Erythropoietin suppresses non-erythroid cell fate options to induce an erythroid lineage bias at all lineage bifurcations between HSCs and committed erythroid progenitors.
Abstract
The erythroid stress cytokine erythropoietin (Epo) supports the development of committed erythroid progenitors, but its ability to act on upstream, multipotent cells remains to be established. We observe that high systemic levels of Epo reprogram the transcriptomes of multi- and bipotent hematopoietic stem/progenitor cells in vivo. This induces erythroid lineage bias at all lineage bifurcations known to exist between hematopoietic stem cells (HSCs) and committed erythroid progenitors, leading to increased erythroid and decreased myeloid HSC output. Epo, therefore, has a lineage instructive role in vivo, through suppression of non-erythroid fate options, demonstrating the ability of a cytokine to systematically bias successive lineage choices in favor of the generation of a specific cell type.
Loss-of-function studies have shown that cytokines have specific roles in maintaining lineage output during steady-state hematopoiesis by promoting the survival and expansion of committed progenitors (Metcalf, 2008). However, further studies have shown that endogenous myeloid cytokine receptors direct lineage choice of stem and progenitor cells (Rieger et al., 2009; Mossadegh-Keller et al., 2013), raising the possibility that this is a general cytokine function.
Elevated levels of hematopoietic cytokines increase the production of specific cell types in response to physiological emergencies such as hypoxia, anemia, or infection, where levels of circulating erythropoietin (Epo) and myeloid colony stimulating factors can increase by several orders of magnitude (Cheers et al., 1988; Watari et al., 1989). During severe anemia, Epo serum levels are elevated up to 1,000-fold (Jelkmann and Wiedemann, 1990), increasing exponentially to the degree of anemia. The Epo–Epo receptor signaling pathway, although dispensable for the formation of erythroid progenitors, is essential for their subsequent proliferation and survival (Wu et al., 1995; Lin et al., 1996). The effect of increased Epo production is therefore generally believed to be due to improved expansion of committed erythroid progenitors (Metcalf, 2008). Multipotent hematopoietic cells have been shown to express functional Epo receptor (Shiozawa et al., 2010), but the effect of elevated systemic Epo levels on non-erythroid hematopoietic lineages and multipotent stem cells and progenitors remains essentially unknown.
We observe here that elevated systemic Epo levels suppress the levels of phenotypic and functional non-erythroid progenitors in the bone marrow, and transplantation of Epo-exposed multipotent stem/progenitor cells results in an erythroid-biased lineage output. Epo therefore acts directly on multipotent hematopoietic cells to alter their progenitor and mature cell output.
RESULTS AND DISCUSSION
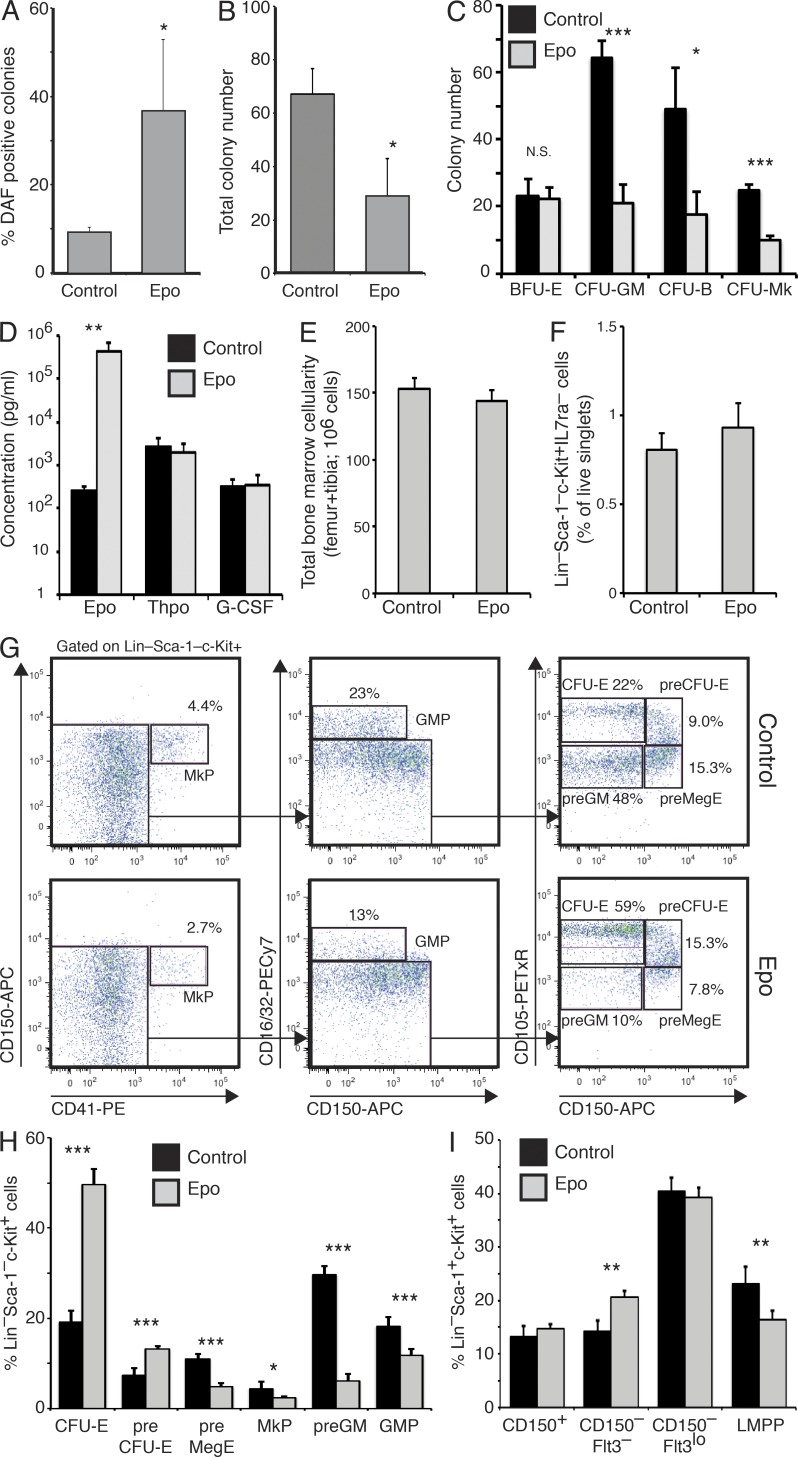
We developed a model where systemic Epo levels were selectively increased through hydrodynamic tail vein injection (Zhang et al., 1999) of a CMV-based Epo expression vector, leading to increased peripheral blood erythrocyte numbers (unpublished data). To determine any preceding effect on erythroid progenitor numbers, we isolated the bone marrow Lin−Sca-1−c-Kit+ population, which contains the myelo-erythroid progenitors (Akashi et al., 2000), including CFU-E and proerythroblasts (Pronk et al., 2007), at 2 d after injection, and plated these cells under conditions permissive for both myeloid and erythroid differentiation. Staining of the developed colonies using 2,7-diaminofluorene (DAF) to detect hemoglobin showed an increase in the proportion of erythroid colonies (Fig. 1 A). However, this was accompanied by a decrease in the total colony number (Fig. 1 B). Plating Lin−Sca-1−c-Kit+ progenitor cells under conditions specifically promoting erythroid (E), megakaryocyte (Mk), B-lymphoid (B), or granulocyte/macrophage (GM) differentiation showed a significant decrease in GM, Mk, and B colony–forming cells, whereas E colony numbers were unchanged (Fig. 1 C). The increase in circulating Epo levels obtained through hydrodynamic injection were comparable to those observed in anemic patients (Jelkmann and Wiedemann, 1990), whereas no change was observed in other lineage-specific cytokines (Thrombopoietin, G-CSF; Fig. 1 D).
Figure 1.
Systemic Epo treatment introduces E bias in myelo-erythroid progenitor and LMPP production. (A) In vitro E differentiation potential of 500 bone marrow Lin−Sca-1−c-Kit+ cells isolated after 2 d of Epo treatment (Epo) or mock treatment (control), measured by 2,7-diaminofluorene (DAF) staining of M3434 methylcellulose cultures after 8 d of culture. Values are mean ± SD, n = 3. One of two representative experiments is shown. (B) Total colonies formed from cells plated in A. (C) Colony-forming potential of 2-d Epo-exposed or control Lin−Sca-1−c-Kit+ bone marrow cells was assayed separately under GM (M3534; 500 cells), E (M3436; 1,000 cells), preB (M3630; 1,000 cells), and Mk (MegaCult; 500 cells) conditions. Values are mean ± SD, n = 3. One of two representative experiments is shown. (D) Bar graph showing serum cytokine levels in mice hydrodynamically injected with pCMV6-Epo or empty pCMV6 vector 2 d after transfection. Values are mean ± SD, n = 8, from two experiments. (E) Bar graphs showing total bone marrow cellularity (femurs and tibiae) in mice hydrodynamically injected with pCMV6-Epo vector (Epo; n = 6) or pCMV6 vector (control; n = 5), as indicated. Analysis was performed 2 d after injection. Values are mean ± SD, n = 5 (control) and 6 (Epo). One of three representative experiments is shown. (F) Bar graphs showing Lin−Sca1−c-Kit+IL7rα− myelo-erythroid progenitor cells from mice in E as percentage of live singlets. (G) Representative flow cytometric analysis of the hematopoietic myelo-erythroid progenitor population from bone marrow of wild-type C57BL/6 mice after 2 d of Epo exposure. The size of gated populations as percentage of the parental population is shown next to each gate. (H) Bar graphs representing the mean size of each myelo-erythroid progenitor population as defined in G, shown as percentage of the total Lin/Sca-1/IL7Rα−c-Kit+ progenitor fraction in 2-d Epo-exposed (Epo) and control mice. Values are mean ± SD, n = 5, from two experiments. (I) Bar graphs represent the frequencies of LSK subpopulations in bone marrow of 2-d Epo-exposed and control mice. Values are mean ± SD, n = 4. One of two representative experiments is shown. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
This observation was at variance with the notion that Epo acts primarily as a proliferation/survival factor of E lineage–committed cells, attributing the relative increase in erythropoiesis, at least in part, to suppression of alternative differentiation programs. Further analysis showed that elevated systemic Epo did not affect bone marrow cellularity (Fig. 1 E) or the proportion of total myeloid progenitors (Fig. 1 F). However, flow cytometry–based progenitor phenotyping (Pronk et al., 2007) showed that committed E progenitors (preCFU-E and CFU-E) were increased, whereas committed GM progenitors (GMP and preGM) and Mk progenitors (MkPs) were decreased (Fig. 1, G and H). Similar skewing of myelo-erythroid progenitor numbers was observed upon injection of recombinant Epo (unpublished data). To determine if lineage skewing occurred in the upstream Lin−Sca-1+c-Kit+ (LSK) hematopoietic stem cell (HSC)/multipotent progenitor (MPP) compartment, this population was analyzed for Flt3 expression, which is normally associated with loss of Mk/E potential and increased lymphoid potential (Adolfsson et al., 2005). We observed that the proportion of Flt3hi lymphoid-primed MPPs (LMPPs; Adolfsson et al., 2005) was decreased upon Epo exposure (Fig. 1 I), consistent with the observed suppression of lymphoid colony-forming cells (Fig. 1 C). Therefore, at all known lineage bifurcations between HSCs and committed E progenitors, elevated Epo levels impaired the formation of progenitor cells lacking E potential (LMPP, preGM/GMP, and MkP), selectively promoting the formation of E committed progenitors (CFU-E and preCFU-E), and this process is initiated within the HSC/MPP compartment.
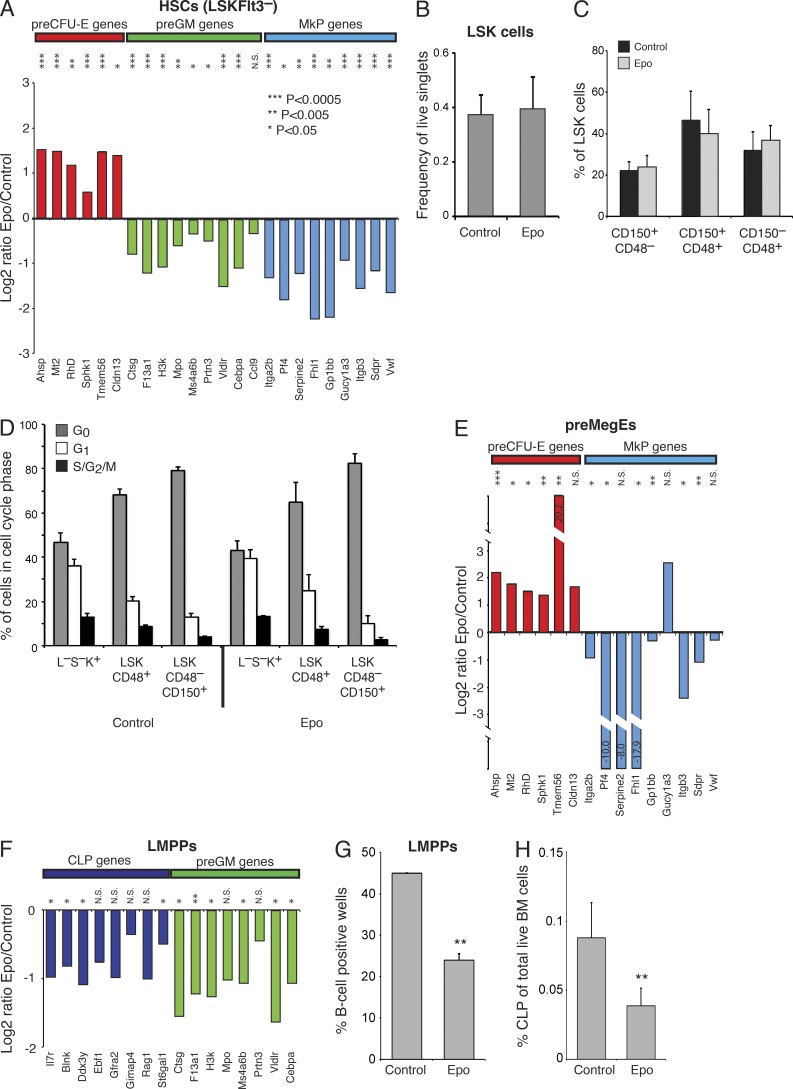
Previous studies have shown that the transcriptional priming of HSCs is an indicator of their lineage potential, and that the priming pattern is regulated by the transcription factors required for subsequent lineage commitment (Bereshchenko et al., 2009; Mancini et al., 2012). To determine whether Epo exposure alters the priming pattern of stem and progenitor cell populations, we mined existing global gene profiling data (Pronk et al., 2007) to identify gene sets highly and selectively expressed in preGMs, preCFU-Es, MkPs, and common lymphoid progenitors (CLPs), representing the first GM, E, Mk, and lymphoid committed progenitors, respectively (Fig. S1). Gene profiling of in vivo Epo-exposed LSKFlt3− cells showed that genes associated with E lineage commitment were up-regulated, whereas those associated with Mk and GM lineage commitment were down-regulated (Fig. 2 A). This occurred without any change to the size or composition of the HSC compartment (Fig. 2, B and C), to the cell cycle status of HSC/MPPs (Fig. 2 D), or to HSC repopulating activity (not depicted). Downstream of HSCs, bipotent preMegEs had up-regulated preCFU-E genes, whereas most MkP gene expression was suppressed (Fig. 2 E), consistent with the observed erythroid bias at the MkP/preCFU-E bifurcation. LMPPs showed general down-regulation of CLP and preGM genes (Fig. 2 F). The decrease in LMPP lymphoid gene expression was accompanied by suppressed LMPP B cell differentiation potential (Fig. 2 G), and a decreased number of downstream CLPs (Fig. 2 H). The effect of Epo on GM and T cell potential of LMPPs remains to be determined. Lineage programming of stem/progenitor cells was therefore regulated by Epo, and correlated with altered lineage potential and progenitor output.
Figure 2.
Epo transcriptionally reprograms HSCs and progenitors in vivo toward an erythroid fate. (A) Bar graphs showing the expression of preCFU-E, preGM, and MkP genes in in vivo, comparing Epo-exposed and control LSKFlt3− cells. Data are shown as log2 of the ratio between Epo-exposed and control cells. Values are means, n = 9, from 3 experiments. (B) Bar graphs showing mean size of Lin−Sca-1+c-Kit+ (LSK) population as percentage of live singlets in mice hydrodynamically injected with pCMV6-Epo vector or pCMV6 control vector on day 2 after injection. Values are mean ± SD, n = 5 (control) and 6 (Epo), from 2 experiments. (C) LSK cells from B were subdivided by CD150 and CD48 expression into MPPs (defined as LSKCD48+CD150+ or LSKCD48+CD150−) and primitive HSCs (defined as LSKCD48−CD150+). The proportion of cells in the indicated fractions is shown as a percentage of the total LSK population. Values are mean ± SD. No significant difference (defined as P < 0.05) was observed between control and Epo-treated populations. (D) MPPs (LSKCD48+) and HSCs (LSKCD48−CD150+) from mice hydrodynamically injected with pCMV6-mEpo vector (Epo, n = 3) or pCMV6 vector (control, n = 3) were analyzed on day 2 after injection for cell cycle status by intracellular staining for Ki67 and DNA content. G0 was defined as Ki67−, G1 as Ki67+/2n DNA content, and S/G2/M as Ki67+/>2n DNA content. Lin−Sca-1−c-Kit+ (L−S−K+) progenitors are included as a positive control for high proliferation rate. Values are mean ± SD, n = 3. One of two representative experiments shown. (E) Expression of preCFU-E and MkP genes in in vivo Epo-exposed and control preMegEs. Data are represented as in A. (F) Expression of CLP and preGM genes in in vivo Epo-exposed and control LMPP. Data are represented as in A. (G) B cell potential of single Epo-exposed and control LSKFlt3hi LMPPs cultured on OP9 stromal cells. Representative FACS profiles of analyzed clone derived from a single cell are shown in Fig. S2 A. Values are mean ± SD, n = 48, from 2 experiments. (H) Bar graphs showing the number of CLPs as percentage of total live BM cells from Epo-exposed and control mice. Values are mean ± SD, n = 5. One of two representative experiments shown. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; N.S., not significant.
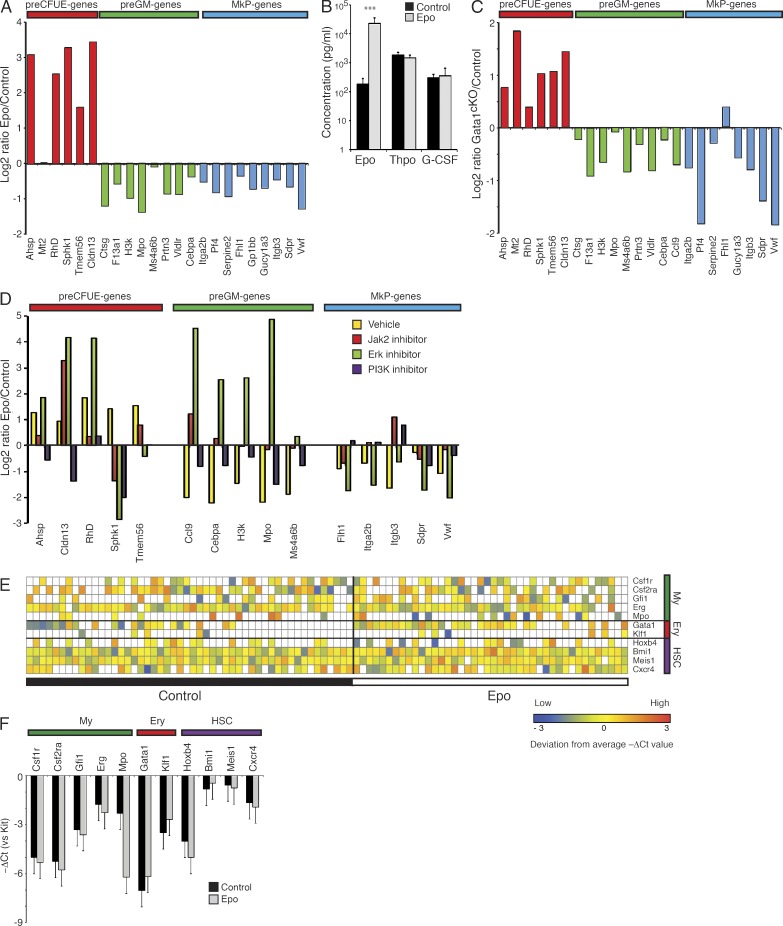
To address if Epo acts directly and instructively on the HSC/MPP compartment, we isolated LSKCD150+Flt3− cells and treated these with recombinant Epo in vitro. We observed a virtually identical pattern of regulation of lineage programming as seen upon in vivo Epo exposure (Fig. 3 A). To determine if endogenous erythropoietic stress would cause similar transcriptional HSC reprogramming, CD45.2 Gata1 conditional null (or control) cells (CD45.2) were co-transplanted with wild-type CD45.1 cells into lethally irradiated CD45.1/2 hosts at a 10:1 ratio, followed by Gata1 deletion through poly(I-C) induction of the Mx1-Cre transgene. This selectively blocks erythropoiesis at the preMegE stage (Mancini et al., 2012), leading to a selective increase in circulating Epo levels (Fig. 3 B) and forcing the wild-type CD45.1 HSCs present along Gata1-deficient CD45.2 HSCs to generate ∼10× more erythrocytes on a per-cell basis. 6 wk later, CD45.1 LSKFlt3− HSC/MPPs were isolated and gene profiled. This showed that CD45.1 LSKFlt3− cells present along Gata1-deficient CD45.2 cells up-regulated erythroid genes and concomitantly suppressed preGM and MkP genes (Fig. 3 C). This divergent regulation was disrupted in the Gata1-deficient CD45.2 cells exposed to the same Epo levels (unpublished data). Transcriptional HSC reprogramming therefore occurs in response to exogenous Epo, to endogenous stress erythropoiesis, and to direct in vitro Epo stimulation and is at least partially GATA-1–dependent.
Figure 3.
Cellular and molecular mechanism of Epo action on HSCs. (A) LSKFlt3−CD150+ cells were cultured in the presence (Epo) or absence (Control) of Epo. After 24 h, cells were harvested and analyzed for gene expression. Data are represented as in Fig. 2 A. Values are means, n = 2, from 2 experiments with triplicate measurements. (B) Bar graph showing different serum cytokines levels in CD45.1/2 mice competitively transplanted with 105 CD45.1 wild-type cells and either 106 CD45.2 Gata1fl/fl; Mx1-Cretg/+ (cKO) or 106 Gata1fl/fl BM cells (Con). Values are mean ± SD, n = 9 (control) and 8 (cKO), from 2 experiments. (C) CD45.1/2 mice were competitively transplanted with 105 CD45.1 wild-type and either 106 CD45.2 Gata1fl/fl; Mx1-Cretg/+ (cKO) or 106 Gata1fl/fl BM cells (Con). Cre recombination was induced by three poly(I-C) injections at 2-d intervals, and CD45.1+CD45.2−LSKFlt3−CD150+ cells were isolated 6 wk after the first poly(I-C) injection and subjected to gene expression profiling. Data are represented as the log2 of the ratio between gene expression in WT/cKO and WT/Control co-transplanted mice after normalization to Hprt. Values are means, n = 2, from 2 experiments with triplicate measurements. (D) LSKFlt3−CD150+ cells were cultured in the presence (Epo) or absence (Control) of Epo. For both conditions, individual cultures were supplied with either vehicle (DMSO) or inhibitors of one of the following kinases: 25 µM Jak2 (AG490), 25 µM PI3K (LY294002), or 25 µM ERK1/2 (PD98059). After 24 h, cells were harvested and analyzed for gene expression. Values are means, n = 2, from 2 experiments with triplicate measurements. (E) Heat map of gene expression in single sorted LSKFlt3− HSCs analyzed by microfluidics-based real time PCR. Expression values were normalized using Kit expression and are shown as deviation from the mean expression value of each individual gene. n = 3 from 3 experiments. (F) Bar graph of mean expression of GM-, E-, and HSC-associated genes in single cells from E. Values represent the mean Δ(ΔCt(gene)−ΔCt(Kit)) for each of the genes indicated, including only cells where expression was detected. ***, P < 0.0005.
There are several signaling pathways that emanate from the Epo receptor, including the phosphoinositol-3 (PI3) kinase/Akt, Erk/MAPK, and JAK–STAT pathways (Richmond et al., 2005). To determine their individual contributions to Epo-induced transcriptional reprogramming, LSKFlt3−CD150+ HSCs were exposed to Epo in vitro in the presence of inhibitors specific for these pathways (Fig. 3 D). We observed that up-regulation of preCFU-E genes was consistently dependent on PI3 kinase activation, whereas JAK/STAT and Erk activation was involved in a gene-specific manner. In contrast, preGM gene expression was not significantly affected by PI3 kinase inhibition, whereas, remarkably, Erk inhibition converted the normal inhibition by Epo to robust up-regulation. The major Epo-activated signaling pathways therefore play distinct and lineage-specific roles in reprogramming of the HSC transcriptome. We have previously shown that loss of C/EBPα function leads to loss of myeloid programming in LSKFlt3−CD150+ HSC/MPPs (Bereshchenko et al., 2009). Direct regulation of myeloid genes by C/EBPα may therefore explain the highly coordinated regulation of preGM genes upon inhibitor treatment.
To further investigate the cellular mechanism underlying Epo-induced transcriptional reprogramming, we performed single cell gene expression profiling of control and Epo exposed LSKFlt3− HSCs. We investigated the expression of GM (Mpo, Erg, Gfi1, Csf1r, and Csf2ra)-, E (Gata1 and Klf1)-, and HSC (Hoxb4, Bmi1, Meis1, and Cxcr4)-associated genes. We observed a 50% increase in proportion of cells expressing one or more E genes (Fig. 3 E). In addition, the average expression of E genes in these cells was increased (1.8-fold, P = 0.10), with a similar decrease in the average expression of GM genes (1.7-fold, P = 0.008; Fig. 3 F). Importantly, we consistently observed coexpression of E genes with one or more GM genes, as well as HSC-associated genes, consistent with the increase in E programming occurring in multipotent stem/progenitor cells, rather than through the expansion of a subpopulation of committed erythroid progenitors.
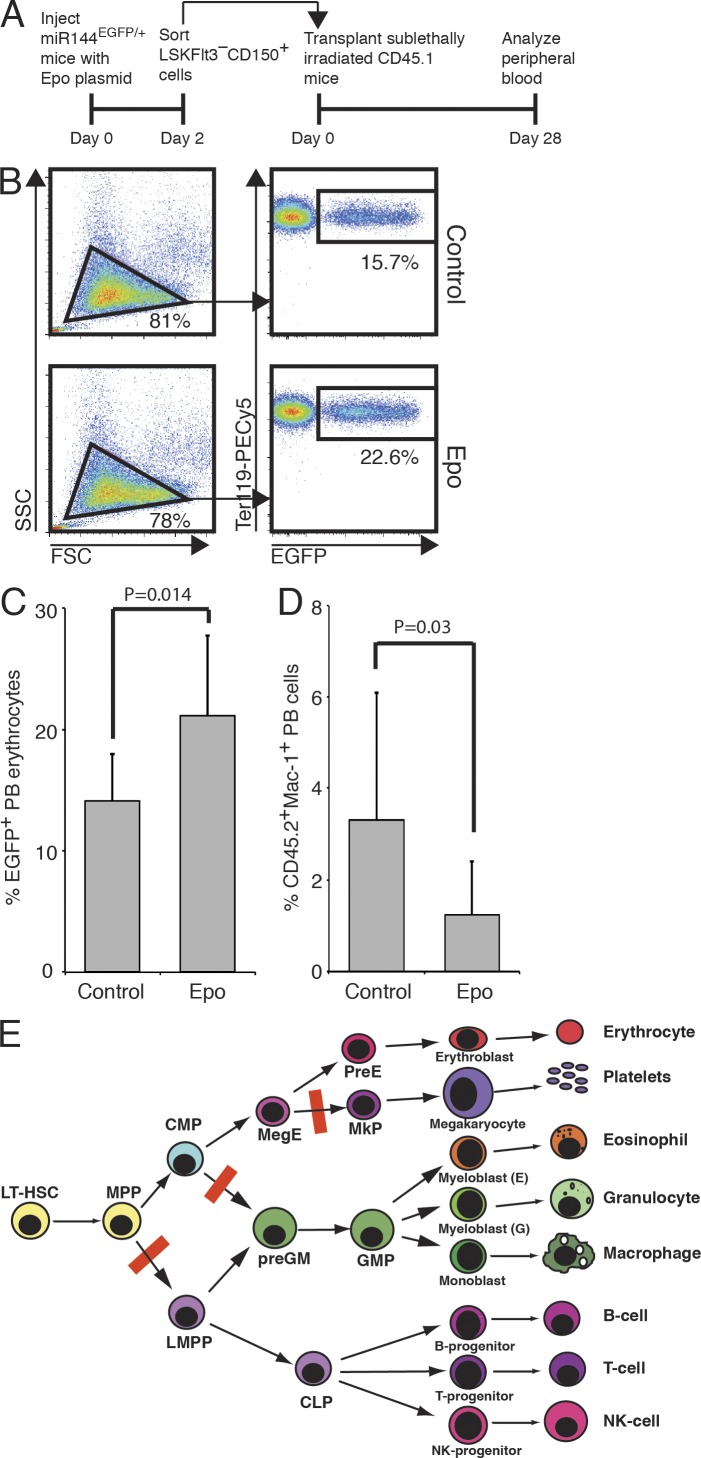
To establish if Epo-induced reprogramming of stem/progenitor cells resulted in skewing of their lineage output, we transplanted control and Epo-exposed HSCs (defined as LSKFlt3−CD150+; CD45.2 allotype) into sublethally irradiated CD45.1 recipients. Transplanted HSCs were isolated from CD45.2 miR144/451eGFP/+ mice. This strain contains an EGFP knockin into a microRNA locus uniformly expressed in E lineage cells, including circulating mature erythrocytes, allowing for EGFP-based tracking of E lineage output (Rasmussen and O’Carroll, 2011; Mancini et al., 2012; Fig. 4 A). Analysis of peripheral blood showed that Epo-exposed HSCs generated significantly higher numbers of erythrocytes (Fig. 4, B and C) and fewer myeloid cells (Fig. 4 D). Overall, a fourfold increase of the erythroid/myeloid output ratio was observed. Exposure of HSC/MPPs to Epo is therefore sufficient to bias lineage output away from the myeloid lineage and toward erythropoiesis, even in the absence of exogenous Epo during their further lineage commitment and differentiation.
Figure 4.
Epo-exposed HSCs generate reduced myeloid lineage output and increased erythroid lineage output. (A) LSKFlt3−CD150+ cells were isolated from Epo-exposed or control miR144/451Egfp/+ mice and transplanted into sublethally irradiated CD45.1 recipients, followed by peripheral blood analysis 28 d after transplantation. (B) Representative flow cytometric analysis of peripheral blood erythrocytes from CD45.1 recipients transplanted in A. The gated population shows the Ter119+EGFP+ donor-derived erythrocytes. The size of gated populations as a percentage of the parental population is shown next to gates. (C and D) Bar graphs representing the mean percentage of donor-derived Ter119+EGFP+ PB erythrocytes and CD45.2+Mac-1+ PB cells, respectively, from mice transplanted in A. Values are mean ± SD, n = 9 (control) and 12 (Epo), from 2 experiments. (E) The overall effect of Epo exposure on the hematopoietic hierarchy: generation of LMPP/CLP, preGM, and MkP progenitors is impaired, leading HSC output to be directed toward the E lineage by blocking alternative exits from the HSC compartment. The result is an erythroid superhighway from the HSC to the preCFU-E.
These results challenge our current view of lineage-specific cytokine action in two ways. First, they show that Epo can act on HSC/MPPs, in addition to E lineage–committed cells, to simultaneously increase E and decrease GM output. Second, we find that this occurs through transcriptional reprogramming, leading to increased E lineage priming and decreased GM, Mk, and lymphoid priming, and the systematic suppression of alternative lymphoid and myeloid lineage options at all lineage bifurcations examined. Up-regulation of erythroid gene expression showed a strong dependence on PI3 kinase signaling. In contrast, Erk signaling was critical for myeloid gene suppression; if Erk activity was blocked, Epo stimulation led to a general up-regulation of myeloid gene expression. This dissociation of lineage reprogramming is consistent with an instructive, rather than selective, effect of Epo on the HSC compartment; Epo exposure could lead to the selective expansion or survival of cells expressing high levels of preCFU-E and low levels of MkP and preGM genes. However, this model is not readily reconciled with the ability of simultaneous Epo stimulation and Erk inhibition to up-regulate CFU-E and preGM genes and suppress MkP genes. Our results therefore show that the high systemic levels of Epo associated with severe anemia can act in a direct and instructive manner on the HSC/MPP compartment, leading to a reduction of GM, Mk, and lymphoid phenotypic progenitors and colony-forming cells. The overall result is the creation of enhanced throughput from HSCs to the E compartment, an “erythroid superhighway,” allowing the focused generation of E-lineage progenitors (Fig. 4 E), upon which Epo may then further act through its well-characterized proliferative and pro-survival effects.
MATERIALS AND METHODS
Mouse lines.
C57BL/6J (CD45.2+) mice and congenic B6.SJL-PtprcaPep3b/BoyJ (CD45.1+ congenic C57BL/6J) mice were bred at the University of Edinburgh. The Gata1 conditional allele (Lindeboom et al., 2003), Mx1-Cre transgenic mice (Kühn et al., 1995), and mir144/451-eGFP reporter mice (Rasmussen and O’Carroll, 2011) were previously described. Mouse strains were backcrossed to C57Bl6/J for six or more generations before use, and mice were aged 8–12 wk at the start of experiments. All animal procedures were approved by the University of Oxford Clinical Medicine Ethical Review Committee and were maintained in accordance with Institutional and UK Home Office guidelines.
Hydrodynamic gene transduction and ELISA.
Mice were tail vein–injected, as described previously (Hirai et al., 2006), in Krebs-Ringer buffer with 1 µg of a pCMV6-Entry vector containing Epo cDNA (OriGene) or empty pCMV6-Entry vector. Epo, Thrombopoietin, and G-CSF serum levels were determined using Quantikine immunoassays (R&D Systems).
Flow cytometry, cell separation, and cell sorting.
Flow cytometry was performed essentially as previously described (Bereshchenko et al., 2009). A list of antibodies used is provided in Table S3. Cells were sorted using FACSAria II cell sorters and analyzed using LSRFortessa (BD). All FACS data were analyzed with FlowJo software (Tree Star).
In vitro culture assays.
Media for colony-forming assays were from STEMCELL Technologies and used according to the manufacturer’s instructions. 500 cells were seeded per culture in M3534, M3434, and Megacult media and 1,000 cells in M3436 and M3630 media. The colonies generated in methylcellulose were scored as BFU-E, CFU-GM, and CFU-B by morphology 7–10 d after seeding, and as CFU-Mk (in MegaCult assay) by acetylcholinesterase staining. To score E colonies in multilineage culture (M3434), colonies were stained with 2,7-diaminofluorene (DAF) staining solution (10 mg/ml DAF in 90% glacial acetic acid, 30% H2O2, and 200 mM Tris-HCl, pH 7.5) and DAF-positive cells were identified as cells with intracellular blue granules. Cells were plated in duplicate or triplicate for each assay. B cell potential of LMPPs was evaluated as previously described (Adolfsson et al., 2005) with B-lineage cells defined as Mac-1−NK1.1−CD19+ cells (Fig. S2 A). For in vitro Epo treatment, BM LSKCD150+Flt3− cells were incubated with 200 ng/ml of recombinant murine Epo (R&D Systems) for 24 h, followed by gene expression analysis. Where indicated, cells were treated with 25 µM Jak2 inhibitor (AG490; EMD Millipore), PI3-kinase inhibitor (LY294002; EMD Millipore), Erk1/2 inhibitor (PD98059; Sigma-Aldrich), or DMSO vehicle.
Multiplex specific target amplification and qPCR analysis.
Multiplexing of specific target amplification of the cDNA samples from various BM populations and qPCR analysis using UPL assays (Roche) were performed as described previously (Sanjuan-Pla et al., 2013). For each gene, the chosen qPCR assay was the most highly ranked by UPL Assay Design Center.
Single cell gene expression analysis.
For single cell LSKFlt3− gene expression analysis, cell capture and target preamplification were performed using the C1 system (Fluidigm) according to the manufacturer’s instructions. Real-time PCR was performed using the BioMark 96.96 Dynamic Array platform (Fluidigm) and DELTAgene assays (Fluidigm; Table S2). In total, >60 single cells from both Epo-exposed and control mice (∼20 cells from each of 3 biological replicates) were analyzed, and those scoring positive for Kit mRNA expression (>85% of cells analyzed) were included in the analysis. ΔCt values relative to Kit (Ct(Kit) − Ct(Gene)) were zero centered for each gene by subtraction of the mean value of all positive cells for the gene. These normalized values were used to generate heat maps, and to calculate the cumulative significance of Epo regulation of E and GM genes (i.e., all E genes without Epo against all E genes with Epo, and the same comparison for GM genes) using Student’s t test.
Bone marrow transplantation.
To evaluate in vivo capacities to generate erythrocytes and myeloid cells, 500 Lin−Sca1+c-Kit+ CD150+ cells from miR144/451eGFP/+ (CD45.2 allotype) after 2 d of Epo exposure (or control) were transplanted into sublethally irradiated CD45.1 recipients. At the indicated time point, blood samples were evaluated by flow cytometry for the levels of GFP+Ter119+ erythrocytes that were identified by their distinct scatter profile. Peripheral blood analysis was performed as described previously (Kirstetter et al., 2006). For Gata1 conditional knockouts, lethal irradiation, bone marrow transplantation, poly(I-C)-induced recombination, and peripheral blood analysis were performed as described previously (Mancini et al., 2012). To evaluate the effect of E stress on lineage priming of HSCs, Gata1-null BM cells (allotype CD45.2) and competitor cells (allotype CD45.1) were transplanted in 10:1 ratio in recipients (CD45.1/CD45.2). The donor and competitor HSC/MPPs (LSKFlt3−) were sorted for gene expression analysis after 6 wk of first poly(I-C) injection.
Lineage-specific gene sets.
Previously published microarray data (Pronk et al., 2007) were normalized and analyzed as previously described (Bereshchenko et al., 2009). Genes differentially expressed between progenitor subsets (CLP, preGM, preMegE, MkP, and preCFU-E) were manually curated to identify those selectively and highly expressed in each of the following populations: CLP, preGM, MkP, and preCFU-E (Table S1).
Statistical analyses.
Statistical significance for all the data was calculated using Student’s t test.
Online supplemental material.
Lineage-specific genes and their normalized expression values are found in Fig. S1 and Table S1. Gating strategies for OP9 cultures and CLPs are shown in Fig. S2. Table S2 contains DELTAGene assays used in single-cell PCR, and Table S3 the list of antibody conjugates used. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131189/DC1.
Supplementary Material
Acknowledgments
This work was supported by a Strategic Award and Program Grant from the Medical Research Council to C. Nerlov, and by a Leukemia and Lymphoma Research Grant to A.J. Mead.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- CLP
- common lymphoid progenitor
- Epo
- erythropoietin
- GM
- granulocyte/macrophage
- GMP
- GM progenitor
- HSC
- hematopoietic stem cell
- LMPP
- lymphoid-primed MPP
- Mk
- megakaryocyte
- MkP
- Mk progenitor
- MPP
- multipotent progenitor
- PI3
- phosphoinositol-3
- preGM
- pre-GM progenitor
References
- Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.J., Thoren L.A., et al. 2005. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 121:295–306 10.1016/j.cell.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Bereshchenko O., Mancini E., Moore S., Bilbao D., Månsson R., Luc S., Grover A., Jacobsen S.E., Bryder D., Nerlov C. 2009. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell. 16:390–400 10.1016/j.ccr.2009.09.036 [DOI] [PubMed] [Google Scholar]
- Cheers C., Haigh A.M., Kelso A., Metcalf D., Stanley E.R., Young A.M. 1988. Production of colony-stimulating factors (CSFs) during infection: separate determinations of macrophage-, granulocyte-, granulocyte-macrophage-, and multi-CSFs. Infect. Immun. 56:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Zhang P., Dayaram T., Hetherington C.J., Mizuno S., Imanishi J., Akashi K., Tenen D.G. 2006. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7:732–739 10.1038/ni1354 [DOI] [PubMed] [Google Scholar]
- Jelkmann W., Wiedemann G. 1990. Serum erythropoietin level: relationships to blood hemoglobin concentration and erythrocytic activity of the bone marrow. Klin. Wochenschr. 68:403–407 10.1007/BF01648581 [DOI] [PubMed] [Google Scholar]
- Kirstetter P., Anderson K., Porse B.T., Jacobsen S.E., Nerlov C. 2006. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat. Immunol. 7:1048–1056 10.1038/ni1381 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Lin C.S., Lim S.K., D’Agati V., Costantini F. 1996. Differential effects of an erythropoietin receptor gene disruption on primitive and definitive erythropoiesis. Genes Dev. 10:154–164 10.1101/gad.10.2.154 [DOI] [PubMed] [Google Scholar]
- Lindeboom F., Gillemans N., Karis A., Jaegle M., Meijer D., Grosveld F., Philipsen S. 2003. A tissue-specific knockout reveals that Gata1 is not essential for Sertoli cell function in the mouse. Nucleic Acids Res. 31:5405–5412 10.1093/nar/gkg723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E., Sanjuan-Pla A., Luciani L., Moore S., Grover A., Zay A., Rasmussen K.D., Luc S., Bilbao D., O’Carroll D., et al. 2012. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J. 31:351–365 10.1038/emboj.2011.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D. 2008. Hematopoietic cytokines. Blood. 111:485–491 10.1182/blood-2007-03-079681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller N., Sarrazin S., Kandalla P.K., Espinosa L., Stanley E.R., Nutt S.L., Moore J., Sieweke M.H. 2013. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 497:239–243 10.1038/nature12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., Bryder D. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Rasmussen K.D., O’Carroll D. 2011. The miR-144/451eGFP allele, a novel tool for resolving the erythroid potential of hematopoietic precursors. Blood. 118:2988–2992 10.1182/blood-2011-04-350728 [DOI] [PubMed] [Google Scholar]
- Richmond T.D., Chohan M., Barber D.L. 2005. Turning cells red: signal transduction mediated by erythropoietin. Trends Cell Biol. 15:146–155 10.1016/j.tcb.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Rieger M.A., Hoppe P.S., Smejkal B.M., Eitelhuber A.C., Schroeder T. 2009. Hematopoietic cytokines can instruct lineage choice. Science. 325:217–218 10.1126/science.1171461 [DOI] [PubMed] [Google Scholar]
- Sanjuan-Pla A., Macaulay I.C., Jensen C.T., Woll P.S., Luis T.C., Mead A., Moore S., Carella C., Matsuoka S., Jones T.B., et al. 2013. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 502:232–236 10.1038/nature12495 [DOI] [PubMed] [Google Scholar]
- Shiozawa Y., Jung Y., Ziegler A.M., Pedersen E.A., Wang J., Wang Z., Song J., Wang J., Lee C.H., Sud S., et al. 2010. Erythropoietin couples hematopoiesis with bone formation. PLoS ONE. 5:e10853 10.1371/journal.pone.0010853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari K., Asano S., Shirafuji N., Kodo H., Ozawa K., Takaku F., Kamachi S. 1989. Serum granulocyte colony-stimulating factor levels in healthy volunteers and patients with various disorders as estimated by enzyme immunoassay. Blood. 73:117–122 [PubMed] [Google Scholar]
- Wu H., Liu X., Jaenisch R., Lodish H.F. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 83:59–67 10.1016/0092-8674(95)90234-1 [DOI] [PubMed] [Google Scholar]
- Zhang G., Budker V., Wolff J.A. 1999. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 10:1735–1737 10.1089/10430349950017734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.