Abstract
Increasing numbers of protein structures are solved each year, but many of these structures belong to proteins whose sequences are homologous to sequences in the Protein Data Bank. Nevertheless, the structures of homologous proteins belonging to the same family contain useful information because functionally important residues are expected to preserve physico-chemical, structural and energetic features. This information forms the basis of our method, which detects RNA-binding residues of a given RNA-binding protein as those residues that preserve physico-chemical, structural and energetic features in its homologs. Tests on 81 RNA-bound and 35 RNA-free protein structures showed that our method yields a higher fraction of true RNA-binding residues (higher precision) than two structure-based and two sequence-based machine-learning methods. Because the method requires no training data set and has no parameters, its precision does not degrade when applied to ‘novel’ protein sequences unlike methods that are parameterized for a given training data set. It was used to predict the ‘unknown’ RNA-binding residues in the C-terminal RNA-binding domain of human CPEB3. The two predicted residues, F430 and F474, were experimentally verified to bind RNA, in particular F430, whose mutation to alanine or asparagine nearly abolished RNA binding. The method has been implemented in a webserver called DR_bind1, which is freely available with no login requirement at http://drbind.limlab.ibms.sinica.edu.tw.
INTRODUCTION
Interactions between proteins and RNA play essential roles for life. For example, protein−RNA interactions mediate RNA metabolic processes such as splicing, polyadenylation, messenger RNA stability, localization and translation (1). Furthermore, many of these RNA-binding proteins are involved in human diseases (2) such as neurological disorders, e.g. TDP-43 (3), ATXN2 (4) and muscular atrophies [SMN (5)]. Consequently, identifying the key amino acid (aa) residues involved in RNA recognition is critical for understanding these important biological processes.
Several methods and servers have been developed to predict RNA-binding residues from the protein 1D sequence or 3D structure. Methods that predict RNA-binding residues using only the protein sequence generally employ machine-learning algorithms such as a neural network (6,7), a Naïve Bayes classifier (8–10), a support vector machine (11–19), random forest (20,21) or decision trees (C4.5 algorithm) (22). These algorithms usually employ aa physico-chemical properties, sequence conservation, the local sequence context, solvent accessibility and secondary structure. Publicly available web servers that implement sequence-based methods include RNABindR (8), Pprint (13), PRINTR (14), PiRaNhA (16), PRBR (21), RISP (23), BindN (11), BindN+ (17) and NAPS (22) for predicting RNA-binding residues. Compared to sequence-based methods, structure-based methods for predicting RNA-binding residues are far fewer (20,24,25) with only a few methods available as web servers, namely, KYG (26) and dRNA-3D (27). The predicted RNA-binding residues can be verified by measuring the RNA-binding affinities of mutant proteins. Hence for an experimentalist, high precision (i.e. high fraction of correctly predicted RNA-binding residues) would be more useful than predicting the entire protein−RNA interface correctly.
In this work, we present a structure-based detection method to identify the most likely RNA-binding residues rather than all RNA-binding and all nonbinding residues. The method is based on evolutionary and physical principles with the following rationale: RNA-binding residues generally possess electropositive atoms that interact with the RNA electronegative atoms or water oxygen atoms. In the absence of RNA or water, these RNA-binding residues would be in an unfavorable electrostatic environment due to the electrostatic repulsion among the electropositive atoms and would therefore be energetically unstable (24,28). On the other hand, RNA-binding residues within the same family are known to be highly conserved (29). They would be expected to preserve not only their physico-chemical features (i.e. aa type and solvent accessibility) but also their energetic features due to their critical functional roles. Hence, solvent-accessible residues that share the highest evolutionary conservation of aa type, as well as structural and energetic features within the same family are predicted to bind RNA. The method was tested on two nonredundant datasets, one containing 81 RNA-bound protein structures (dataset I) and the other with 35 RNA-free structures (dataset II). It was also tested on CPEB3, an important nucleocytoplasm-shuttling RNA-binding protein, and the predictions were experimentally verified. Since the method should work for other polyanions, it was also tested on a set of 83 DNA-bound protein structures taken from our previous work (30). The method, as described in the next section, has been implemented in a webserver called DR_bind1.
MATERIALS AND METHODS
Datasets
Dataset I
To create dataset I, all available ≤3 Å X-ray structures of RNA-bound proteins were obtained from the May 2012 release of the Protein Data Bank (PDB) (31). For protein structures belonging to the same class, architecture, topology and homologous (CATH) superfamily (32), the structure with the best resolution was selected as the representative one. If any of these representative proteins share >30% sequence identity, the protein with the longer sequence was kept, while the others were discarded. This yielded 81 RNA-bound protein structures with distinct CATH codes, which are listed alphabetically according to the PDB code in Supplementary Table S1. All these proteins have conservation data in the ConSurf-DB database (http://consurfdb.tau.ac.il/) (33).
Dataset II
Dataset II was derived from dataset I by searching each of the 81 RNA-bound proteins for proteins sharing ≥90% sequence identity with RNA-free structure(s) using the SAS database (http://www.ebi.ac.uk/thornton-srv/databases/sas/). The root-mean-square deviation of the Cα atoms (Cα-RMSD) in the RNA-free structure from those in the RNA-bound structure was computed using the SSAP program (34). If multiple RNA-free structures were found, we chose the structure with the largest Cα-RMSD as the representative one since the purpose of dataset II is to evaluate the effect of protein conformational changes on the RNA-binding residue prediction. This yielded 35 RNA-free structures that deviate from the respective RNA-bound structures with RMSDs ranging from 0.35 to 8.87 Å. Supplementary Table S1 lists these proteins along with their RMSDs and sequence identities between the RNA-bound and corresponding free proteins, which were computed using global alignment with ClustalW1.83 (35).
Searching for homologous proteins
The SAS database was used to search all sequences in the PDB that are homologous to each protein in dataset I/II. For proteins in dataset II, the homologous proteins found were excluded if their structures contain RNA. Since sequences corresponding to the RNA-bound and free protein structures share ≥90% sequence identity (see above), homologous proteins sharing ≥90% sequence identity were deemed to be similar and grouped together using CD-HIT (36), and the longest protein was selected as representative of that group. If a homologous protein representative shared <30% pairwise sequence identity with the target protein sequence in dataset I/II, it was excluded as proteins belonging to the same family generally exhibit pairwise residue identities ≥30% (37).
Definition of true RNA-binding residues
A residue was considered to bind RNA if it contains ≥1 nonhydrogen atoms within van der Waals contact (≤4.0 Å) or hydrogen-bonding distance (≤3.5 Å) to the nonhydrogen atom of its binding partner directly or indirectly via a bridging water molecule(s). The hydrogen bonds and van der Waals contacts were computed using HBPLUS (38).
Definition of solvent-accessible residues
An aa X is considered to be solvent accessible if the percent ratio of its relative solvent-accessible surface area is ≥15% (39) computed by NACCESS (40).
Electrostatic ranking of each residue
Given the 3D structure of a l-residue protein, all Asp/Glu residues were deprotonated, while Arg/Lys residues were protonated; His residues were protonated if both side chain nitrogen atoms were within hydrogen-bonding distance to an acceptor atom, or deprotonated if the side chain nitrogen was not within hydrogen-bonding distance of an acceptor atom. l mutant structures were generated by mutating each Ala, Asn, Asp, Cys, Gly, Ser, Thr or Val in the wild-type (wt) sequence to Asp− and the other residues to Glu− using SCWRL (41). To relieve bad contacts resulting from the sidechain replacement, each mutant structure i was energy minimized with heavy constraints on all heavy atoms, and the resulting structure was used to compute the gas-phase (ε = 1) electrostatic energy of the mutant (mut) protein relative to that of the wt protein (Eelecmut,i – Eelecwt). The corresponding difference in an ‘extended reference’ state, where the residues do not interact with one another, was computed as E′elecD/E – E′eleci. All energy calculations were performed using the AMBER (42) program with the all-hydrogen-atom AMBER force field (43). The change in the gas-phase electrostatic energy upon mutation of aa i to Asp−/Glu−, ΔΔEeleci, is given by:
| (1) |
A negative ΔΔEeleci means that residue i is electrostatically stabilized upon mutation to an Asp− /Glu− and would likely bind to the electronegative RNA atoms (see ‘Introduction’ section). Hence, residues with the top 10% most negative <ΔΔEelec>i values were assigned Rankele = 10, residues with the next 10% most negative <ΔΔEelec>i values were assigned Rankele = 9, while the least likely RNA-binding residues were assigned Rankele = 1.
Evolutionary ranking of each residue
For a given protein, the conservation score of residue i, Ci, was obtained from the ConSurf-DB database (29,44). The Ci score is an integer number ranging from 9 for a slowly evolving, conserved residue to 1 for a rapidly evolving, highly variable residue.
Cleft assignment of each residue
Given the 3D protein structure, the 10 largest clefts were found using SURFNET (45), where cleft 1 is the biggest and cleft 10 is the smallest. If any atom of a residue was assigned as a constituent of the cleft by the SURFNET program, then this residue was regarded as a component of the cleft. When atoms of a residue were assigned to two different clefts, the residue was assigned to the larger of the two clefts. Residues not in any of these 10 clefts were assigned to cleft 11.
Detecting RNA-binding residues
Given the structures of protein X and its homologs, RNA-binding residues were detected as follows: for each residue in protein X, the sum of Rankele and C was computed. Let Max denote the largest value of Rankele + C in protein X. Based on the structure of protein X, n residues that are solvent accessible with Rankele + C = Max were identified. If n is <3, we included m solvent-accessible residues in van der Waals contacts to these n residues with Rankele + C = Max – 1. If n + m is still <3, then Rankele + C was successively decreased by one until n + m is ≥ 3. Max was then redefined as the value of Rankele + C for which n + m is ≥ 3. Let N denote n or n + m.
Next, the structure of protein X was aligned with that of each homologous protein representative using the MASPCI program (46) to determine the correspondence between the N residues of protein X and the respective residues in the homologous proteins. N′ residues of the N residues of protein X were selected if their corresponding residues in any of the homologous proteins were also solvent accessible with Rankele + C ≥Max. If N′ = 0, then the original N residues of protein X were chosen. The N′ or N residues were grouped according to their cleft number, and the cleft containing the most residues was predicted to be the RNA-binding site. If two or more clefts contained the same number of residues, then the residues comprising these clefts were predicted to bind RNA.
Detecting RNA-binding residues in human CPEB3
The above RNA-binding residue method was used to predict the unknown RNA-binding residues in the C-terminal RNA-binding domain (RBD) of human CPEB3 (hCPEB3) using the NMR structure (2dnl-A) of hCPEB3 RNA recognition motif 1 (RRM1)-binding domain (residues 426–532). First, the SAS database was used to search all sequences in the PDB that were homologous to the 2dnl-A. This yielded three representative homologous proteins (1whw-A, 1wi8-A and 2dhg-A), which share 35%, 33% and 31% sequence identity with 2dnl-A, respectively.
Based on the 2dnl-A structure, residues P469 and F474 in the hCPEB3 RBD were found to be solvent accessible with a maximum Rankele + C value of 18: F474 has Rankele = 9 and C = 9, while P469 has Rankele = 10 and C = 8 (no residues have Rankele = 10 and C = 9). Since n = 2, we searched for solvent-exposed residues within van der Waals contacts of P469 and F474, and found two with Rankele + C = 17, namely, F430 with Rankele = 9 and C = 8 and D456 with Rankele = 10 and C = 7. Among F430, D456, P469 and F474, only two residues, F430 and F474, have corresponding residues in the homologous proteins that were also solvent accessible with Rankele + C ≥ 17. The residues corresponding to F430 in 1whw-A (F41) and 2dhg-A (F99) were both solvent exposed with Rankele = 10 and C = 7 or 8. The residues corresponding to F474 in 1whw-A (F83) and 2dhg-A (F141) were also solvent exposed with Rankele = 10 and C = 9. Hence, F430 in cleft #1 and F474 in cleft #8 in the hCPEB3 RBD were both predicted to bind RNA.
To compare with DR_bind1, two RNA-binding residues were also predicted using two structure-based methods, KYG (http://cib.cf.ocha.ac.jp/KYG/) (26) and OPRA (25), based on the 2dnl-A structure and two sequence-based methods, BindN+ (http://bioinfo.ggc.org/bindn+/) (17) and Pprint (http://www.imtech.res.in/raghava/pprint/index.html) (13) based on the 2dnl-A sequence. The two residues predicted to bind RNA are those with the most positive KYG, BindN+ or Pprint scores and the most negative OPRA values.
Performance evaluation
The performance of our method was evaluated by computing the numbers of (i) correctly predicted RNA-binding residues (TP), (ii) correctly predicted non-RNA-binding residues (TN), (iii) wrongly predicted RNA-binding residues (FP) and (iv) wrongly predicted non-RNA-binding residues (FN). These numbers were then used to compute the following performance measures:
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Verifying RNA-binding residues in CPEB3
To verify the RNA-binding residues predicted by DR_bind1 (F430, F474), KYG (R449, G432), OPRA (R449, R514), BindN+ (R427, S465) and Pprint (K460, D456), we constructed single alanine-substituted mutants (see Supplementary Methods) and tested the RNA-binding activity by UV-cross-linking RNA-binding assay and western blotting. Twenty microliter reactions containing 4 × 104 cpm of labeled RNA, 50 µg heparin, 1 µg yeast tRNA and 10 µl of 293T cell lysate were kept on ice for 10 min, and then irradiated with 1200 J of UV (254 nm) light for 10 min. The UV-cross-linked samples were treated with 200 ng of ribonuclease A at 37°C for 10 min and resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). The radioactive signals were monitored by the phosphorimager Typhoon FLA 4100 system (GE Healthcare). Two microliters of cell lysates mixed with 20 µl 1× Laemmli sample buffer were separated on SDS–PAGE and then transferred to PVDF membrane for western blotting using myc antibody. The immunoblotted signals, analyzed by the ImageJ software, represented the expression levels of various CPEB3 mutants. The normalized RNA-binding ability was calculated by dividing the specific RNA-binding signal (i.e. after subtracting the background signal in the mock-transfected lysate) with the expression level of mutant CPEB3.
RESULTS
Comparison with KYG, OPRA, BindN+ and Pprint using default settings
DR_bind1 was tested on 81 RNA-bound structures (dataset I, Table 1) as well as 35 unbound-bound RNA-binding protein structures (dataset II, Table 2) to assess the effect of protein conformational changes upon binding RNA. Using the same datasets, its performance was compared with the performance of two structure-based methods, KYG (26) and OPRA (25) using the default prediction mode, and two sequence-based methods, BindN+ (17) and Pprint (13) using the default specificity settings. These methods were chosen because they had been shown to outperform previous RNA-binding residue prediction methods (47) and were available for testing. Their results were compared with the results using DR_bind1 based on the dataset I structures in Table 1 and dataset II structures in Table 2.
Table 1.
Performance of DR_bind1 based on 81 RNA-bound protein structures compared to that of KYG, OPRA, BindN+ or Pprint using default settings
| DR_bind1 | KYG | OPRA | BindN+ | Pprint | |
|---|---|---|---|---|---|
| TP | 166 | 1820 | 1021 | 2235 | 2516 |
| FP | 75 | 2916 | 1018 | 1868 | 3534 |
| TN | 14 628 | 11 787 | 13 685 | 12 835 | 11 169 |
| FN | 2892 | 1238 | 2037 | 823 | 542 |
| Sensitivity | 0.05 | 0.60 | 0.33 | 0.73 | 0.82 |
| Specificity | 0.99 | 0.80 | 0.93 | 0.87 | 0.76 |
| Precision | 0.69 | 0.38 | 0.50 | 0.54 | 0.42 |
| Accuracy | 0.83 | 0.77 | 0.83 | 0.85 | 0.77 |
| MCC | 0.16 | 0.34 | 0.31 | 0.54 | 0.46 |
Table 2.
Performance of DR_bind1 based on 35 RNA-free protein structures compared to that of KYG, OPRA, BindN+ or Pprint using default settingsa
| DR_bind1 | KYG | OPRA | BindN+ | Pprint | |
|---|---|---|---|---|---|
| TP | 47 (64) | 457 (528) | 179 (224) | 554 (601) | 673 (735) |
| FP | 46 (45) | 1699 (1688) | 440 (549) | 1007 (1001) | 1773 (1798) |
| TN | 8307 (8522) | 6654 (6879) | 7913 (8018) | 7346 (7566) | 6580 (6769) |
| FN | 903 (967) | 493 (503) | 771 (807) | 396 (430) | 277 (296) |
| Sensitivity | 0.05 (0.06) | 0.48 (0.51) | 0.19 (0.22) | 0.58 (0.58) | 0.71 (0.71) |
| Specificity | 0.99 (0.99) | 0.80 (0.80) | 0.95 (0.94) | 0.88 (0.88) | 0.79 (0.79) |
| Precision | 0.51 (0.59) | 0.21 (0.24) | 0.29 (0.29) | 0.35 (0.38) | 0.28 (0.29) |
| Accuracy | 0.90 (0.89) | 0.76 (0.77) | 0.87 (0.86) | 0.85 (0.85) | 0.78 (0.78) |
| MCC | 0.13 (0.17) | 0.20 (0.23) | 0.16 (0.17) | 0.37 (0.39) | 0.34 (0.35) |
aNumbers with and without parentheses are based on the RNA-bound and free protein structures, respectively.
Since providing an experimentalist with a set of predicted RNA-binding residues containing few false positives (i.e. high precision) would be more useful than a comprehensive set with many false positives, DR_bind1 aimed to detect the most likely RNA-binding residues rather than all RNA-binding residues. Hence, DR_bind1 predicted fewer RNA-binding residues (TP + FP = 241) than KYG (4736), OPRA (2039), BINDN+ (4103) and Pprint (6050). Because DR_bind1 predicted an order of magnitude less RNA-binding residues than the other methods, it yielded relatively large FN and thus much lower sensitivity (0.05) and MCC (0.16) values. However, its precision (0.69) is higher than the precision of KYG (0.38), OPRA (0.50), BindN+ (0.54) and Pprint (0.42). Using the default prediction mode in KYG and OPRA and the default specificity settings in BindN+ and Pprint, the accuracy of DR_bind1 (0.83) is comparable to OPRA (0.83) and BindN+ (0.85), but is higher than that of KYG or Pprint (0.77).
Dependence on protein conformational change upon binding RNA
To assess how the performance of the structure-based methods would be affected by protein conformational changes that accompany RNA binding, the performance measures derived from the free structures were compared with those derived from the respective RNA-bound structures (numbers in parentheses in Table 2). Protein conformational changes upon RNA binding do not seem to significantly affect the performance of DR_bind1: even though the RMSD of the RNA-free structure from the respective RNA-bound structure may be as large as 9 Å (see Supplementary Table S1), the sensitivity, specificity, accuracy, derived from the RNA-bound and respective free structures are nearly identical, while the precision and MCC values decrease slightly (by 0.08 and 0.04, respectively) when the free structures were used instead of the bound ones. For the other structure-based methods, KYG and OPRA, the performance measures [Equations (2–6)] derived from the RNA-bound and respective free structures do not differ by more than 0.03. Because the protein sequences of the RNA-bound and respective free structures may not always be identical (see ‘Materials and Methods’ section), they yield slightly different precision and MCC values for the two sequence-based methods.
Dependence on the dataset composition
Ribosomal proteins consist of roughly half the proteins in dataset I (41/81) and a fifth of the proteins in dataset II (7/35). Interestingly, the percentage number of RNA-binding residues in ribosomal proteins is three to four times more than that in nonribosomal proteins: 35% of residues in ribosomal proteins bind RNA, whereas only 9% of residues in nonribosomal proteins bind RNA. To determine if the different RNA-binding residue prediction methods perform equally well for the two types of RNA-binding proteins, they were tested on the 41 ribosomal proteins in dataset I and separately on the remaining 40 nonribosomal proteins. All the methods showed significantly higher precision for ribosomal proteins than for nonribosomal proteins (numbers in parentheses in Table 3): the precision for ribosomal proteins is greater than that for nonribosomal proteins by 0.31 (DR_bind1), 0.43 (KYG), 0.44 (OPRA), 0.37 (BindN+) and 0.30 (Pprint).
Table 3.
Performance of DR_bind1 based on 41 ribosomal (or 40 nonribosomal) RNA-bound protein structures compared to that of KYG, OPRA, BindN+ or Pprint using default settingsa
| DR_bind1 | KYG | OPRA | BindN+ | Pprint | |
|---|---|---|---|---|---|
| TP | 102 (64) | 1334 (486) | 931 (90) | 1679 (556) | 1782 (734) |
| FP | 19 (56) | 812 (2104) | 593 (425) | 730 (1138) | 1406 (2128) |
| TN | 3673 (10 955) | 2880 (8907) | 3099 (10 586) | 2962 (9873) | 2286 (8883) |
| FN | 1883 (1009) | 651 (587) | 1054 (983) | 306 (517) | 203 (339) |
| Sensitivity | 0.05 (0.06) | 0.67 (0.45) | 0.47 (0.08) | 0.85 (0.52) | 0.90 (0.68) |
| Specificity | 0.99 (0.99) | 0.78 (0.81) | 0.84 (0.96) | 0.80 (0.90) | 0.62 (0.81) |
| Precision | 0.84 (0.53) | 0.62 (0.19) | 0.61 (0.17) | 0.70 (0.33) | 0.56 (0.26) |
| Accuracy | 0.66 (0.91) | 0.74 (0.78) | 0.71 (0.88) | 0.82 (0.86) | 0.72 (0.80) |
| MCC | 0.15 (0.16) | 0.44 (0.18) | 0.33 (0.06) | 0.63 (0.34) | 0.50 (0.33) |
aNumbers with and without parentheses were derived from 40 nonribosomal and 41 ribosomal RNA-bound protein structures, respectively.
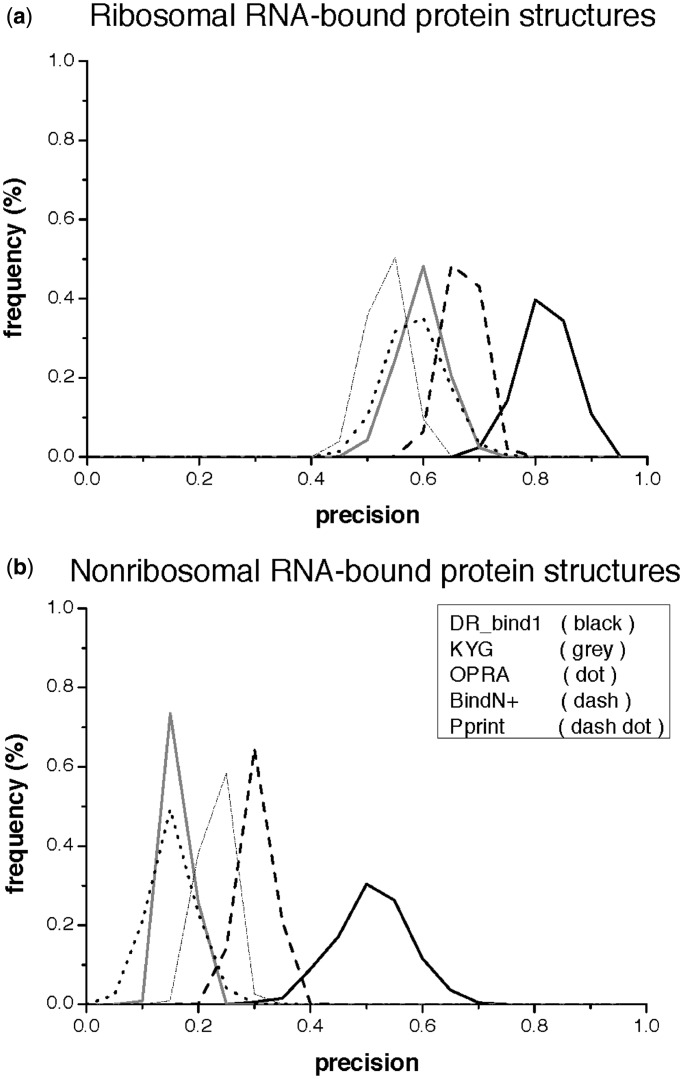
To further examine the performance sensitivity of the various methods on the dataset composition (proportion of ribosomal/nonribosomal proteins), we randomly chose 20 ribosomal and 20 nonribosomal RNA-bound protein structures, and computed the precision obtained by each of the methods; this was repeated 1000 times. Figure 1a and b shows the frequency distribution of the precision values derived from ribosomal and nonribosomal RNA-bound protein structures, respectively. Since DR_bind1 requires no training dataset, its precision is less dependent on the dataset composition than the precision of KYG, OPRA, BindN+ or Pprint. DR_bind1 yielded precision values derived from ribosomal protein structures (0.70–0.95) that partially overlap with those derived from nonribosomal protein structures (0.30–0.70). In contrast, the other methods yielded precision values derived from ribosomal protein structures that do not overlap with those derived from nonribosomal protein structures: KYG yielded precision values ranging from 0.45 to 0.70 for ribosomal proteins that are much higher than those for nonribosomal proteins (0.10–0.20). OPRA yielded precision values ranging from 0.45 to 0.75 for ribosomal proteins and 0.05–0.35 for nonribosomal ones, while BindN+ and Pprint, respectively, yielded precision values ranging from 0.55 to 0.75 and 0.45–0.65 for ribosomal proteins but 0.20–0.40 and 0.15–0.30 for nonribosomal ones.
Figure 1.
Frequency distribution of the precision values derived from ribosomal (top) and nonribosomal (bottom) RNA-bound protein structures using DR_bind1 (black curves), KYG (gray curves), OPRA (dotted curves), BindN+(dashed curves) and Pprint (dashed dot curves). (a) Ribosomal RNA-bound protein structures. (b) Nonribosomal RNA-bound protein structures.
Comparison with KYG, OPRA, BindN+ and Pprint for the same number of predictions
To evaluate how the performance of KYG, OPRA, BindN+ and Pprint for ribosomal/nonribosomal proteins would change if their sensitivities/specificities were comparable to DR_bind1’s sensitivity/specificity, they were compared to the performance of DR_bind1 for the same number of predictions. Thus, if DR_bind1 predicted m RNA-binding residues for protein X, then we chose the same number (m) of RNA-binding residues for KYG, OPRA, BindN+ or Pprint. We chose m residues with the most positive KYG, BindN+ or Pprint scores or the most negative OPRA values. For example, using the 1di2-A protein structure, DR_bind1 predicted three RNA-binding residues, but KYG predicted 16. To compare with DR_bind1, the 16 residues predicted by KYG were ranked according to their scores from the most positive to the most negative, and the top three residues with scores of 1.81, 1.64 and 1.17 were deemed to be the RNA-binding residues predicted by KYG.
When KYG, OPRA, BindN+ and Pprint yielded the same number of predictions (same TP + FP) as DR_bind1, their sensitivity, specificity and accuracy values became similar or identical to those of DR_bind1 (Table 4). Notably their MCC values are now less than the MCC value of DR_bind1, in contrast to their values when the number of predictions greatly exceeded DR_bind1 (see Table 3). Although the precision values of KYG, OPRA, BindN+ or Pprint for the same number of predictions as DR_bind1 (Table 4) has increased by ∼2–20% compared to their values using default settings (Table 3), they are still less than the precision of DR_bind1: for ribosomal proteins, the precision of DR_bind1 (0.84) is higher than that obtained by KYG (0.68) or OPRA (0.63) or the two sequence-based methods (0.80 or 0.74). For nonribosomal proteins, the precision of DR_bind1 (0.53) is also higher than that of KYG (0.28), OPRA (0.22), BindN+ (0.49) or Pprint (0.40).
Table 4.
Performance of DR_bind1 based on 41 ribosomal (or 40 nonribosomal) RNA-bound protein structures compared to that of KYG, OPRA, BindN+ or Pprint for the same number of predictions made by DR_bind1a
| DR_bind1 | KYG | OPRA | BindN+ | Pprint | |
|---|---|---|---|---|---|
| TP | 102 (64) | 82 (33) | 76 (26) | 97 (59) | 90 (48) |
| FP | 19 (56) | 39 (87) | 45 (94) | 24 (61) | 31 (72) |
| TN | 3673 (10 955) | 3653 (10 924) | 3647 (10 917) | 3668 (10 950) | 3661 (10 939) |
| FN | 1883 (1009) | 1903 (1040) | 1909 (1047) | 1888 (1014) | 1895 (1025) |
| Sensitivity | 0.05 (0.06) | 0.04 (0.03) | 0.04 (0.02) | 0.05 (0.05) | 0.05 (0.04) |
| Specificity | 0.99 (0.99) | 0.99 (0.99) | 0.99 (0.99) | 0.99 (0.99) | 0.99 (0.99) |
| Precision | 0.84 (0.53) | 0.68 (0.28) | 0.63 (0.22) | 0.80 (0.49) | 0.74 (0.40) |
| Accuracy | 0.66 (0.91) | 0.66 (0.91) | 0.66 (0.91) | 0.66 (0.91) | 0.66 (0.91) |
| MCC | 0.15 (0.16) | 0.10 (0.07) | 0.09 (0.05) | 0.14 (0.14) | 0.12 (0.11) |
aNumbers with and without parentheses were derived from 40 nonribosomal and 41 ribosomal RNA-bound protein structures, respectively.
Difference between the RNA-binding residues predicted by DR_bind1 and other methods
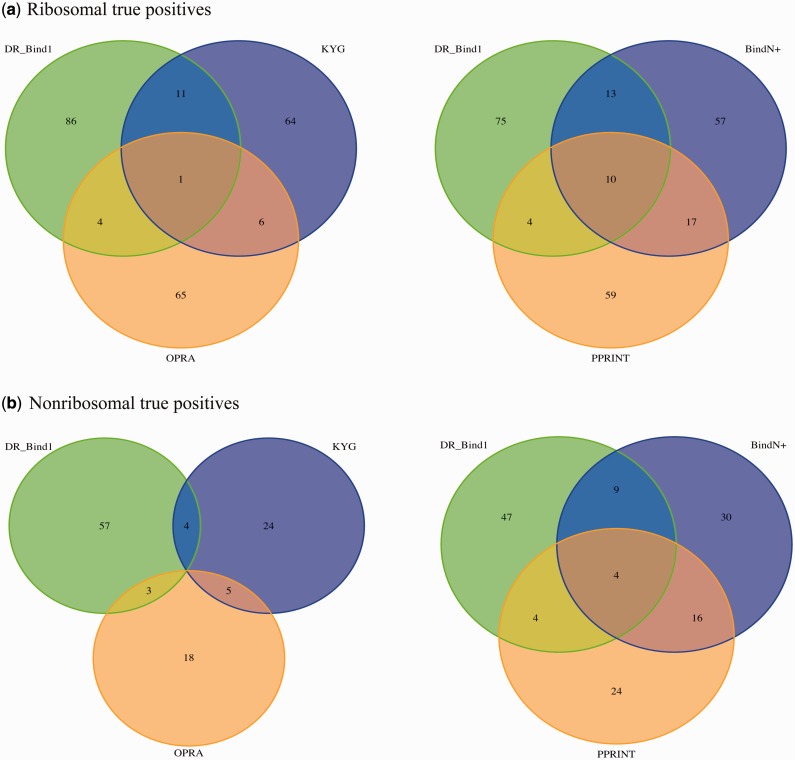
Does DR_bind1 predict the same RNA-binding residues as KYG, OPRA, BindN+ or Pprint for the same number of predictions (Table 4)? To answer this question, we compared the true positives predicted by DR_bind1 with those predicted by KYG, OPRA, BindN+ or Pprint and identified those RNA-binding residues correctly predicted by DR_bind1 that were not predicted by the other methods. The results in Figure 2 show that each method could yield true positives that are not found by other methods. For example, in nonribosomal proteins, DR_bind1, KYG, OPRA, BindN+ and Pprint correctly predicted 64, 33, 26, 59 and 48 RNA-binding residues, respectively. Among the 102 correctly predicted ribosomal RNA-binding residues by DR_bind1, 12, 5, 23 and 14 are also predicted by KYG, OPRA, BindN+ or Pprint, respectively, with 66 true positives predicted only by DR_bind1 (Figure 2a). Likewise, among the 64 correctly predicted nonribosomal RNA-binding residues by DR_bind1, 4, 3, 13 and 8 are also predicted by KYG, OPRA, BindN+ and Pprint, respectively, while 44 true positives were ‘missed’ by the other methods (Figure 2b). The numbers of unique true positives predicted by DR_bind1, KYG, OPRA, BindN+ and Pprint are, respectively, 66, 48, 51, 49 and 44 in ribosomal proteins and 44, 17, 16, 24 and 22 in nonribosomal proteins.
Figure 2.
Venn diagram showing four sets of true positives predicted by DR_bind1, KYG, OPRA, BindN+ and Pprint. (a) Ribosomal true positives. (b) Nonribosomal true positives.
Performance of DR_bind1 compared with BindN+ for ‘novel’ proteins
For the same number of predictions made by DR_bind1, the precision of BindN+ is close to that of DR_bind1 (see Table 4). However, BindN+ requires a training dataset, PRINR25 (11), so its precision may drop if it were used to predict RNA-binding residues in ‘novel’ proteins whose sequences are not homologous to those in its training dataset. Hence, BindN+ was used to predict the RNA-binding residues of 17 proteins in dataset I (referred to as dataset I_17) whose sequences share <30% sequence identity with the sequences in PRINR25. For the same number of RNA-binding residues predicted by DR_bind1, the precision (0.47) and MCC (0.12) values of BindN+ in predicting the RNA-binding residues in dataset I_17 becomes significantly less than those of DR_bind1 (0.74 and 0.22) (Supplementary Table S2).
How would DR_bind1 perform for a protein with no homologous structures? To address this question, DR_bind1 was used to detect RNA-binding residues based solely on the target protein structure without using any homologous structures. The results in the second column of Table 5 show that when homologous structures were removed, the precision of DR_bind1 based on 40 nonribosomal RNA-bound protein structures remained the same as that in Table 4 (0.53), while that based on 41 ribosomal RNA-bound protein structures dropped from 0.84 to 0.82. Notably, even if homologous structures were not available, the precision of DR_bind1 is still higher than that obtained by the other methods.
Table 5.
Performance of DR_bind1 based on 41 ribosomal (or 40 nonribosomal) RNA-bound protein structures compared to that of dRNA-3Da
| Homolog structures | DR_bind1 |
dRNA-3D |
||
|---|---|---|---|---|
| Noneb | No complexc | Best complexd | Second best complexe | |
| TP | 110 (74) | 101 (58) | 1950 (873) | 1295 (627) |
| FP | 24 (66) | 22 (54) | 173 (321) | 463 (681) |
| TN | 3668 (10 945) | 3670 (10 957) | 3519 (10 690) | 3229 (10 330) |
| FN | 1875 (999) | 1884 (1015) | 35 (200) | 690 (446) |
| Sensitivity | 0.06 (0.07) | 0.05 (0.05) | 0.98 (0.81) | 0.65 (0.58) |
| Specificity | 0.99 (0.99) | 0.99 (1) | 0.95 (0.97) | 0.87 (0.94) |
| Precision | 0.82 (0.53) | 0.82 (0.52) | 0.92 (0.73) | 0.74 (0.48) |
| Accuracy | 0.67 (0.91) | 0.66 (0.91) | 0.96 (0.96) | 0.80 (0.91) |
| MCC | 0.15 (0.17) | 0.15 (0.15) | 0.92 (0.75) | 0.54 (0.48) |
aNumbers with and without parentheses were derived from 40 nonribosomal and 41 ribosomal RNA-bound protein structures, respectively.
bNumbers were derived without free/complex structures of homologs.
cNumbers were derived without complex structures of homologs.
dNumbers were derived based on the best matching complex structure.
eNumbers were derived based on the second best matching complex structure.
Performance of DR_bind1 compared with dRNA-3D for proteins with homologous protein–RNA complex structures
Unlike the above methods, dRNA-3D (27) requires protein–RNA complex structures in predicting RNA-binding residues. In dRNA-3D, the target protein structure is structurally aligned with known protein−RNA complex structures, and if structural similarity is above a given threshold, it replaces the template protein structure to yield its complex structure; if the lowest binding energy between the target protein and template RNA computed using a knowledge-based energy function is below a given threshold, the corresponding protein−RNA structure is used to predict all RNA-binding residues. If no templates can be found to satisfy the structural similarity and binding energy thresholds, the test protein is predicted to be a non-RNA-binding one.
In contrast to dRNA-3D, DR_bind1 does not require protein-RNA complex structures: when structures of the test protein homologs in complex with RNA were removed, the resulting performance measures in Table 5 (third column) differ from those in Table 4 (second column) by ≤0.02. However, the precision of DR_bind1 is lower than that of dRNA-3D (by 0.10 and 0.21 for ribosomal and nonribosomal proteins, respectively) using the best template. The high precision obtained by dRNA-3D is because 71 of the 81 test proteins share > 90% sequence identity with the respective proteins from the best templates. However, only 12 of the 81 test proteins share >90% sequence identity with the respective proteins from the second-best template. If the RNA-binding residues were predicted using the second-best template, the precision of dRNA-3D dropped significantly (by 0.18 and 0.25 for ribosomal and nonribosomal proteins, respectively), indicating that its precision is sensitive to the sequence identity between test and template proteins.
Verification of the predicted RNA-binding residues in hCPEB3
To test the precision of DR_bind1, KYG, OPRA, BindN+ and Pprint, the five methods were used to predict the RNA-binding residues in hCPEB3, as described in the ‘Materials and Methods’ section. Based on the representative structure of the hCPEB3 RBD (2dnl-A) and representative homologous structures, DR_bind1 predicted two RNA-binding residues, namely, F430 and F474. The two most probable RNA-binding residues predicted by KYG, OPRA, BindN+ and Pprint are (R449, G432), (R514, R449), (R427, S465) and (K460, D456), respectively. Interestingly, based on the 2dnl-A structure, dRNA-3D predicted the hCPEB3 RBD as a non-RNA-binding protein, but the 22 predicted binding residues based on the best template (1b7f-A) encompass the RNA-binding residues predicted by DR_bind1, OPRA (R514) and Pprint.
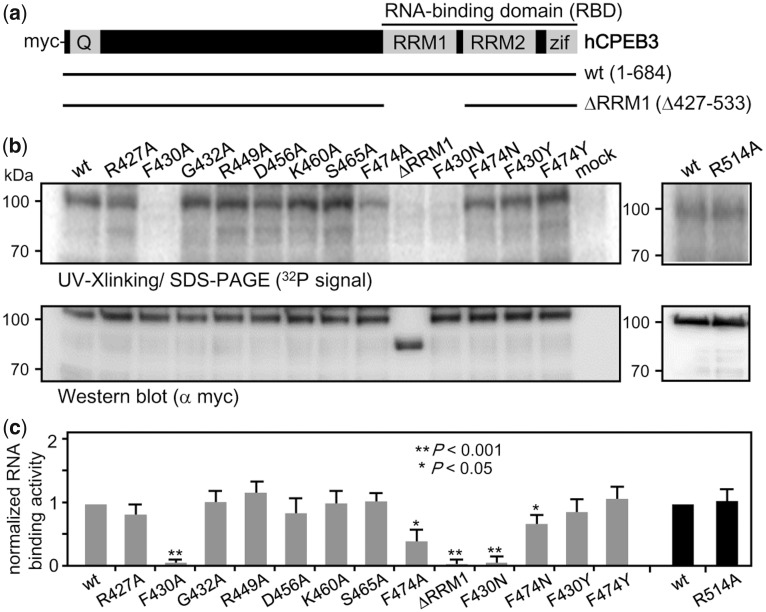
To experimentally verify the predicted RNA-binding residues, single alanine-substituted mutants were constructed to assess their contributions to RNA interaction (see Supplementary Methods). Figure 3a shows the myc-tagged wt and RRM1-deleted mutant CPEB3 used as the positive and negative controls for RNA binding, respectively (48). The RNA binding and expression of the CPEB3 mutants were examined by UV-cross-linking RNA-binding assay and western blotting, respectively (Figure 3b). The normalized RNA-binding ability (i.e. the ratio of RNA-binding signal versus the expression level) of these alanine-substituted mutants from three independent experiments was analyzed and the difference in binding RNA as compared to wt CPEB3 was evaluated using the Student’s t-test (Figure 3c). Among the alanine-substituted mutants, only F430A and F474A mutants were defective in RNA binding like the RRM1-deleted mutant CPEB3. To ensure that such a defect was not caused by protein conformational changes due to replacing phenylalanine with the much smaller alanine, additional F430N and F474N mutants were constructed and tested for RNA binding (Figure 3b and c). Although the F474N mutant interacted with the RNA better than the F474A mutant, its RNA-binding ability was still impaired. In contrast, the F430 residue is crucial for RNA binding, as the F430N mutant remained defective in RNA binding like the F430A mutant. To assess if the aromatic rings of F430 and F474 are important in binding RNA, they were retained by mutating the Phe sidechains to tyrosines (Figure 3b and c). Both F430Y and F474Y mutants bound to the RNA like wt CPEB3, suggesting the aromatic ring is important for stabilizing the interaction with RNA.
Figure 3.
Experimental evaluation of the predicted RNA-interacting aa residues in CPEB3. (a) Salient features of CPEB3 showing the N-terminal glutamine-rich region (Q) and the C-terminal RBD composed of two RRMs and zinc fingers (Zif). The myc-tagged wt and the RRM1-deleted (ΔRRM1) hCPEB3 are shown. All point mutations are located in the RRM1 domain. (b) The 293T lysates containing wt or various mutant CPEB3 proteins were cross-linked with the radiolabeled 1904 RNA probe for RNA-binding assay or used for western blotting with myc antibody. (c) The normalized RNA-binding abilities of various CPEB3 mutants were expressed relative to the wt CPEB3, which was arbitrarily set to 1. Gray and black bars indicate that the two sets of experiments were conducted separately. The data from three independent experiments were expressed as mean ± standard deviation. One and two asterisks denote the statistical significance, *P < 0.05 and **P < 0.001, respectively, from the Student’s t-test.
Application of DR_bind1 to predict DNA-binding residues
The method implemented in DR_bind1 should in principle be able to detect DNA-binding residues, which, like RNA-binding residues, would be expected to preserve their aa type, solvent accessibility and energetic features (30,49) due to their critical functional roles. Hence, DR_bind1 was tested on 83 DNA-bound structures taken from our previous work (30). The results in Supplementary Table S3 show that the precision of DR_bind1 in detecting DNA-binding residues (0.68) is similar to that for RNA-binding residues (0.69), while the accuracy (0.90) and MCC (0.22) are higher than those in Table 1.
DISCUSSION
The novelty of this work lies in predicting RNA-binding residues on the basis that these functionally important residues would preserve not only their aa type but also their structural and energetic features within the same protein family. DR_bind1 requires as input the structure and conservation scores of the target protein and yields as output, RNA-binding residues that share evolutionary conserved structural and energetic features in the same family. The key advantage of DR_bind1 is that it requires no training data set and it has no parameters, hence the precision of DR_bind1 is less dependent on the nature of the target (test) protein than that of KYG, OPRA, BindN+ or Pprint (see Figure 1). In contrast, machine-learning methods such as BindN+ require training datasets, hence their precision values drop significantly when applied to ‘novel’ sequences that are nonhomologous to the sequences in the training data sets (Supplementary Table S2). For such ‘novel’ proteins, DR_bind1 generally yields higher precision than the structure-based methods, KYG and OPRA, and sequence-based methods, BindN+ and Pprint for the same number of RNA-binding residues predicted by DR_bind1. It is complementary to these structure/sequence-based methods, as its predicted RNA-binding residues generally differ from the top-scoring residues by KYG, OPRA, BindN+ or Pprint. For non-novel proteins with homologous protein–RNA complex structures dRNA-3D (27), which employs the latter structures in predicting RNA-binding residues, may yield better precision than DR_bind1, but it is not clear which of the predicted residues should be experimentally tested first. The key limitation of DR_bind1 is that it requires conservation scores of the target protein like most methods such as BindN+ as well as structures of homologous proteins. This limitation, however, would be alleviated by the increasing number of sequences and free protein structures solved each year, most of which are not truly novel but share ≥30% sequence identity to known proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Institute of Biomedical Sciences, Academia Sinica. This work was funded by Academia Sinica and National Science Council, Taiwan [NSC 95-2113-M-001-038, NSC 95-2113-M-001-001 to C.L. and NSC 99-2311-B-001-020-MY3 to Y.H.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Laura Pérez-Cano and Juan Fernández-Recio for sending us their OPRA program as well as Laura Pérez-Cano and Miguel Romero for help in using OPRA.
REFERENCES
- 1.Tuschl T. Functional genomics: RNA sets the standard. Nature. 2003;421:268–272. doi: 10.1038/421220a. [DOI] [PubMed] [Google Scholar]
- 2.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 5.Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum. Mol. Genet. 1999;8:775–782. doi: 10.1093/hmg/8.5.775. [DOI] [PubMed] [Google Scholar]
- 6.Keil M, Exner TE, Brickmann J. Pattern recognition strategies for molecular surfaces: III. Binding site prediction with a neural network. J. Comput. Chem. 2004;25:779–789. doi: 10.1002/jcc.10361. [DOI] [PubMed] [Google Scholar]
- 7.Jeong E, Chung I, Miyano S. A neural network method for identification of RNA-interacting residues in protein. Genome Inform. Ser. Workshop Genome Inform. 2004;15:105–116. [PubMed] [Google Scholar]
- 8.Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578–W584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maetschke SR, Yuan Z. Exploiting structural and topological information to improve prediction of RNA-protein binding sites. BMC Bioinformatics. 2009;10:341. doi: 10.1186/1471-2105-10-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towfic F, Caragea C, Gemperline DC, Dobbs D, Honavar V. Struct-NB: predicting protein-RNA binding sites using structural features. Int. J. Data Min. Bioinform. 2010;4:21–43. doi: 10.1504/ijdmb.2010.030965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng CW, Su EC, Hwang JK, Sung TY, Hsu WL. Predicting RNA-binding sites of proteins using support vector machines and evolutionary information. BMC Bioinformatics. 2008;9:S6. doi: 10.1186/1471-2105-9-S12-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M, Gromiha MM, Raghava G. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins Struct. Funct. Bioinf. 2008;71:189–194. doi: 10.1002/prot.21677. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Xue Z, Shen G, Xu J. PRINTR: prediction of RNA binding sites in proteins using SVM and profiles. Amino Acids. 2008;35:295–302. doi: 10.1007/s00726-007-0634-9. [DOI] [PubMed] [Google Scholar]
- 15.Huang YF, Chiu LY, Huang CC, Huang CK. Predicting RNA-binding residues from evolutionary information and sequence conservation. BMC Genomics. 2010;11(Suppl. 4):S2. doi: 10.1186/1471-2164-11-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami Y, Spriggs RV, Nakamura H, Jones S. PiRaNhA: a server for the computational prediction of RNA-binding residues in protein sequences. Nucleic Acids Res. 2010;38:W412–W416. doi: 10.1093/nar/gkq474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Huang C, Yang M, Yang J. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst. Biol. 2010;4:S3. doi: 10.1186/1752-0509-4-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun MR, Byun Y, Han K. Predicting RNA-binding sites in proteins using the interaction propensity of amino acid triplets. Protein Pept Lett. 2010;17:1102–1110. doi: 10.2174/092986610791760388. [DOI] [PubMed] [Google Scholar]
- 19.Choi S, Han K. Prediction of RNA-binding amino acids from protein and RNA sequences. BMC Bioinformatics. 2011;13(Suppl.):S7. doi: 10.1186/1471-2105-12-S13-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ZP, Wu LY, Wang Y, Zhang XS, Chen L. Prediction of protein-RNA binding sites by a random forest method with combined features. Bioinformatics. 2010;26:1616–1622. doi: 10.1093/bioinformatics/btq253. [DOI] [PubMed] [Google Scholar]
- 21.Ma X, Guo J, Wu J, Liu H, Yu J, Xie J, Sun X. Prediction of RNA-binding residues in proteins from primary sequence using an enriched random forest model with a novel hybrid feature. Proteins. 2011;79:1230–1239. doi: 10.1002/prot.22958. [DOI] [PubMed] [Google Scholar]
- 22.Carson MB, Langlois R, Lu H. NAPS: a residue-level nucleic acid-binding prediction server. Nucleic Acids Res. 2010;38:W431–W435. doi: 10.1093/nar/gkq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong J, Jiang P, Lu ZH. RISP: a web-based server for prediction of RNA-binding sites in proteins. Comput. Methods Programs Biomed. 2008;90:148–153. doi: 10.1016/j.cmpb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Chen YC, Lim C. Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res. 2008;36:e29. doi: 10.1093/nar/gkn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Cano L, Fernandez-Recio J. Optimal protein-RNA area, OPRA: a propensity-based method to identify RNA-binding sites on proteins. Proteins. 2010;78:25–35. doi: 10.1002/prot.22527. [DOI] [PubMed] [Google Scholar]
- 26.Kim OTP, Yura K, Go N. Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Res. 2006;34:6450–6460. doi: 10.1093/nar/gkl819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Yang Y, Zhou Y. Structure-based prediction of RNA-binding domains and RNA-binding sites and application to structural genomics targets. Nucleic Acids Res. 2011;39:3017–3025. doi: 10.1093/nar/gkq1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YC, Lim C. Common physical basis of macromolecule-binding sites in proteins. Nucleic Acids Res. 2008;36:7078–7087. doi: 10.1093/nar/gkn868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YC, Wright JD, Lim C. DR.bind: a web server for predicting DNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res. 2012:1–8. doi: 10.1093/nar/gks481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Iype L, Jain S, Fagan P, Marvin J, et al. The Protein Data Bank. Acta Crystallogr. D. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 32.Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, Lee D, et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: Pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor WR, Orengo CA. Protein structure alignment. J. Mol. Biol. 1989;208:1–22. doi: 10.1016/0022-2836(89)90084-3. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 37.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of protein database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 38.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 39.Joseph AP, Valadié H, Srinivasan N, de Brevern AG. Local structural differences in homologous proteins: specificities in different SCOP classes. PLoS One. 2012;7:e38805. doi: 10.1371/journal.pone.0038805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubbard SJ, Thornton JM. Department of Biochemistry and Molecular Biology. London: University College; 1993. [Google Scholar]
- 41.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Case DA, Darden T, Cheatham Iii TE, Simmerling C, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 43.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 44.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 46.Ilinkin I, Ye J, Janardan R. Multiple structure alignment and consensus identification for proteins. BMC Bioinformatics. 2010;11:71. doi: 10.1186/1471-2105-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puton T, Kozlowski L, Tuszynska I, Rother K, Bujnicki JM. Computational methods for prediction of protein–RNA interactions. J. Struct. Biol. 2012;179:261–268. doi: 10.1016/j.jsb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Chao HW, Lai YT, Lu YL, Lin CL, Mai W, Huang YS. NMDAR signaling facilitates the IPO5-mediated nuclear import of CPEB3. Nucleic Acids Res. 2012;40:8484–8498. doi: 10.1093/nar/gks598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen YC, Wu CY, Lim C. Predicting DNA-binding sites on proteins from electrostatic stabilization upon mutation to Asp/Glu and evolutionary conservation. Proteins Struct. Funct. Bioinf. 2007;67:671–680. doi: 10.1002/prot.21366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.