde (Thermo Fisher Scientific) for 10 min at room temperature and blocked with Dulbecco's phosphate-buffered saline (DPBS)-Triton 100-X solution supplemented with 5% bovine serum albumin (BSA) (Life Technologies). Slides were incubated at room temperature for 1 h with primary antibody diluted in DPBS-Triton X-100 solution supplemented with 1% BSA. After several washes with DPBS, slides were incubated with secondary antibody (A11001, goat anti-mouse Alexa Fluor 488, Life Technologies) diluted in DPBS-Triton X-100 solution supplemented with 1% BSA at room temperature for 1 h. Cells were washed several times with DPBS. ProLong Gold antifade reagent containing 4′,6-diamidino-2-phenylindole stain was added to each slide and covered with a cover slip and incubated overnight at room temperature in the dark. The following day, slides were sealed and analyzed with a Zeiss 510 Meta multiphoton confocal microscope. The following were the primary antibodies used: Anti-Hs6st1 (ab106195, Abcam), anti-Hs2st (ab108541, Abcam), anti-Ndst2 (AP5759b, Abgent) and anti-58K Golgi protein (ab23932, Abcam).
Isolation, purification and enzymatic depolymerization of GAGs
Isolation and purification from mastocytoma cells relied on established techniques (Zhang et al. 2006; Yang et al. 2011). Briefly, the cell samples were individually subjected to proteolysis at 55°C with 10% (w/v) of actinase E (20 mg/mL in HPLC grade water, Kaken Biochemicals, Tokyo, Japan) for 20 h. After proteolysis, particulates were removed from the resulting solutions by passing each through a 0.22-μm membrane syringe filter. Samples were then concentrated using Microcon YM-10 centrifugal filter units (10 kDa MW cutoff, Millipore) by centrifugation at 12,000 × g and washed with 15 mL of distilled water to remove peptides. The retentate was collected and lyophilized. Samples were dissolved in 0.5 mL of 8 M urea containing 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (pH 8.3). A Vivapure Mini Q H spin column (Viva science, Edgewood, NJ) was prepared by equilibrating with 200 μL of 8 M urea containing 2% CHAPS (pH 8.3). To remove any remaining proteins, the clarified, filtered samples were loaded onto and run through the equilibrated Vivapure Mini Q H spin columns under centrifugal force (700 × g). The columns were then washed with 200 μL of 8 M urea containing 2% CHAPS at pH 8.3, followed by five washes with 200 μL of 200 mM NaCl. GAGs were released from the spin column by washing three-times with 50 μL of 2.7 M NaCl and desalted using YM-10 spin columns. Elution with 0.5, 1 and 2 M of NaCl was done and separate fractions were collected to determine the ratio of nonsulfated, moderately sulfated and highly sulfated GAGs. Finally, the GAGs were lyophilized.
The recovered GAGs were next exhaustively depolymerized using polysaccharide lyases. Chondroitin lyase ABC (5 m-units) and chondroitin lyase ACII (2 m-units) in 10 μL of 0.1% BSA were added to an ∼5 μg GAG sample in 25 μL of distilled water and incubated at 37°C for 10 h. The enzymatic products were recovered by centrifugal filtration at 13,000 × g. CS/DS disaccharides that passed through the filter were freeze-dried for LC-MS analysis. GAGs remaining in the retentate were collected by reversing the filter and centrifuging at 13,000 × g, followed by incubation with 10 m-units of HP lyases I–III at 37°C for 10 h. The products were recovered by centrifugal filtration using a YM-10 spin column, and the disaccharides were collected in the flow-through and freeze-dried. Cloning, overexpression in Escherichia coli and purification of the recombinant HP lyase I (EC 4.2.2.7), HP lyase II (no EC assigned) and HP lyase III (EC 4.2.2.8) from Flavobacterium heparinum were all performed as described (Yoshida et al. 2002; Shaya et al. 2006).
Derivatization of unsaturated disaccharides with 2-aminoacridone (AMAC)
The freeze-dried biological sample containing GAG-derived disaccharides (∼5 μg) or a mixture of 17 disaccharide standards (5 μg/per each disaccharide or 0.5 nmol/per each disaccharide) was added to 10 μL of 0.1 M 2-aminoacridone (AMAC) solution in acetic acid (AcOH)/dimethyl sulfoxide (DMSO) (3:17, v/v) and mixed by vortexing for 5 min. Next, 10 μL of 1 M NaBH3CN was added to the reaction mixture and incubated at 45°C for 4 h (Kitagawa et al. 1995). Finally, the AMAC-tagged disaccharide mixtures were diluted to various concentrations (0.5–100 ng) using 50% (v/v) aqueous DMSO, and LC-MS analysis was performed.
LC-MS disaccharide composition analysis of HS/HP
LC-MS analyses were performed on an Agilent 1200 LC/MSD instrument (Agilent Technologies, Inc. Wilmington, DE) equipped with a 6300 ion trap and a binary pump. The column used was a Poroshell 120 C18 column (2.1 × 150 mm, 2.7 μm, Agilent) at 45°C. Eluent A was 80 mM ammonium acetate solution and eluent B was methanol. Solution A and 15% solution B were flowed (150 μL/min) through the column for 5 min followed by a linear gradients from 15 to 30% solution B from 5 to 30 min. The column effluent entered the electrospray ionization (ESI)-MS source for continuous detection by MS. The electrospray interface was set in negative ionization mode with a skimmer potential of −40.0 V, a capillary exit of −40.0 V and a source temperature of 350°C, to obtain the maximum abundance of the ions in a full-scan spectrum (150–1200 Da). Nitrogen (8 L/min, 40 psi) was used as a drying and nebulizing gas (Yang et al. 2012).
Quantification analysis of AMAC-labeled disaccharides was performed using calibration curves constructed by separation of increasing amounts of unsaturated disaccharide standards (0.1, 0.5, 1, 5, 10, 20, 50, 100 ng/each disaccharide or 0.02, 0.03, 0.05, 0.1, 0.2, 0.3 nM/each disaccharide). Linearity was assessed based on the amount of disaccharide and peak intensity in UV255 nm, MS total ion chromatography and extract ion chromatography.
Tetrasaccharide mapping
For tetrasaccharide analysis, the HP lyase II (40 mU in 20 μL of 25 mM Tris, 500 mM NaCl, 300 mM imidazole buffer (pH 7.4)) was added to 50–100 μg of GAG sample in 40 μL of distilled water and incubated at 35°C for 10 h. The products of enzymatic degradation were freeze-dried for further LC-MS analysis.
LC-MS analyses were performed on an Agilent 1200 LC/MSD instrument (Agilent Technologies, Inc. Wilmington, DE) equipped with a 6300 ion trap and a binary pump followed by a UV detector equipped with a high-pressure cell. The column used was a Poroshell 120 C18 column (2.1 × 100 mm, 2.7 μm, Agilent). Eluent A was water/acetonitrile (85:15) v/v and eluent B was water/acetonitrile (35:65) v/v. Both eluents contained 12 mM tributylamine and 38 mM NH4OAc with pH adjusted to 6.5 with acetic acid. 100% Solution A was applied for 2 min followed by a linear gradient from 2 to 40 min (0–30% solution B) all at a flow rate of 150 μL/min. The column effluent entered the source of the ESI-MS for continuous detection by MS. The electrospray interface was set in negative ionization mode with a skimmer potential of −40.0 V, a capillary exit of −40.0 V and a source temperature of 350°C, to obtain the maximum abundance of the ions in a full-scan spectrum (200–1500 Da). Nitrogen (8 l/min, 40 psi) was used as a drying and nebulizing gas.
Anti-factor Xa assay
Antithrombin and factor Xa were from the HemosIL Heparin kit (Instrumentation Laboratory, Bedford, MA) and reconstituted according to the manufacturer's instructions. The chromogenic substrate, Arg-Gly-Arg-pNA, was either from the HemosIL Heparin kit or from Hyphen Biomed (Neuville-Sur-Oise, France). Tris, NaCl and HP were reagent grade or better from Sigma Chemical Co. (St. Louis, MO).
In a 96-well plate (Costar), 100 μL reactions in 0.9% NaCl 50 mM Tris pH 8.4 [Tris-buffered saline (TBS)] containing 5 μL (5 mIU) antithrombin solution, 2 µL (0.0272 nkat) factor Xa solution and the HP samples were covered and incubated for 1 h on a rocking platform. The buffer from the HemosIL kit was not used since detergents present caused interference in this assay method. Twenty microliters of a 0.75 mg/mL solution of the chromogenic substrate in TBS was then added to each well. The rate of hydrolysis of the substrate was monitored by measuring the absorbance at 410 nm over the course of 1 h in a Tecan Infinite M200 plate reader operating at its maximum sampling rate. The rate curves were linearized by taking into account the decreasing substrate concentration as the reaction progressed. The concentrations of standard HP samples were then plotted against the reciprocal of their rates of hydrolysis and the slope used to determine analyte concentrations.
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
This study was supported by the National Institutes of Health in the form of grants HL62244, GM38060, HL094463, HL096972 (R.J.L.), GM090127 (S.S.), GM93131 and GM33063 (J.E.) and a research contract from Aventis (J.E.).
Conflict of interest
None declared.
Abbreviations
AT, antithrombin III; BSA, bovine serum albumin; CHO, Chinese hamster ovary; CS, chondroitin sulfate; DMSO, dimethyl sulfoxide; ESI, electrospray ionization; Ext, exostosin; GAG, glycosaminoglycan; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; GlcNS, N-sulfoglucosamine; HP, heparin; HS, heparan sulfate; Hs2st, heparan sulfate 2-O-sulfotransferase; Hs3st, heparan sulfate 3-O-sulfotransferase; Hs3st1, heparan sulfate 3-O-sulfotransferase-1; Hs6st, heparan sulfate 6-O-sulfotransferase; IdoA, l-iduronic acid; LC, liquid chromatography; MS, mass spectrometry; MST, murine mastocytoma; MW, molecular weight; Ndst, N-deacetylase/N-sulfotransferase; PG, proteoglycan; RT–PCR, reverse transcription–polymerase chain reaction; SDS, sodium dodecyl sulphate.
Supplementary Material
Acknowledgements
The authors are grateful to Eric R. Gamache for proofreading and offering helpful suggestions for this manuscript.
References
- Baik JY, Gasimli L, Yang B, Datta P, Zhang F, Glass CA, Esko JD, Linhardt RJ, Sharfstein ST. Metabolic engineering of Chinese hamster ovary cells: Towards a bioengineered heparin. Metabol Eng. 2012;14:81–90. doi: 10.1016/j.ymben.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Ann Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Billett HH, Scorziello BA, Giannattasio ER, Cohen HW. Low molecular weight heparin bridging for atrial fibrillation: Is VTE thromboprophylaxis the major benefit? J Thromb Thrombolysis. 2010;30:479–485. doi: 10.1007/s11239-010-0470-8. [DOI] [PubMed] [Google Scholar]
- Dagalv A, Holmborn K, Kjellen L, Abrink M. Lowered expression of heparan sulfate/heparin biosynthesis enzyme N-deacetylase/n-sulfotransferase 1 results in increased sulfation of mast cell heparin. J Biol Chem. 2011;286:44433–44440. doi: 10.1074/jbc.M111.303891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty DB, Knott HW, Hoyt JL, Koepke JA. Heparin dose for accurate anticoagulation in cardiac surgery. J Cardiovasc Surg. 1979;20:597–604. [PubMed] [Google Scholar]
- Esko JD, Kimata K, Lindahl U. Proteoglycans and sulfated glycosaminoglycans. In: Varki A., et al., editors. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press: 2009. pp. 229–248. [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Feyerabend TB, Li JP, Lindahl U, Rodewald HR. Heparan sulfate C5-epimerase is essential for heparin biosynthesis in mast cells. Nat Chem Biol. 2006;2:195–196. doi: 10.1038/nchembio777. [DOI] [PubMed] [Google Scholar]
- Forsberg E, Pejler G, Ringvall M, Lunderius C, Tomasini-Johansson B, Kusche-Gullberg M, Eriksson I, Ledin J, Hellman L, Kjellen L. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- Fu L, Li G, Yang B, Onishi A, Li L, Sun P, Zhang F, Linhardt RJ. Structural L. characterization of pharmaceutical heparins prepared from different animal tissues. J Pharm Sci. 2013;102:1447–1457. doi: 10.1002/jps.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth J, Hagen P, Hirsch EI. Transplantable mastocytoma in the mouse containing histamine, heparin, 5-hydroxy tryptamine. Proc Soc Exp Biol Med. 1957;95:824–828. doi: 10.3181/00379727-95-23375. [DOI] [PubMed] [Google Scholar]
- Gustafson GT, Pihl E. Staining of mast cell acid glycosaminoglycans in ultrathin sections by ruthenium red. Nature. 1967;216:697–698. doi: 10.1038/216697a0. [DOI] [PubMed] [Google Scholar]
- Hassell JR, Robey PG, Barrach HJ, Wilczek J, Rennard SI, Martin GR. Isolation of a heparan sulfate-containing proteoglycan from basement membrane. Proc Natl Acad Sci USA. 1980;77:4494–4498. doi: 10.1073/pnas.77.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst RE, Nakamura N, West SS. Isolation and characterization of glycosaminoglycans from the Furth murine mastocytoma. Prep Biochem. 1978;8:37–56. doi: 10.1080/00327487808068217. [DOI] [PubMed] [Google Scholar]
- Katz HR, Austen KF, Caterson B, Stevens RL. Secretory granules of heparin-containing rat serosal mast cells also possess highly sulfated chondroitin sulfate proteoglycans. J Biol Chem. 1986;261:13393–13396. [PubMed] [Google Scholar]
- Kitagawa H, Kinoshita A, Sugahara K. Microanalysis of glycosaminoglycan-derived disaccharides labeled with the fluorophore 2-aminoacridone by capillary electrophoresis and high-performance liquid chromatography. Anal Biochem. 1995;232:114–121. doi: 10.1006/abio.1995.9952. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Morimoto K, Shimizu T, Takahashi M, Kurosawa H, Shirasawa T. Association of EXT1 and EXT2, hereditary multiple exostoses gene products, in Golgi apparatus. Biochem Biophys Res Commun. 2000;268:860–867. doi: 10.1006/bbrc.2000.2219. [DOI] [PubMed] [Google Scholar]
- Kolset SO, Pejler G. Serglycin: A structural and functional chameleon with wide impact on immune cells. J Immunobiol. 2011;187:4927–4933. doi: 10.4049/jimmunol.1100806. [DOI] [PubMed] [Google Scholar]
- Langenecker SA, Felfernig M, Werba A, Mueller CM, Chiari A, Zimpfer M. Anticoagulation with prostacyclin and heparin during continuous venovenous hemofiltration. Crit Care Med. 1994;22:1774–1781. [PubMed] [Google Scholar]
- Lidholt K, Eriksson I, Kjellen L. Heparin proteoglycans synthesized by mouse mastocytoma contain chondroitin sulphate. Biochem J. 1995;311:233–238. doi: 10.1042/bj3110233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt RJ. Heparin: Structure and activity. J Med Chem. 2003;46:2551–2554. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Gunay NS. Production and chemical processing of low molecular weight heparins. Semin Thromb Hemost. 1999;25(Suppl 3):5–16. [PubMed] [Google Scholar]
- Montgomery RI, Lidholt K, Flay NW, Liang J, Vertel B, Lindahl U, Esko JD. Stable heparin-producing cell lines derived from the Furth murine mastocytoma. Proc Natl Acad Sci USA. 1992;89:11327–11331. doi: 10.1073/pnas.89.23.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogren S, Lindahl U. Degradation of heparin in mouse mastocytoma tissue. Biochem J. 1971;125:1119–1129. doi: 10.1042/bj1251119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci USA. 2008;105:4751–4756. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaya D, Tocilj A, Li Y, Myette J, Venkataraman G, Sasisekharan R, Cygler M. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J Biol Chem. 2006;281:15525–15535. doi: 10.1074/jbc.M512055200. [DOI] [PubMed] [Google Scholar]
- Shworak NW, Liu J, Fritze LM, Schwartz JJ, Zhang L, Logeart D, Rosenberg RD. Molecular cloning and expression of mouse and human cDNAs encoding heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J Biol Chem. 1997;272:28008–28019. doi: 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- Theoharides TC, Cochrane DE. Critical role of mast cells in inflammatory diseases and the effect of acute stress. J Neuroimmunol. 2004;146:1–12. doi: 10.1016/j.jneuroim.2003.10.041. [DOI] [PubMed] [Google Scholar]
- Uhlin-Hansen L, Kusche-Gullberg M, Berg E, Eriksson I, Kjellen L. Mouse mastocytoma cells synthesize undersulfated heparin and chondroitin sulfate in the presence of brefeldin A. J Biol Chem. 1997;272:3200–3206. doi: 10.1074/jbc.272.6.3200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kovanen PT. Heparin proteoglycans released from rat serosal mast cells inhibit proliferation of rat aortic smooth muscle cells in culture. Circ Res. 1999;84:74–83. doi: 10.1161/01.res.84.1.74. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Tappen BR, Ly M, Zhao W, Canova LP, Guan H, Linhardt RJ. Heparin mapping using heparin lyases and the generation of a novel low molecular weight heparin. J Med Chem. 2011;54:603–610. doi: 10.1021/jm101381k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Chang Y, Weyers AM, Sterner E, Linhardt RJ. Disaccharide analysis of glycosaminoglycan mixtures by ultra-high-performance liquid chromatography-mass spectrometry. J Chromatogr A. 2012;1225:91–98. doi: 10.1016/j.chroma.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Weyers A, Baik JY, Sterner E, Sharfstein S, Mousa SA, Zhang F, Dordick JS, Linhardt RJ. Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis. Anal Biochem. 2011;415:59–66. doi: 10.1016/j.ab.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E, Arakawa S, Matsunaga T, Toriumi S, Tokuyama S, Morikawa K, Tahara Y. Cloning, sequencing, and expression of the gene from bacillus circulans that codes for a heparinase that degrades both heparin and heparan sulfate. Biosci Biotechnol Biochem. 2002;66:1873–1879. doi: 10.1271/bbb.66.1873. [DOI] [PubMed] [Google Scholar]
- Young JD, Liu CC, Butler G, Cohn ZA, Galli SJ. Identification, purification, and characterization of a mast cell-associated cytolytic factor related to tumor necrosis factor. Proc Natl Acad Sci USA. 1987;84:9175–9179. doi: 10.1073/pnas.84.24.9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sun P, Munoz E, Chi L, Sakai S, Toida T, Zhang H, Mousa S, Linhardt RJ. Microscale isolation and analysis of heparin from plasma using an anion-exchange spin column. Anal Biochem. 2006;353:284–286. doi: 10.1016/j.ab.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.