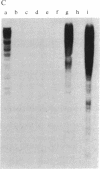
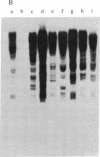
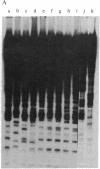
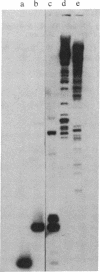
Abstract
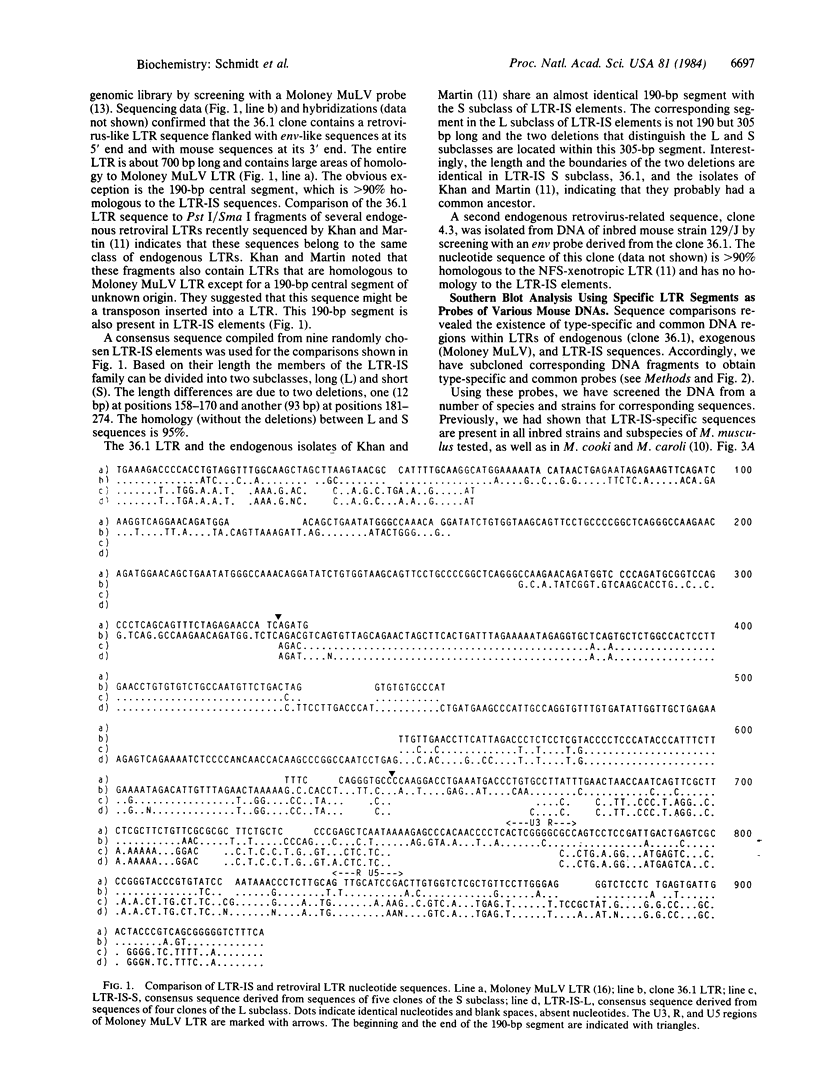
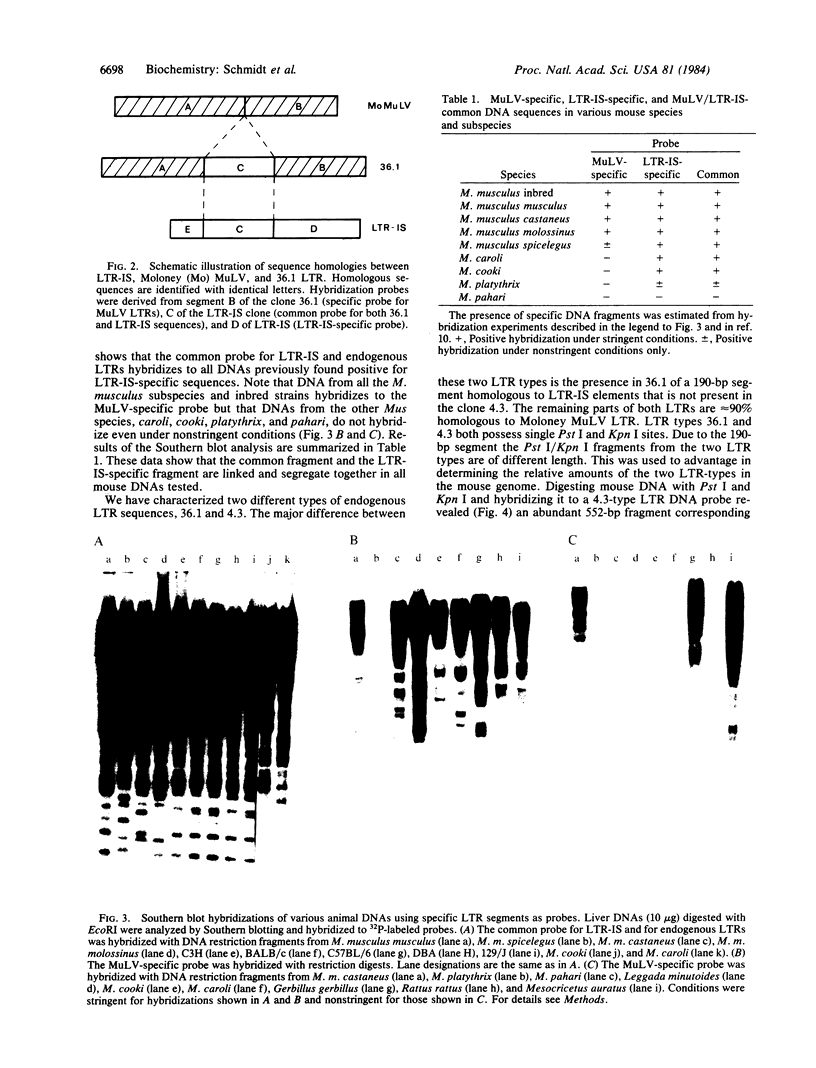
Two endogenous retroviral long terminal repeats (LTRs) were sequenced and compared to LTR-IS (a family of insertion-element-like sequences with structural features of solitary retroviral LTRs) and to Moloney murine leukemia virus DNA. The sequence comparisons revealed that the major difference between these two endogenous LTRs is a 190-base-pair segment which is also present in LTR-IS elements. Hybridization analysis of DNAs from several mouse species using specific probes shows linkage of the 190-base-pair segment to a LTR-IS specific fragment. It is concluded that the major class of endogenous LTRs has been generated by recombination between exogenous retroviral LTRs and LTR-IS sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Cloyd M. W., Linemeyer D. L., Lander M. R., Rands E., Lowy D. R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature. 1982 Jan 7;295(5844):25–31. doi: 10.1038/295025a0. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Khan A. S., Martin M. A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci U S A. 1983 May;80(9):2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung M. L., Hartley J. W., Rowe W. P., Hopkins N. H. Large RNase T1-resistant oligonucleotides encoding p15E and the U3 region of the long terminal repeat distinguish two biological classes of mink cell focus-forming type C viruses of inbred mice. J Virol. 1983 Jan;45(1):275–290. doi: 10.1128/jvi.45.1.275-290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Neel B. G., Hayward W. S., Robinson H. L., Fang J., Astrin S. M. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell. 1981 Feb;23(2):323–334. doi: 10.1016/0092-8674(81)90128-8. [DOI] [PubMed] [Google Scholar]
- Ou C. Y., Boone L. R., Yang W. K. A novel sequence segment and other nucleotide structural features in the long terminal repeat of a BALB/c mouse genomic leukemia virus-related DNA clone. Nucleic Acids Res. 1983 Aug 25;11(16):5603–5620. doi: 10.1093/nar/11.16.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Courtneidge S. A., Crittenden L. B., Fadly A. M., Bishop J. M., Varmus H. E. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981 Feb;23(2):311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Roblin R., Young J. M., Mural R. J., Bell T. E., Ihle J. N. Molecular cloning and characterization of murine leukemia virus-related DNA sequences from C3H/HeN mouse DNA. J Virol. 1982 Jul;43(1):113–126. doi: 10.1128/jvi.43.1.113-126.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J., Faller D. V., Hopkins N. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc Natl Acad Sci U S A. 1978 Jan;75(1):495–499. doi: 10.1073/pnas.75.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P. Leukemia virus genomes in the chromosomal DNA of the mouse. Harvey Lect. 1978;71:173–192. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tlsty T. D., Albertini A. M., Miller J. H. Gene amplification in the lac region of E. coli. Cell. 1984 May;37(1):217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Wirth T., Glöggler K., Baumruker T., Schmidt M., Horak I. Family of middle repetitive DNA sequences in the mouse genome with structural features of solitary retroviral long terminal repeats. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3327–3330. doi: 10.1073/pnas.80.11.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Schmidt M., Baumruker T., Horak I. Evidence for mobility of a new family of mouse middle repetitive DNA elements (LTR-IS). Nucleic Acids Res. 1984 Apr 25;12(8):3603–3610. doi: 10.1093/nar/12.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]