Abstract
To identify high-affinity interactions between long-chain α-neurotoxins and nicotinic receptors, we determined the crystal structure of the complex between α-btx (α-bungarotoxin) and a pentameric ligand-binding domain constructed from the human α7 AChR (acetylcholine receptor) and AChBP (acetylcholine-binding protein). The complex buries ~2000 Å2 (1 Å = 0.1 nm) of surface area, within which Arg36 and Phe32 from finger II of α-btx form a π-cation stack that aligns edge-to-face with the conserved Tyr184 from loop-C of α7, while Asp30 of α-btx forms a hydrogen bond with the hydroxy group of Tyr184. These inter-residue interactions diverge from those in a 4.2 Å structure of α-ctx (α-cobratoxin) bound to AChBP, but are similar to those in a 1.94 Å structure of α-btx bound to the monomeric α1 extracellular domain, although compared with the monomer-bound complex, the α-btx backbone exhibits a large shift relative to the protein surface. Mutational analyses show that replacing Tyr184 with a threonine residue abolishes high-affinity α-btx binding, whereas replacing with a phenylalanine residue maintains high affinity. Comparison of the α-btx complex with that coupled to the agonist epibatidine reveals structural rearrangements within the binding pocket and throughout each subunit. The overall findings high-light structural principles by which α-neurotoxins interact with nicotinic receptors.
Keywords: crystal structure, molecular recognition, neurotoxin, nicotinic acetylcholine receptor
INTRODUCTION
Snake venom α-neurotoxins exert their lethal consequences through high affinity, slowly reversible binding to AChRs (acetylcholine receptor) at the neuromuscular junction [1]. These twin properties have made α-neurotoxins invaluable reagents to quantify AChR number [2] and track cellular and subcellular locations [3]. High-affinity binding of α-neurotoxins is thus a striking example of evolution-driven molecular recognition.
The prototypical α-neurotoxin α-btx (α-bungarotoxin) is a small basic protein with a three-finger scaffold held together by intramolecular disulfide bonds. The structure of α-btx was determined by X-ray crystallography and NMR, alone and in complex with peptides from a region of the AChR ligand-binding site known as loop C [4-6], a structural element essential for agonist and antagonist binding [7-9]. The ability of α-btx to form complexes with loop C peptides suggested that loop C was a primary site for α-btx binding in the intact AChR. Furthermore, in animals resistant to α-btx, N-linked glycosylation within loop C eliminates high-affinity binding [10-12], and studies of site-directed mutations showed that loop C is the most important of the multiple binding site loops in conferring high affinity for α-neurotoxins [13,14]. The crystal structure of the related α-ctx (α-cobratoxin) bound to the pentameric AChBP (acetylcholine-binding protein) revealed the orientation of the toxin backbone relative to the ligand-binding site [15], and showed that loop C lodged between fingers I and II of α-ctx. However, the resolution of 4.2 Å (1 Å = 0.1 nm) limited the accuracy with which inter-residue interactions could be analysed. The low sequence homology between AChBP and AChR further limits interpretation of α-neurotoxin-induced structural changes as they relate to AChR function. The structure of α-btx bound to a monomeric extracellular domain from the muscle α1 subunit defined the inter-residue contacts, owing to the resolution of 1.94 Å [16], but the absence of the complementary subunit of the ligand-binding site may have altered the orientation of the toxin and the inter-residue interactions. Comparison of the α-btx–α1 and α-ctx–AChBP complexes reveals marked differences in both the orientation of the toxin backbone relative to the protein surface and the inter-residue interactions. Thus the molecular scale interactions between α-neurotoxins and their nicotinic receptor targets remain unresolved.
To identify inter-residue interactions that stabilize α-neurotoxins bound to their nicotinic receptor targets, we determined the crystal structure of α-btx bound to an α7 receptor ligand-binding domain chimaera [17]. Based on inter-residue interactions deduced from the structure, mutational analyses disclosed essential interactions required for high-affinity binding of α-btx to the α7 AChR.
EXPERIMENTAL
Construction, protein expression and purification of the α7/AChBP chimaera
The α7/AChBP chimaera was expressed in the yeast Pichia pastoris and purified as described previously [17]. To assess the stability of the complex between α-btx and α7/AChBP, the protein was bound to an anti-FLAG M2 agarose affinity gel (Sigma) in 50 mM potassium phosphate buffer, pH 6.0, and 0.15 M NaCl and incubated with an excess of 125I-labelled α-btx (PerkinElmer) for 1 h at 4 °C. After removing unbound toxin, the gel was diluted into phosphate buffer at 21 °C, and the amount of 125I-labelled α-btx retained by the α7/AChBP-bound gel was measured at specified times.
Sample preparation for crystallization
α-btx (Invitrogen) was mixed with the α7/AChBP chimaera (~1 mg/ml) at a molar ratio of 3:1 in 50 mM potassium phosphate buffer, pH 6.0, and 0.15 M NaCl. The sample was then treated with Endo Hf (New England Biolabs) overnight at 4 °C, followed by removal of Endo Hf using amylose resin and then size-exclusion chromatography using Superdex 200. Endo Hf treatment was necessary for crystal production even though part of the sugar chain was still visible in the final structure, perhaps because it was partially protected from Endo Hf digestion by bound α-btx. Thus trimming the terminal braches of the glycan chains on the protein was apparently critical to crystallization.
Crystallization
Crystals of the α-btx–α7/AChBP complex were grown at 18–24 °C using the hanging-drop vapour diffusion method by mixing protein (2–3 mg/ml, 1–2 μl) with an equal volume of reservoir solution. Large rod-like crystals were obtained with a reservoir solution containing 0.1 M potassium phosphate buffer, pH 6.0, 20% PEG4000, 0.2% NaN3, 0.1 mM Triton X-100 and 10 mM spermidine. Spermidine and Triton X-100 greatly improved the crystal quality and reduced mosaicity.
Data collection and structure determination
Data were collected at the ALS (Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA, U.S.A.) beam line 8.2.2 and the APS (Advanced Photon Source, Argonne National Laboratory, Chicago, IL, U.S.A.) beam line 23ID-B at 100 K. To improve resolution, before flash cooling, the crystals were dehydrated overnight in crystallization buffer containing 50% (v/v) glycerol. The data were processed using HKL2000 [18]. The crystals belonged to the space group P6522, with the unit cell dimensions a = 142.150 Å, b = 142.150 Å and c= 518.135 Å. There is one pentamer in each asymmetric unit. The structure was solved by molecular replacement with the program Molrep [19] using the α7/AChBP chimaera pentamer (PDB code 3SQ6) and α-btx (PDB code 2QC1) as partial search models. The model for the crystal structure was rebuilt using Coot [20] and refined with CNS [21]. The initial electron density indicated that all five subunits within the pentamer had nearly identical structures except for loop C, which, together with the bound toxin molecule, adopts slightly different orientations in the different crystal-packing environments. Therefore NCS (non-crystallographic symmetry) restraints were applied only to the main body of the α7/AChBP pentamer and excluded residues 180 through 193, as well as α-btx. Crystallographic analysis and refinement statistics are summarized in Table 1. The structural co-ordinates reported will appear in the PDB under accession code 4HQP.
Table 1. Data collection and refinement statistics.
Numbers in parentheses represent the highest resolution shell. NAG, N-acetylglucosamine.
| Parameter | α7/AChBP chimaera–α-btx |
|---|---|
| Data collection | |
| Space group | P6522 |
| Unit cell | |
| a , b , c (Å) | 142.150, 142.150, 518.135 |
| α, β, γ (°) | 90.000, 90.000, 120.000 |
| Resolution (Å) | 50–3.5 (3.63–3.50) |
| R sym | 0.071 (0.672) |
| I/σ (I) | 15.2 (2.0) |
| Completeness (%) | 98.4 (98.2) |
| Redundancy | 3.6 (3.4) |
| Number of unique reflections | 38979 |
| Refinement | |
| Rwork/Rfree (%) | 0.311/0.321 |
| Number of non-H atoms | |
| Protein | 10980 |
| Ligands | |
| NAG | 112 |
| Average B-factors (Å2) | |
| Protein | 159.7 |
| Ligands | |
| NAG | 168.6 |
| RMSDs | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.00 |
| Ramachandran plot | |
| Favoured region (%) | 88.0 |
| Allowed region (%) | 10.2 |
| Outlier region (%) | 1.8 |
Binding of 125I-labelled α-btx to AChRs expressed in HEK (human embryonic kidney)-293 cells
cDNA encoding the α7/5HT3A (5-hydroxytryptamine type 3A) chimaeric receptor was transfected into HEK-293 cells using the calcium phosphate precipitation method as described previously [22]. Mutations were generated using the QuikChange® site-directed mutagenesis kit (Stratagene) and were confirmed by sequencing the entire coding region. Transfected cells were harvested by gentle agitation in PBS, centrifuged at 1000 g for 1 min and resuspended in potassium Ringer’s solution (140 mM KCl, 5.4 mM NaCl, 1.8 mM CaCl2, 1.7 mM MgCl2 and 25 mM Hepes, pH 7.4). Cell suspensions were mixed with the concentrations of 125I-labelled α-btx (PerkinElmer) specified and incubated for 4 h at 21 °C before separating bound complexes from the free toxin using a Brandel M-48T cell harvester. Non-specific binding could not be determined by incubation in the presence of an excess of competitive ligand, because the mutant AChRs impaired binding of small-molecule agonists and antagonists. Also, unlabelled α-btx was not suitable because it can displace the chemically similar 125I-labelled α-btx from non-specific sites, and its affinity is markedly reduced by the Y184T mutation. Thus non-specific binding was determined by applying identical procedures to cells transfected with an equal amount of the pRBG4 vector containing the muscle AChR δ-subunit cDNA, which induces protein expression but does not form AChRs. At a saturating concentration of 30 nM 125I-labelled α-btx, non-specific binding was 3.4 ± 1.0% (±S.D., n = 7) of that obtained for the control α7/5HT3A receptor. After subtracting non-specific binding, the amount of bound 125I-labelled α-btx was expressed as a percentage of the maximum binding obtained for the control non-mutant AChR, and the Hill equation was fitted to determinations of percent occupancy against 125I-labelled α-btx using GraphPad Prism 5 software.
RESULTS
Crystallization of the complex between α-btx and the α7/AChBP chimaera
The α7/AChBP ligand-binding domain shares 64% sequence identity and 71% similarity with the human α7 AChR [17]; all residues in loop C and those that line the aromatic-rich binding cavity originate from native α7. Under stoichiometric binding conditions, a single complex forms with five α-btx molecules bound to one molecule of the α7/AChBP pentamer. Measurements of radiolabelled α-btx dissociation from α7/AChBP show that the complex is highly stable (Supplementary Figure S1 at http://www.biochemj.org/bj/454/bj4540303add.htm). The complex remained stable throughout purification by gel filtration and crystallization, and afforded crystals that diffracted to a resolution of 3.5 Å (I/σ cut off of 2). The structure was solved by molecular replacement using the structures of the α7/AChBP chimaera and α-btx bound to the muscle α1 extracellular domain as partial search models (PDB codes 3SQ6 and 2QC1). The asymmetric unit contained one α7/AChBP pentamer with five molecules of α-btx bound, which allowed five-fold NCS averaging in all regions except loop C and α-btx (see the Experimental section). The average B-factor is higher than expected for a 3.5 Å structure, and may reflect slight disordering within the crystal owing to partial deglycosylation, radiation damage or a combination of the two. Nevertheless, the refined structural model is supported by well-defined electron density (Supplementary Figure S2 at http://www.biochemj.org/bj/454/bj4540303add.htm), and enabled structural and mutational analyses. Statistics of data collection and structural refinement are given in Table 1.
Overall structure
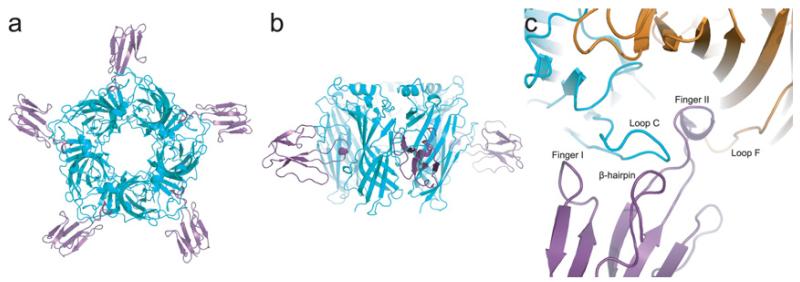
The structure of the complex is shaped like a star with the five α-btx molecules forming the vertices (Figure 1a). Diagonal pairs of α-btx molecules are separated by ~110 Å, whereas the height of the complex, ~55 Å, mirrors that of the apo protein because the toxin binds to the middle of the pentamer barrel (Figure 1b). Finger II of α-btx inserts deeply into the region of the subunit interface that forms the ligand-binding site, displacing loop C of the principal subunit away from an axis through the central lumen (Figure 1c). Loop C lodges between fingers I and II of α-btx, and contributes to a large region of contact where the two protein surfaces complement one another precisely. Formation of the complex buries ~1440 Å2 of surface area on the principal subunit and ~590 Å2 on the complementary subunit. When the structure is viewed from the side, the long axis of α-btx is normal to the pentamer barrel; whereas, when viewed from the top, the α-btx molecules tilt in a clockwise direction (Figures 1a and 1b).
Figure 1. Overall structure of the complex between α-btx (purple) and the α7/AChBP chimaera (cyan).
PDB code 4HQP. (a) Top view. (b) Side view. (c) Close-up of the view in (a) showing α-btx (purple) and the principal (cyan) and complementary (orange) subunits of the α7/AChBP chimaera.
Comparison with previous structures
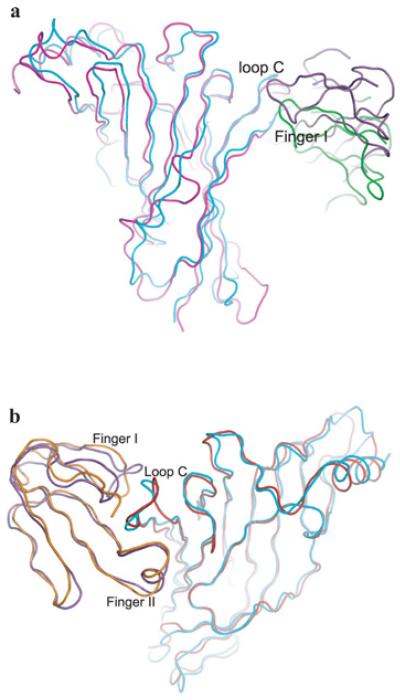
The structure of the complex diverges from that of α-btx bound to the monomeric α1 extracellular domain [19] (PDB code 2QC1), with the main chain of α-btx exhibiting a large rigid body shift relative to the protein surface (Figure 2a). Possible reasons for this include the presence of both principal and complementary subunits and the absence of toxin–sugar interactions in the structure. After superimposing the main chains of α7/AChBP and the monomeric α1 extracellular domain, the RMSD was 1.71 Å, indicating similar conformations despite the sequence differences. However, when the superimposition is viewed from the top, α-btx rotates approximately 20 ° clockwise in the α7/AChBP chimaera compared with the α1 complex, and when viewed from the side the outermost ends of α-btx tilt up approximately 30 ° in the α7/AChBP chimaera compared with the α1 complex, and the tip of toxin finger II translates 4.5 Å away from the surface of the complementary subunit. Superimposition of the main chains of α-btx in the two complexes yields an RMSD of 0.51 Å, indicating nearly identical protein backbones.
Figure 2. Comparison of α-neurotoxin complexes with α7/AChBP, α 1 and AChBP.
(a) Structural superimposition of α7/AChBP (cyan) bound to α-btx (purple) and α1 (magenta)bound to α-btx (green) (PDB code 2QC1). (b) Structural superimposition of α7/AChBP (cyan)bound to α-btx (purple) and AChBP (red) bound to α-ctx (orange) (PDB code 1YI5).
On the other hand, the orientations of the protein main chains of α-btx and α7/AChBP are similar to those in the complex between α-ctx and AChBP [15] (PDB code 1YI5) (Figure 2b). After superimposing the main chains of α7/AChBP and AChBP, the RMSD is 1.42 Å, and that between α-btx and α-ctx is 1.23 Å, indicating that the two complexes are similar at the level of the protein backbone.
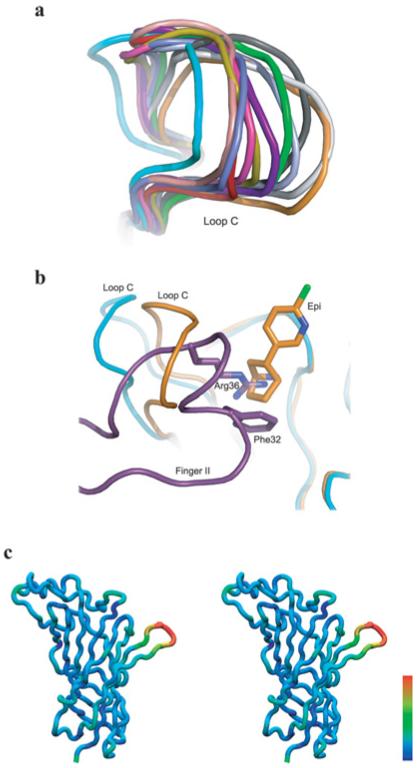
Comparison of α7/AChBP with bound α-btx against that with the bound agonist epibatidine reveals structural rearrangements of the protein backbone. The most significant change occurs in loop C, which exhibits an opened-up conformation in all five subunits within the asymmetric unit. This uniformly opened-up conformation contrasts with the wide range of conformations of loop C in the apo structure of α7/AChBP, and with the uniformly closed-in conformation of loop C in the structure with bound epibatidine [17] (Figure 3a). Superimposition of the α-btx- and epibatidine-bound structures reveals that finger II ofα-btx approaches the location of the azabicyclo moiety of epibatidine within the principal face of the ligand-binding site (Figure 3b). In particular, the guanidinium group of Arg36 from the tip ofα-btx finger II occupies the same location as the bridging nitrogen atom within the azabicyclo moiety of epibatidine. Additional changes of the protein backbone are evident distal to loop C, including the region between strandβ9 and theβ8-β9 loop, and that between helix α1 and strand β1 (see Figure 3c and Supplementary Material S1 at http://www.biochemj.org/bj/454/bj4540303add.htm). The co-ordinated nature of these changes suggests that they are not artifacts of limited resolution, which would produce random changes. Thus ligands of opposing functional efficacy stabilize distinct conformations of α7/AChBP.
Figure 3. Conformations of α7/AChBP without and with bound ligands.
(a) Comparison of loop C from α7/AChBP with bound α–btx (cyan), bound epibatidine (orange) and without bound ligands (different colour for each conformation; PDB code 3SQ9). (b)Super imposition of the α7/AChBP chimaera(cyan) with bound α-btx (purple) and the α7/AChBP chimaera (orange) with bound epibatidine (PDB code 3SQ6) showing the spatial relationship between Arg36 of α-btx and epibatidine. (c) Stereo view of a subunit from the complex between α7/AChBP and α-btx in which residue RMSDs between the α-btx and epibatidine conformations are colour coded on the protein backbone; the colour scale is linear with RMSD, with the range from blue to red spanning RMSDs from 0.53 to 6.05Å.
Inter-residue interactions
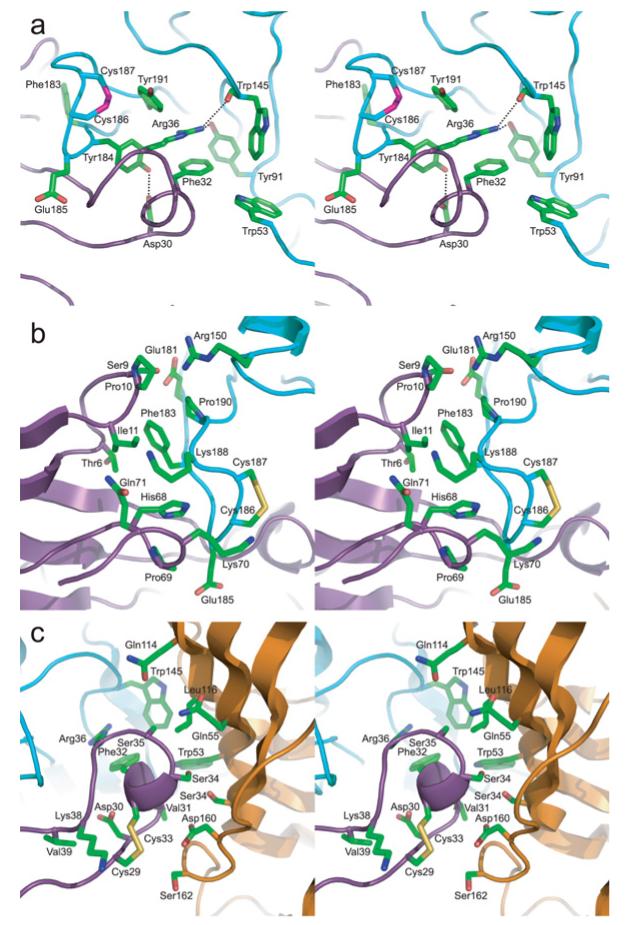
Arg36 and Phe32 from finger II of α-btx form a π-cation stack, which aligns edge-to-face with the aromatic ring of Tyr184 from loop C of α7/AChBP, while Tyr184 is anchored by a hydrogen bond between its hydroxy group and Asp30 of α-btx (Figure 4a). Furthermore, Tyr191 from the C-terminal end of loop C interacts with both Arg36 and Tyr184, while Arg36 forms hydrogen bonds with the hydroxy group of Tyr91 of loop A and the main-chain carbonyl group of Trp145 of loop B (Figure 4a). Finally, Arg182 from loop C establishes a π-cation interaction with Tyr184 and also an electrostatic interaction with Asp30 of α-btx (Supplementary Figure S3 at http://www.biochemj.org/bj/454/bj4540303add.htm). Overall, the contacts between residues at the principal face of α7/AChBP and α-btx are densely clustered around Tyr184 and are highly inter-connected, suggesting that this region may tolerate limited structural changes and thus may be decisive in stabilizing the complex.
Figure 4. Inter-residue interactions between α-btx and the α7/AChBP chimaera.
(a) Stereo view of key residue side chains between the tip of finger II of α-btx (purple) and the principal subunit of the α7/AChBP chimaera (cyan). (b) Stereo view of key residues between finger I of α-btx (purple) and the backside of loop C of the α7/AChBP chimaera (cyan). (c) Stereo view of key residues between finger II of α-btx (purple) and the complementary subunit of the α7/AChBP chimaera (orange).
In addition to interacting with aromatic residues deep within the ligand-binding pocket, α-btx contacts the part of loop C exposed on the protein surface (Figure 4b). Phe183, immediately adjacent to Tyr184, inserts into a hydrophobic pocket between fingers I and II of α-btx. In this region, Thr6, Pro10, Ile11 and Val40 of α-btx form the hydrophobic pocket into which Phe183 inserts, while Lys38 of α-btx engages in electrostatic interaction with Glu185 of α7/AChBP. Furthermore, main-chain amide groups, from Phe183 to Glu185 of α7/AChBP, form hydrogen bonds with main-chain carbonyl groups from Lys38 to Val40 of α-btx (Supplementary Figure S3). Additional residues at the tip of finger I of α-btx, Pro10 and Ile11, interact with Lys188 and Pro190 from loop C, while residues at the C-terminus of α-btx, His68, Pro69 and Lys70, contact the main chain and side chains of Glu185, Cys186 and Cys187 at the tip of loop C (Figure 4b). Overall, the contact region between loop C of α7/AChBP and fingers I and II and the C-terminus of α-btx exhibits remarkable chemical and shape complementarity.
Finger II of α-btx also approaches the complementary subunit of the ligand-binding site of α7/AChBP (Figure 4c). Phe32 of α-btx extends towards loop D from the complementary subunit where it contacts Trp53, which also aligns edge-to-face with Trp145 in loop B of the principal subunit. Additional residues at the tip of toxin finger II, Cys29, Asp30, Val31, Phe32, Cys33, Ser34 and Ser35, contact residues of the complementary subunit, including Ser34, Trp53, Gln55, Gln114, Leu116, Asp160 and Ser162. Inter-residue contacts between α-btx and α7/AChBP are listed in Supplementary Table S1 (at http://www.biochemj.org/bj/454/bj4540303add.htm).
Comparison with the α-btx/α1 structure
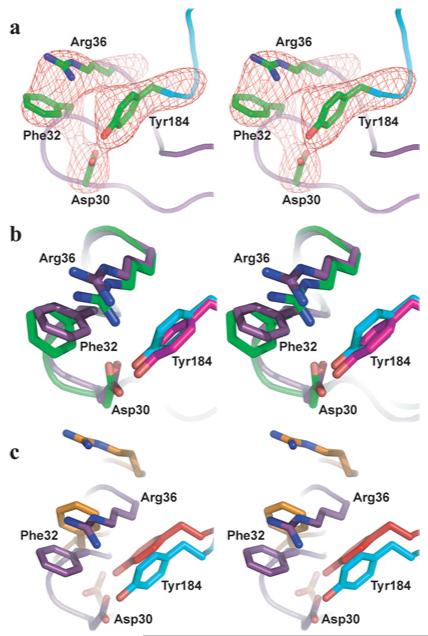
In the complex between α-btx and α7/AChBP, the side chains of the four interacting residues Asp30, Phe32, Arg36 and Tyr184 are well defined by the electron density (Figure 5a). The spatial arrangement of this tetrad is similar to that of the equivalent tetrad in the complex between α-btx and the monomeric α1 extracellular domain obtained by X-ray diffraction at a resolution of 1.94 Å [16] (Figure 5b). This similarity of residue side chains occurs despite the large rigid body shift of α-btx compared with its α1 binding partner. Presumably the absence of the complementary subunit, together with the mobility of loop C, allowed residues from α-btx to achieve a close register with the equivalent target residues in the monomeric α1.
Figure 5. The interaction tetrad Tyr184, Arg36, Phe32 and Asp30.
(a) Electron density of the complex between α-btx and α7/AChBP in the region of the tetrad. (b) Superimposition of the tetrad in the complex between α-btx and α7/AChBP (Arg36, Phe32 and Asp30 are purple; Tyr184 is cyan) with that of the complex between α-btx and α1 (Arg36, Phe32 and Asp30 are green; Tyr184 is magenta). (c) Superimposition of the tetrad from the complex between α-btx and α7/AChBP (colours as in b) with that of the complex between α-ctx and AChBP (residues equivalent to Arg36, Phe32 and Asp30 are orange; residue equivalent to Tyr184 is red).
Comparison with the α-ctx–AChBP structure
The spatial arrangement of Asp30, Phe32, Arg36 and Tyr184 in the current structure diverges markedly from that of equivalent residues in the complex between α-ctx and AChBP deduced from X-ray diffraction at a resolution of 4.2 Å [15] (Figure 5c). In the α-ctx–AChBP complex, Arg33 of α-ctx, equivalent to Arg36 of α-btx, lodges against the complementary subunit, rather than within the aromatic box of the principal subunit. As a result, a large distance separates Arg33 and Tyr185 of AChBP, unlike the equivalent Arg36 and Tyr184 in the current structure, whereas Phe31 of α-ctx approaches Tyr185 and occupies the centre of the aromatic-binding pocket of AChBP. Furthermore, Arg33 of α-ctx and Tyr89 of AChBP are separated by a large distance, in contrast with the close proximity of the equivalent Arg36 and Tyr91 in the current structure that allows formation of a hydrogen bond (Figure 4a).
Mutational analyses
The structure of the complex between α-btx and α7/AChBP suggests that the main stabilizing interactions centre around finger II of α-btx. The π-cation stack formed by Arg36 and Phe32 extends from finger II of α-btx, where it occupies the same position as the cationic moiety of the agonist, and is surrounded by all five canonical aromatic residues of the AChR ligand-binding site. This arrangement of aromatic residues around Arg36 and Phe32 suggests that one or more of these residues contribute to α-btx binding.
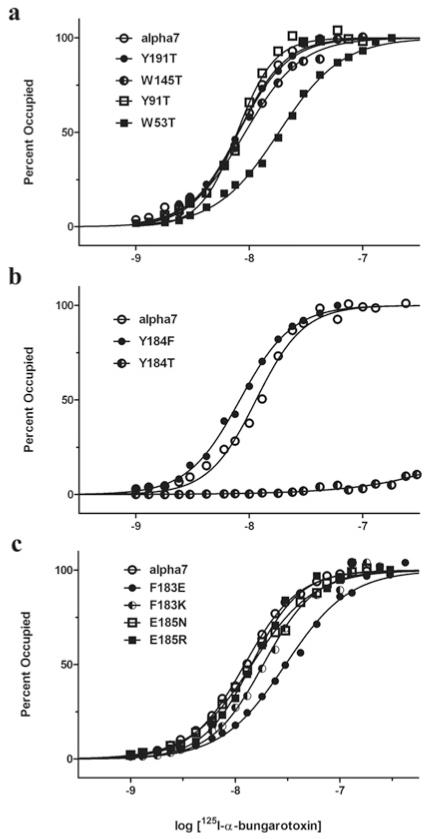
To determine which of the five canonical aromatic residues contribute to α-btx binding, threonine was replaced at equivalent positions in an AChR chimaera comprising the α7 sequence throughout the ligand-binding domain and the 5HT3A sequence in the membrane and cytoplasmic domains [22]. This chimaeric receptor expresses robustly in HEK cells, in contrast with the very low expression of the native α7 AChR. We then measured the concentration dependence of radiolabelled α-btx binding to intact HEK cells expressing each mutant AChR. For consistency with residue numbering in α7/AChBP, residue positions in the α7/5HT3A receptor are numbered according to those in the crystal structure. α-btx binds with high affinity to four of the five mutant AChRs (Figure 6a and Table 2). However, for the Y184T mutant specific binding is undetectable at α-btx concentrations lower than 30 nM, which saturates the control α7/5HT3A chimaeric AChR (Figure 6b). However, at concentrations of α-btx greater than 100 nM, specific binding, although low, is detectable, indicating that affinity is markedly reduced (Table 2). On the other hand, replacing Tyr184 with a phenylalanine residue maintains high-affinity α-btx binding. Together with our structural data, these results show that the essential interaction is that between the aromatic ring of Tyr184 of α7 and the Arg36/Phe32 pair of α-btx, whereas the hydrogen bond between Tyr184 and Asp30 of α-btx appears dispensable. Thus, of the five canonical aromatic residues of the AChR ligand-binding site, only one, Tyr184 in loop C, is essential for high-affinity binding of α-btx.
Figure 6. Origin of high-affinity α-btx binding determined by steady-state 125I-labelled-α-btx binding.
(a) Mutational analyses of four of the five conserved aromatic residues show retention of high-affinity binding after replacing with a threonine residue. (b) Mutational analyses of Tyr184 show retention of high-affinity binding after replacing with a phenylalanine residue, but loss of high-affinity binding after replacing with a threonine residue. (c) Mutational analyses of Phe183 and Glu185 that flank Tyr184. Binding of 125I-labelled α-btx to α7/5HT3A AChRs expressed by HEK-293 cells is described in the Experimental section. Curves are fits of Hill equation to the data (Table 2).
Table 2. 125I-labelled α-btx binding to mutant α7/5HT3A receptors.
Mutants were examined in three separate experiments, each of which included determinations for the control α7/5HT3A receptor. For the mutants, fitted parameters±S.E.M. follow in the rows below the corresponding control.
| Mutation | Kd (nM) | n H | Kmut/Kwt |
|---|---|---|---|
| α7/5HT3A | 8.1±0.5 | 2.0±0.12 | 1.0 |
| W53T | 18.2±0.8 | 1.6±0.04 | 1.8 |
| Y91T | 7.8±0.5 | 2.6±0.19 | 0.96 |
| W145T | 9.2±0.5 | 1.8±0.06 | 1.1 |
| Y191T | 8.1±0.5 | 1.9±0.1 | 1.0 |
| α7/5HT3A | 11.5±1.1 | 2.1±0.12 | 1.0 |
| Y184F | 8.3±0.3 | 1.8±0.06 | 0.72 |
| Y184T | 4166±3180 | 0.82±0.1 | 362 |
| α7/5HT3A | 12.2±0.5 | 1.7±0.05 | 1.0 |
| F183E | 30.1±2.2 | 1.4±0.06 | 2.5 |
| F183K | 18.5±1.2 | 1.7±0.08 | 1.5 |
| E185N | 14.4±0.8 | 1.5±0.05 | 1.2 |
| E185R | 14.2±0.8 | 1.9±0.05 | 1.2 |
Phe183 and Glu185 flank the essential Tyr184, extend into crevices on the surface of α-btx and engage in multiple inter-residue interactions (Figures 4a and 4b). To determine whether these flanking residues contribute to high-affinity α-btx binding, we replaced Phe183 with a glutamate or lysine residue, and Glu185 with an asparagine or arginine residue and measured α-btx binding to the mutant AChRs. α-btx binds with high affinity to each of these mutant receptors, with only the F183E mutant exhibiting a not-able, but still modest decrease in affinity (Figure 6c and Table 2).
DISCUSSION
We present the crystal structure of α-btx bound to a pentameric α7 ligand-binding domain chimaera. Structures of α-neurotoxins bound to peptides from loop C of the AChR [5,6], to the monomeric α1 extracellular domain [16] and to the pentameric AChBP [15] have been determined, but a consensus on the key stabilizing interactions has remained elusive. Advantages of the current structure include the presence of both the principal and complementary subunits that form the ligand-binding site, native α7 residues throughout loop C and lining the surface of the binding pocket, and resolution sufficient to localize residue side chains. Furthermore, through a combination of structural and mutational analyses, a single focal stabilizing interaction has been identified. Comparison of structures with either bound α-btx or the agonist epibatidine reveals structural rearrangements within the binding pocket and throughout each subunit. The overall findings reveal structural principles of molecular recognition of AChRs by three-finger α-neurotoxins, and illustrate the range of structural rearrangements of the AChR ligand-binding domain produced by an agonist and peptide antagonist.
α-btx and the agonist epibatidine arrest α7/AChBP in distinctly different conformations. Binding of α-btx maintains loop C in a uniformly opened-up conformation that approaches the most opened-up conformation observed in the apo structure [17]. Binding of epibatidine, however, maintains loop C in a uniformly closed-in conformation. Comparison of the α-btx- and epibatidine-bound structures shows that as loop C changes from opened-up to closed-in, the contiguous strand β9 and the juxtaposed β8–β9 loop splay apart (Supplementary Material). These local changes appear to propagate throughout the subunit and extend as far away as the protein backbone between helix α1 and strand β1. Thus the structures with bound α-btx and epibatidine provide a model for the range of conformational changes of the AChR ligand-binding domain.
The backbone of α-btx is held rigid by multiple intramolecular disulfide bonds, but residue side chains remain conformationally dynamic. In the NMR structure of α-btx, the stacked conformation is relatively rare [5,6], and in the crystal structure [4] the Phe32 and Arg36 side chains do not stack, unlike in the current structure. Thus the current structure suggests that the stacked conformation of Phe32 and Arg36 of α-btx and the opened-up conformation of loop C in α7/AChBP are selected from a range of conformations.
Fingers I and II and the C-terminus of α-btx envelop loop C from the principal face of the ligand-binding site, establishing a large region of contact in which the two protein surfaces complement one another precisely. In contrast, only a few residues within the contact region are critical for high-affinity binding. Mutation of only one residue, the conserved Tyr184, eliminates high-affinity binding. A previous study used a competition assay based on the initial rate of radiolabelled α-btx binding, and concluded that replacing alanine for the residue equivalent to Tyr184 prevented AChR expression [14]. However, by directly measuring radiolabelled α-btx binding over a range of concentrations at steady state, we find that high concentrations of α-btx exhibit low, but detectable, specific binding, indicating markedly reduced affinity. Previously, co-expression of the Y184T mutant with non-mutant α7/5HT3A subunits resulted in cell-surface heteropentamers that exhibited single channel currents in response to agonists [23]. Thus the subunit with the Y184T mutation incorporates into cell-surface pentameric AChRs and markedly reduces the affinity for α-btx.
The structure of α-ctx bound to the pentameric AChBP (PDB code 1YI5) was deduced from X-ray diffraction at a resolution of 4.2 Å [15]. The protein backbone of the toxin and its orientation relative to the pentamer surface are similar to those in the current structure. However, in the α-ctx–AChBP structure the inter-residue contacts diverge markedly from those in the current complex between α-btx and α7/AChBP. α-ctx residues Phe29 and Arg33, equivalent to Phe32 and Arg36 in α-btx, were splayed apart, with the guanidium group of Arg33 lodged against the complementary subunit, and Phe29 localized to the centre of the aromatic-rich cavity of the principal subunit. By contrast, in the structure described in the present study, the Arg36 side chain of α-btx stacks upon that of Phe32 and the pair localizes within the aromatic cavity in the principal subunit where the guanidium group of Arg36 occupies the same location as the cationic nitrogen of the agonist. The increased resolution in the current structure probably accounts for differences in inter-residue contacts between the two structures. Furthermore, interactions between Tyr184 of α7 and residues at the tip of finger II of α-btx are fully congruent with mutational analyses showing that Tyr184 is uniquely essential for high-affinity binding of α-btx.
The structure of α-btx bound to the monomeric α1 subunit extracellular domain was determined by X-ray diffraction at a resolution of 1.94 Å [16]. Compared with the structure described in the present study, the protein main chain of the toxin showed a rigid body shift relative to the surface of the α1 subunit. This shift may be due to the absence of the complementary subunit, the ability of loop C to flex upon ligand binding, replacement α1-Trp149 with arginine or a combination of all three [16]. However, despite these differences, the key interacting tetrad, α1 residue Tyr190 and α-btx residues Phe32, Arg36 and Asp30, is similar to that in the current structure. Thus the complex between α-btx and α7/AChBP described in the present study provides a consensus on both the orientation of the protein main chain of α-btx relative to the protein surface and the inter-residue contacts that stabilize long-chain α-neurotoxins bound to nicotinic receptors. The findings provide a foundation for future studies of the structural basis of the high affinity and target-selective interaction between α-neurotoxins and nicotinic receptors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the ALS BCSB (Berkeley Center for Structural Biology) staff members C. Ralston, P. Zwart, C. Bertoldo, A. Rozales and K. Royal, and the APS staff members J. Brunzelle and N. Sanishvili for help with data collection, the USC (University of Southern California) NanoBiophysics Core Facility where preliminary crystallographic data were collected, and Dr H.-L. Wang for help in making Figure 3(c).
FUNDING
This work was supported by the National Institutes of Health [grant numbers NS031744 (to S.M.S.) and GM064642 (to L.C.)].
Abbreviations used
- AChBP
acetylcholine-binding protein
- AChR
acetylcholine receptor
- α-btx
α-bungarotoxin
- α-ctx
α-cobratoxin
- HEK
human embryonic kidney
- 5HT3A
5-hydroxytryptamine type 3A
- NCS
non-crystallographic symmetry
Footnotes
The structural co-ordinates reported will appear in the PDB under accession code 4HQP.
REFERENCES
- 1.Chang CC, Lee CY. Isolation of neurotoxins from the venom of Bungaris multicinctus and their modes of neuromuscular blocking action. Arch. Int. Pharmacodyn. Ther. 1963;144:241–257. [PubMed] [Google Scholar]
- 2.Anderson MJ, Cohen MW. Nerve-induced and spontaneous redistribution of acetylcholine receptors on cultured muscle cells. J. Physiol. 1977;268:757–773. doi: 10.1113/jphysiol.1977.sp011880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber M, Changeux JP. Binding of Naja nicricollis (3H)α-toxin to membrane fragments from Electropheris and Torpedo electric organs. I. Binding of the tritiated α-neurotoxin in the absence of effector. Mol. Pharmacol. 1974;10:1–14. [PubMed] [Google Scholar]
- 4.Love RA, Stroud RM. The crystal structure of α-bungarotoxin at 2.5Å resolution: relation to solution structure and binding to acetylcholine receptor. Protein Eng. 1986;1:37–46. doi: 10.1093/protein/1.1.37. [DOI] [PubMed] [Google Scholar]
- 5.Scherf T, Balass M, Fuchs S, Katchalski-Katzir E, Anglister J. Three-dimensional structure of the complex of α-bungarotoxin with a library-derived peptide. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6059–6064. doi: 10.1073/pnas.94.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng H, Moise L, Grant MA, Hawrot E. The solution structure of the complex formed between α-bungarotoxin and an 18-mer cognate peptide derived from the α1 subunit of the nicotinic acetylcholine receptor from Torpedo californica. J. Biol. Chem. 2001;276:22930–22940. doi: 10.1074/jbc.M102300200. [DOI] [PubMed] [Google Scholar]
- 7.Sine SM. The nicotinic receptor ligand binding domain. J. Neurobiol. 2002;53:431–446. doi: 10.1002/neu.10139. [DOI] [PubMed] [Google Scholar]
- 8.Taylor P, Talley TT, Radic Z, Hansen SB, Hibbs RE, Shi J. Structure-guided drug design: conferring selectivity among neuronal nicotinic receptor and acetylcholine-binding protein subtypes. Biochem. Pharmacol. 2007;74:1164–1171. doi: 10.1016/j.bcp.2007.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson AJ, Lester HA, Lummis SCR. The structural basis of function in Cys-loop receptors. Quart. Rev. Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 10.Neuman D, Barchan D, Fridkin M, Fuchs S. Analysis of ligand binding to the synthetic dodecapeptide 185-196 of the acetylcholine receptor α subunit. Proc. Natl. Acad. Sci. U.S.A. 1986;83:9250–9253. doi: 10.1073/pnas.83.23.9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barchan D, Kachalsky S, Neumann D, Vogel Z, Ovadia M, Kochva E, Fuchs S. How the mongoose can fight the snake: the binding site of the mongoose acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7717–7721. doi: 10.1073/pnas.89.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krienkamp H, Sine SM, Maeda R, Taylor P. Glycosylation sites selectively interfere with α-toxin binding to the nicotinic acetylcholine receptor. J. Biol. Chem. 1994;269:8108–8114. [PubMed] [Google Scholar]
- 13.Levandoski MM, Lin Y, Moise L, McLaughlin JT, Cooper E, Hawrot E. Chimeric analysis of a neuronal nicotinic acetylcholine receptor reveals amino acids conferring sensitivity to α-bungarotoxin. J. Biol. Chem. 1999;274:26113–2619. doi: 10.1074/jbc.274.37.26113. [DOI] [PubMed] [Google Scholar]
- 14.Fruchardt-Gaillard C, Gilquin B, Antil-Delbeke S, Le Novere N, Tamiya T, Corringer PJ, Changeux JP, Menez A, Servent D. Experimentally based model of a complex between a snake toxin and the α7 nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3216–3221. doi: 10.1073/pnas.042699899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. The crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR a1 bound to α-bungarotoxin at 1.94Å resolution. Nat. Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Huang S, Bren N, Noridomi K, Dellisanti CD, Sine SM, Chen L. Crystal structures of the ligand-binding domain of a pentameric α7 nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 2011;14:1253–1259. doi: 10.1038/nn.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Bouzat C, Bartos M, Corradi J, Sine SM. Binding-pore interface of homomeric Cys-loop receptors governs open channel lifetime and rate of desensitization. J. Neurosci. 2008;28:7808–7819. doi: 10.1523/JNEUROSCI.0448-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rayes D, De Rosa MJ, Sine SM, Bouzat C. Number and locations of agonist binding sites required to activate homomeric Cys-loop receptors. J. Neurosci. 2009;29:6022–6032. doi: 10.1523/JNEUROSCI.0627-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.