Abstract
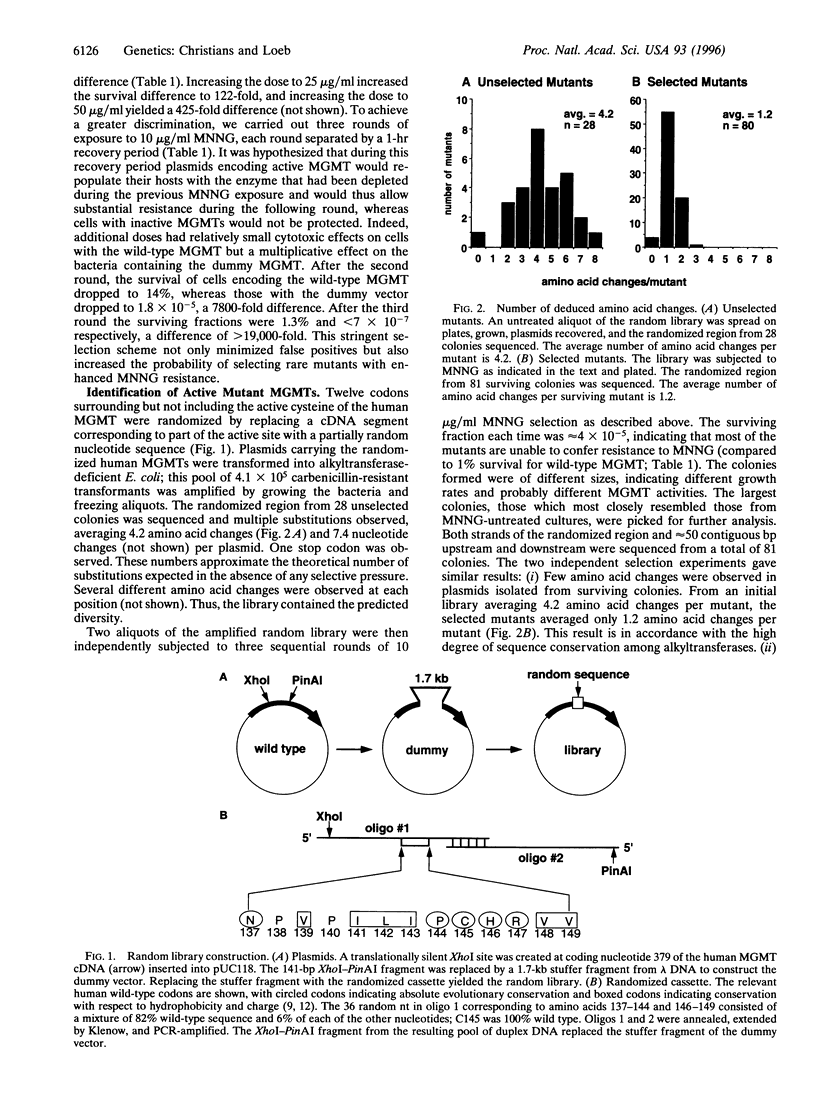
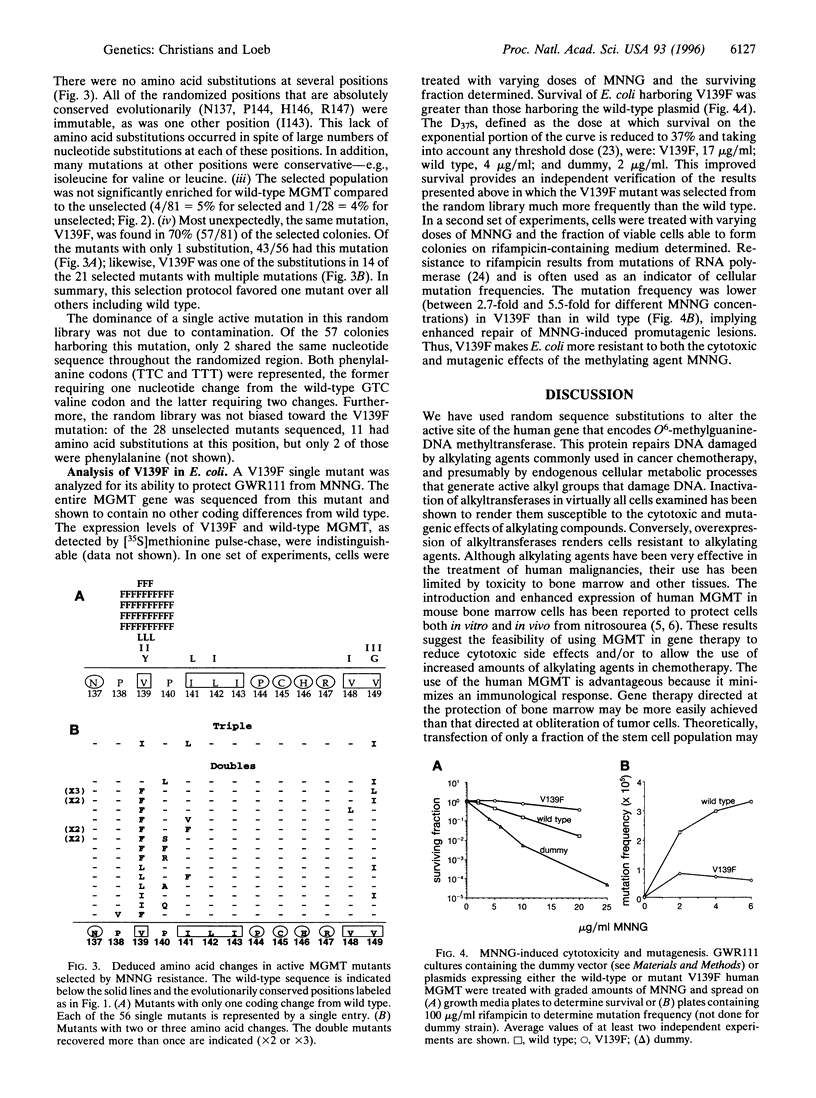
DNA repair alkyltransferases protect organisms against the cytotoxic, mutagenic, and carcinogenic effects of alkylating agents by transferring alkyl adducts from DNA to an active cysteine on the protein, thereby restoring the native DNA structure. We used random sequence substitutions to gain structure-function information about the human O6-methylguanine-DNA methyltransferase (EC 2.1.1.63), as well as to create active mutants. Twelve codons surrounding but not including the active cysteine were replaced by a random nucleotide sequence, and the resulting random library was selected for the ability to provide alkyltransferase-deficient Escherichia coli with resistance to the methylating agent N-methyl-N'-nitro-N-nitrosoguanidine. Few amino acid changes were tolerated in this evolutionarily conserved region of the protein. One mutation, a valine to phenylalanine change at codon 139 (V139F), was found in 70% of the selected mutants; in fact, this mutant was selected much more frequently than the wild type. V139F provided alkyltransferase-deficient bacteria with greater protection than the wild-type protein against both the cytotoxic and mutagenic effects of N-methyl-N'-nitro-N-nitrosoguanidine, increasing the D37 over 4-fold and reducing the mutagenesis rate 2.7-5.5-fold. This mutant human alkyltransferase, or others similarly created and selected, could be used to protect bone marrow cells from the cytotoxic side effects of alkylation-based chemotherapeutic regimens.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allay J. A., Dumenco L. L., Koc O. N., Liu L., Gerson S. L. Retroviral transduction and expression of the human alkyltransferase cDNA provides nitrosourea resistance to hematopoietic cells. Blood. 1995 Jun 1;85(11):3342–3351. [PubMed] [Google Scholar]
- Black M. E., Loeb L. A. Identification of important residues within the putative nucleoside binding site of HSV-1 thymidine kinase by random sequence selection: analysis of selected mutants in vitro. Biochemistry. 1993 Nov 2;32(43):11618–11626. doi: 10.1021/bi00094a019. [DOI] [PubMed] [Google Scholar]
- Bobola M. S., Blank A., Berger M. S., Silber J. R. Contribution of O6-methylguanine-DNA methyltransferase to monofunctional alkylating-agent resistance in human brain tumor-derived cell lines. Mol Carcinog. 1995 Jun;13(2):70–80. doi: 10.1002/mc.2940130203. [DOI] [PubMed] [Google Scholar]
- Chueh L. L., Nakamura T., Nakatsu Y., Sakumi K., Hayakawa H., Sekiguchi M. Specific amino acid sequences required for O6-methylguanine-DNA methyltransferase activity: analyses of three residues at or near the methyl acceptor site. Carcinogenesis. 1992 May;13(5):837–843. doi: 10.1093/carcin/13.5.837. [DOI] [PubMed] [Google Scholar]
- Day R. S., 3rd, Ziolkowski C. H., Scudiero D. A., Meyer S. A., Lubiniecki A. S., Girardi A. J., Galloway S. M., Bynum G. D. Defective repair of alkylated DNA by human tumour and SV40-transformed human cell strains. Nature. 1980 Dec 25;288(5792):724–727. doi: 10.1038/288724a0. [DOI] [PubMed] [Google Scholar]
- Dube D. K., Loeb L. A. Mutants generated by the insertion of random oligonucleotides into the active site of the beta-lactamase gene. Biochemistry. 1989 Jul 11;28(14):5703–5707. doi: 10.1021/bi00440a001. [DOI] [PubMed] [Google Scholar]
- Dumenco L. L., Allay E., Norton K., Gerson S. L. The prevention of thymic lymphomas in transgenic mice by human O6-alkylguanine-DNA alkyltransferase. Science. 1993 Jan 8;259(5092):219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- Erickson L. C., Laurent G., Sharkey N. A., Kohn K. W. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumour cells. Nature. 1980 Dec 25;288(5792):727–729. doi: 10.1038/288727a0. [DOI] [PubMed] [Google Scholar]
- Fass D. N., Hewick R. M., Knutson G. J., Nesheim M. E., Mann K. G. Internal duplication and sequence homology in factors V and VIII. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1688–1691. doi: 10.1073/pnas.82.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick A. M., Fahl W. E. Forced evolution of glutathione S-transferase to create a more efficient drug detoxication enzyme. Proc Natl Acad Sci U S A. 1995 Aug 29;92(18):8140–8144. doi: 10.1073/pnas.92.18.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. Promoters selected from random DNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7405–7409. doi: 10.1073/pnas.83.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K., Kawate H., Chueh L. L., Hayakawa H., Sekiguchi M. Requirement of the Pro-Cys-His-Arg sequence for O6-methylguanine-DNA methyltransferase activity revealed by saturation mutagenesis with negative and positive screening. Mol Gen Genet. 1994 May 25;243(4):379–389. doi: 10.1007/BF00280468. [DOI] [PubMed] [Google Scholar]
- Kanugula S., Goodtzova K., Edara S., Pegg A. E. Alteration of arginine-128 to alanine abolishes the ability of human O6-alkylguanine-DNA alkyltransferase to repair methylated DNA but has no effect on its reaction with O6-benzylguanine. Biochemistry. 1995 May 30;34(21):7113–7119. doi: 10.1021/bi00021a024. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Mol C. D., Arvai A. S., Slupphaug G., Kavli B., Alseth I., Krokan H. E., Tainer J. A. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995 Mar 24;80(6):869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Mol C. D., Kuo C. F., Thayer M. M., Cunningham R. P., Tainer J. A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995 Mar 23;374(6520):381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- Moore M. H., Gulbis J. M., Dodson E. J., Demple B., Moody P. C. Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 1994 Apr 1;13(7):1495–1501. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz T., Mackay W., Glassner B. J., Williams D. A., Samson L. Retrovirus-mediated expression of a DNA repair protein in bone marrow protects hematopoietic cells from nitrosourea-induced toxicity in vitro and in vivo. Cancer Res. 1995 Jun 15;55(12):2608–2614. [PubMed] [Google Scholar]
- Munir K. M., French D. C., Loeb L. A. Thymidine kinase mutants obtained by random sequence selection. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4012–4016. doi: 10.1073/pnas.90.9.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuru Y., Matsukuma S., Nemoto N., Sugano H., Sekiguchi M., Ishikawa T. O6-methylguanine-DNA methyltransferase protects against nitrosamine-induced hepatocarcinogenesis. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6468–6472. doi: 10.1073/pnas.90.14.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Struhl K. An efficient method for generating proteins with altered enzymatic properties: application to beta-lactamase. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9094–9098. doi: 10.1073/pnas.86.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. W., Kim S. T., Sancar A., Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995 Jun 30;268(5219):1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- Pegg A. E., Dolan M. E., Moschel R. C. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- Rafferty J. A., Tumelty J., Skorvaga M., Elder R. H., Margison G. P., Douglas K. T. Site-directed mutagenesis of glutamic acid 172 to glutamine completely inactivated human O6-alkylguanine-DNA-alkyltransferase. Biochem Biophys Res Commun. 1994 Feb 28;199(1):285–291. doi: 10.1006/bbrc.1994.1226. [DOI] [PubMed] [Google Scholar]
- Rebeck G. W., Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991 Mar;173(6):2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch K. M., Chen L., Verdine G. L., Lipscomb W. N. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995 Jul 14;82(1):143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. On base flipping. Cell. 1995 Jul 14;82(1):9–12. doi: 10.1016/0092-8674(95)90046-2. [DOI] [PubMed] [Google Scholar]
- Savva R., McAuley-Hecht K., Brown T., Pearl L. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995 Feb 9;373(6514):487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- Takano K., Nakabeppu Y., Sekiguchi M. Functional sites of the Ada regulatory protein of Escherichia coli. Analysis by amino acid substitutions. J Mol Biol. 1988 May 20;201(2):261–271. doi: 10.1016/0022-2836(88)90137-4. [DOI] [PubMed] [Google Scholar]
- Tano K., Shiota S., Collier J., Foote R. S., Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci U S A. 1990 Jan;87(2):686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wibley J. E., McKie J. H., Embrey K., Marks D. S., Douglas K. T., Moore M. H., Moody P. C. A homology model of the three-dimensional structure of human O6-alkylguanine-DNA alkyltransferase based on the crystal structure of the C-terminal domain of the Ada protein from Escherichia coli. Anticancer Drug Des. 1995 Jan;10(1):75–95. [PubMed] [Google Scholar]
- di Mauro E., Synder L., Marino P., Lamberti A., Coppo A., Tocchini-Valentini G. P. Rifampicin sensitivity of the components of DNA-dependent RNA polymerase. Nature. 1969 May 10;222(5193):533–537. doi: 10.1038/222533a0. [DOI] [PubMed] [Google Scholar]


