Abstract
High-throughput sequencing technologies, also known as next-generation sequencing (NGS) technologies, have revolutionized the way that genomic research is advancing. In addition to the static genome, these state-of-art technologies have been recently exploited to analyze the dynamic transcriptome, and the resulting technology is termed RNA sequencing (RNA-seq). RNA-seq is free from many limitations of other transcriptomic approaches, such as microarray and tag-based sequencing method. Although RNA-seq has only been available for a short time, studies using this method have completely changed our perspective of the breadth and depth of eukaryotic transcriptomes. In terms of the transcriptomics of teleost fishes, both model and non-model species have benefited from the RNA-seq approach and have undergone tremendous advances in the past several years. RNA-seq has helped not only in mapping and annotating fish transcriptome but also in our understanding of many biological processes in fish, such as development, adaptive evolution, host immune response, and stress response. In this review, we first provide an overview of each step of RNA-seq from library construction to the bioinformatic analysis of the data. We then summarize and discuss the recent biological insights obtained from the RNA-seq studies in a variety of fish species.
Introduction
Transcriptomics refers to the study of the complete set of transcripts in a specific cell, tissue, or organism for a given developmental stage or physiological condition (Wang et al., 2009). This complete set of transcripts is known as a transcriptome, including protein-coding messenger RNA (mRNA) and non-coding RNA [ncRNA: ribosomal RNA (rRNA), transfer RNA (tRNA), and other ncRNAs] (Lindberg and Lundeberg, 2010; Okazaki et al., 2002). Unlike the relatively stable genome, the transcriptome varies with developmental stage, physiological condition, and external environment. Transcriptome analysis is a powerful tool for dissecting the relationship between genotype and phenotype, leading to a better understanding of the underlying pathways and mechanisms controlling cell fate, development, and disease progression. The aims of transcriptomics are not limited to the quantification of change in expression level for each gene among different transcriptome samples but include also the mapping and annotation of the transcriptome and the determination of the functional structure of each gene in the genome (Costa et al., 2010; Ruan et al., 2004; Wang et al., 2009).
The complexity of transcriptome determines the reliance on high-throughput tools for transcriptome studies. Over the years, several technologies, either hybridization- or sequence-based, have been developed to survey transcriptomes in a high-throughput manner. Hybridization-based technologies usually rely on incubation of fluorescently-labeled cDNA with probes fixed onto a solid surface (microarray). Since its first application in 1995 (Schena et al., 1995), microarray has been widely used in transcriptomics. Tilling microarray, an updated microarray with probes representing the genome at a high density, has also been generated and can interrogate transcriptomes at a relatively high resolution and can even discover novel transcripts (Cheng et al., 2005; David et al., 2006; Kampa et al., 2004). The use of microarray in fish biological studies has been more than one decade and well reviewed elsewhere (Douglas, 2006; Hook, 2010; Nielsen and Pavey, 2010; Zhang et al., 2009). Microarray technology, however, suffers from some intrinsic limitations (Bradford et al., 2010; Fu et al., 2009; Marioni et al., 2008; ’t Hoen et al., 2008), including dependence on the existing knowledge of genomic sequence, signal saturation for certain transcripts with a high abundance, and high background noise due to nonspecific hybridization. Aside from these intrinsic disadvantages, specific problems exist for fish transcriptomic studies when using microarray: only a few commercial microarray platforms are available, and they are specifically designed for model species such as zebrafish (Danio rerio) (Douglas, 2006); the construction of customized microarray or tiling array relies on the preexisting knowledge about EST sequences or genomic sequences, which is usually not available for many fish species (Douglas, 2006; Zhang et al., 2009).
In contrast to microarray, sequence-based technologies directly determine the cDNA sequence. These approaches rely on the generation of featured EST tags that correspond to the fragments of those transcripts in a sample and their subsequent concatenation prior to cloning and sequencing (Fullwood et al., 2009). Different tag-based sequencing methods, such as serial analysis of gene expression (SAGE) (Matsumura et al., 2005; Velculescu et al., 1995), polony multiplex analysis of gene expression (PMAGE) (Kim et al., 2007), and massively parallel signature sequencing (MPSS) (Brenner et al., 2000), have already been developed. SAGE has so far been used in three transcriptomic studies in zebrafish regarding oogenesis (Knoll-Gellida et al., 2006), sexual dimorphism (Zheng et al., 2013), and stress response to toxicant (Cambier et al., 2009). Most of these tag-based sequencing methods, however, are based on expensive Sanger sequencing; these methods are labor-consuming and time-consuming for the cloning step (Morozova et al., 2009); it is often impossible to precisely annotate the tags unless the whole genome information is available (Costa et al., 2010); only a portion of transcripts are analyzed (Harbers and Carninci, 2005); and differential isoform and allelic expressions are usually indistinguishable (Wang et al., 2009).
Recently, the advent of low-cost (NGS) technologies, paralleling the sequencing process, has led to the generation of a new method for both mapping and quantifying transcriptome, known as RNA-seq. RNA-seq is free from almost all the limitations of the methods mentioned above (Table 1). RNA-seq has only been available for several years; however, this method is already revolutionizing the field of transcriptomics, improving our understanding of genome expression and regulation. Importantly, RNA-seq, combined with other state-of-the-art omics technologies, has been applied to analyze the detailed integrative personal omics profile for evaluating disease risk and monitoring disease progression for personalized treatment (Chen et al., 2012; Roychowdhury et al., 2011). Besides, RNA-seq has already been applied to a substantial amount of fish biology studies (Table 2).
Table 1.
Comparison of RNA-Seq with Other Methods for Surveying Transcriptome*
| Tilling microarray | Tag-based sequencing | RNA-Seq | |
|---|---|---|---|
| Principle | Hybridization | Sanger sequencing | Parallelized high-throughput sequencing |
| Resolution of data | Several to 100 bp | Single base | Single base |
| Sensitivity | Low | Moderate | High |
| Throughput | High | Low | High |
| Turnaround time | Long | Long | Short |
| Required amount of RNA samples | High | High | Low |
| Cost per sample (excluding equipments) | High | High | Relatively low |
| Reliance on existing genomic sequence | Yes | No | No |
| Linear dynamic range of expression levels | <2 orders of magnitude | Not practical | Limited only by sequencing depth |
| Discovery of unknown transcribed regions | Limited | Yes | Yes |
| Detection of differences in isoformic and allelic expressions | Limited | Limited | Yes |
| Detection of mutations | Limited | Yes | Yes |
| Determination of splicing sites | Limited | Yes | Yes |
| Identification of UTRs | Limited | Limited | Yes |
Table 2.
Representatives of Publications that Have Had Fish Transcriptomes Studied by RNA-Seq
| Major application | Fish species | Sequecing platform | Reference |
|---|---|---|---|
| Transcriptome mapping and genome annotation | Poecilia reticulate | Roche 454 | Fraser et al., 2011 |
| Danio rerio | Illumina | Collins et al., 2012 | |
| Ictalurus punctatus | Illumina | Liu et al., 2012b | |
| Nothobranchius furzeri | Illumina | Petzold et al., 2013 | |
| Novel transcript discovery | Labeo rohita, Hamilton | Illumina | Robinson et al., 2012 |
| Danio rerio | Illumina | Pauli et al., 2012 | |
| Oncorhynchus mykiss | Illumina | Palstra et al., 2013 | |
| Salmo salar | Illumina | Kure et al., 2013 | |
| Detection of RNA splicing | Danio rerio | AB SOLiD | Aanes et al., 2011; |
| SNP discovery | Ictalurus furcatus | Illumina | Liu et al., 2011 |
| Cyprinus carpio | Illumina | Xu et al., 2012 | |
| Oncorhynchus mykiss | Illumina | Salem et al., 2012 | |
| Quantification of gene expression | Danio rerio | AB SOLiD | Vesterlund et al., 2011 |
| Lates calcarifer | Roche 454 | Xia et al., 2013 | |
| Lateolabrax japonicus | Illumina | Xiang et al., 2010 | |
| Fundulus heteroclitus | Roche 454 | Oleksiak et al., 2011 | |
| Salmo trutta | Illumina | Uren Webster et al., 2013 | |
| Fundulus grandis | Illumina | Garcia et al., 2012 | |
| Perca flavescens | Roche 454 | Pierron et al., 2011 | |
| Melanotaenia duboulayi | Illumina | Smith et al., 2013 | |
| Coregonus clupeaformis spp., Salmonidae | Roche 454 | Jeukens et al., 2010 | |
| Gasterosteus aculeatus | Illumina | Greenwood et al., 2012 | |
| Astyanax mexicanus | Roche 454 | Gross et al., 2013 |
In this review, we present an overview of RNA-seq method from data generation to bioinformatic analysis, discuss the challenges for RNA-seq, and then review the biological insights already gained from RNA-seq for a variety of fish species. We apologize to all those investigators whose articles were not cited due to space constraints.
Overview of RNA-seq
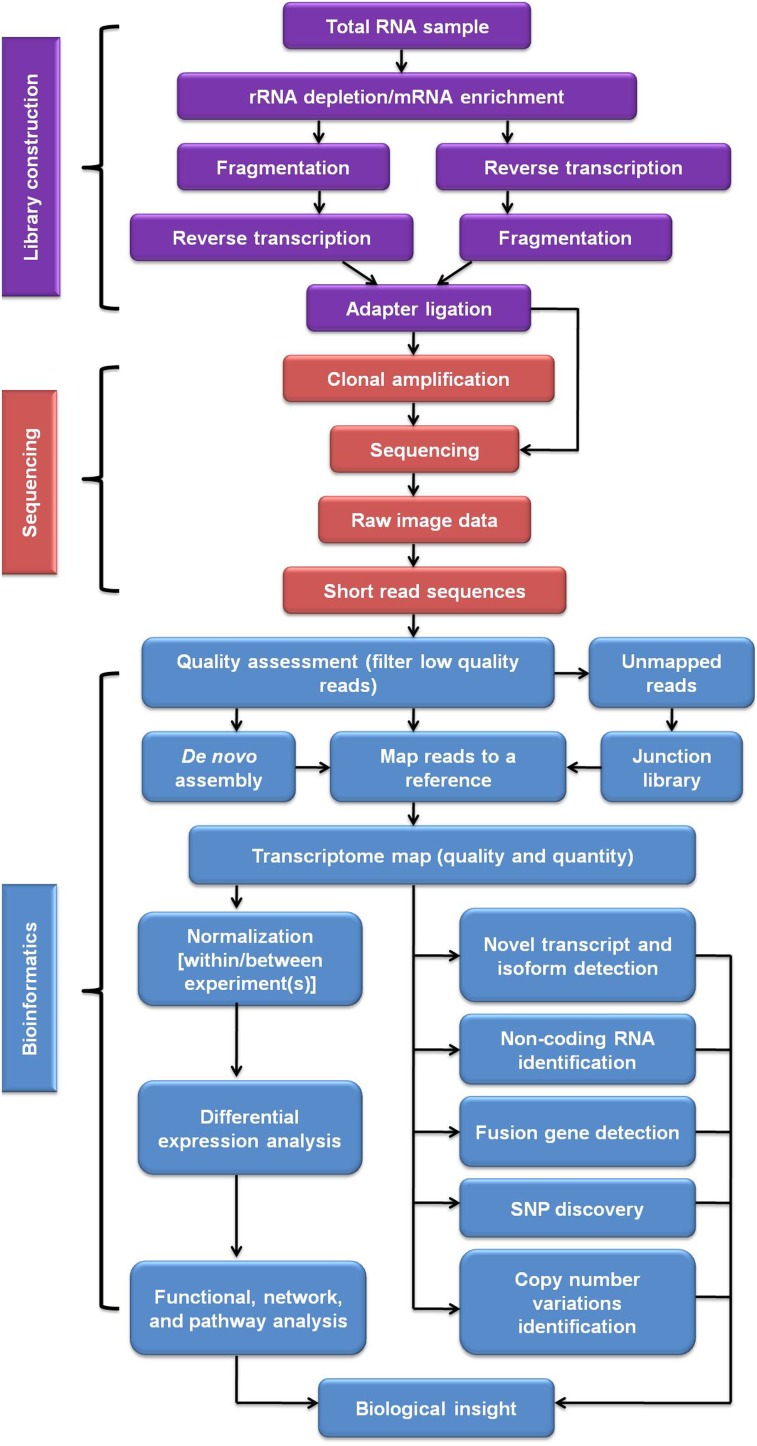
RNA-seq, also known as whole transcriptome shotgun sequencing (WTSS), employs the NGS technologies to sequence cDNA directly from a RNA sample of interest (Morozova et al., 2009; Wilhelm et al., 2008). Transcriptome analysis by RNA-seq is a three-step method, including library construction, sequencing on a specific NGS platform, and bioinformatic analysis (Fig. 1). In the remainder of this section, we will explain each step of a typical RNA-seq experiment and discuss the accompanying challenges and the possible solutions.
FIG. 1.
Illustration of RNA-Seq data analysis pipeline.
Library construction
The library preparation is a key step for RNA-seq as it determines to a large extent how accurately the final sequencing data reflects the original transcriptome. The first procedure in this step is to collect appropriate samples, usually tissues, to be analyzed. One key concern here is the invariably heterogeneous nature of tissues. This is because tissues typically contain tens or hundreds of unique cell types, and thus, transcriptomic analysis of a tissue confounds the real transcriptomic profiles of its constituent cell types (Islam et al., 2011; Shapiro et al., 2013). One solution to this problem is to analyze single cells rather than cell populations. In fact, various single-cell isolation methods have been developed and already coupled with RNA-seq technology in practice (Shapiro et al., 2013). This single-cell RNA-seq lets many previously impossible applications in both basic and clinical research become possible, such as characterization of the initial differentiation events in embryogenesis (Tang et al., 2009; 2010), the investigation of tumor heterogeneity (Dalerba et al., 2011), transcriptomic analysis of rare, transiently existing adult stem cells (Lister et al., 2011), and others.
Following sample collection, total RNA is usually prepared via organic extraction and/or absorption onto silica-membranes of spin columns. Next, RNA is converted to a library of cDNA fragments. Although total RNA can be directly used, total RNA has to be fractionated in most cases. This is because the rRNA, making up more than 80% of total cellular RNA (Lindberg and Lundeberg, 2010), is usually not the research focus, and its presence greatly reduces the useful transcript coverage in the following sequencing step. Total RNA samples are therefore processed either by direct selection of poly(A) RNA or by selective removal of rRNA (ribo-depletion) (Costa et al., 2010). Oligo(dT)-based mRNA purification procedure, widely used in eukaryotes, takes advantage of the presence of a poly(A) tail at the 3′ of eukaryotic mRNA. A large fraction of non-ribosomal RNAs (both coding and noncoding) in eukaryotes, however, lacks the poly(A) tail and is therefore missed (Jacquier, 2009). When compared to the poly(A) RNA selection, the ribo-depletion method is preferred since it enriches all nonribosomal RNA species, including nonpoly(A) mRNA, preprocessed RNA, tRNA, and other ncRNAs with known or unknown functions (Lindberg and Lundeberg, 2010). Although many different rRNA depletion methods have been developed, the two most popular ones are (He et al., 2010; Wilhelm and Landry, 2009): (1) hybridization capture of rRNA by the biotin-labeled anti-rRNA probes, followed by removal with streptavidin-coated magnetic beads; and (2) selective degradation of rRNA by a 5′-3′ exonuclease that specifically recognizes rRNA with a 5′ phosphate.
A double-stranded cDNA library is then prepared via either RNA fragmentation (RNA hydrolysis or nebulization) prior to the reverse transcription or reverse transcription first followed by cDNA fragmentation (DNase I treatment or sonication) (Roberts et al., 2011; Wang et al., 2009). The purpose of fragmentation is to reach the desired length for NGS technologies (Metzker, 2009). Each of these two fragmentation methods causes a different bias in final results. Particularly, cDNA fragmentation usually generates an under-representation of the 5′ of the transcripts in the data, while RNA fragmentation allows a good representation of the transcript body but causes depleted transcript ends (Mortazavi et al., 2008; Nagalakshmi et al., 2008). Additionally, the process of reverse transcription can also complicate final RNA-seq data due to the tendency of reverse transcriptase to generate spurious second-strand cDNA and the artificial chimeric transcripts introduced by template switching (Ozsolak and Milos, 2011b).
After the generation of fragmented cDNA, sequencing adapters are ligated to both ends of the fragments. During this process, information about the orientation of transcripts is completely lost. Fortunately, strand-directionality information can be maintained by converting cytidine into uridine with sodium bisulfate (He et al., 2008); the resulting C–T transition position then labels the coding strand of each transcript. Other approaches that maintain the strand specificity are involved with how the adaptors are ligated to the cDNA fragments, and these methods are well reviewed elsewhere (Costa et al., 2010; Marguerat and Bähler, 2010). Like the process of fragmentation, adaptor ligation may also introduce coverage non-uniformity since the fidelities of DNA ligases cannot be guaranteed (Faulhammer et al., 2000).
Sequencing
The sequencing step relies on the NGS technologies. The three most popular, massively parallel NGS platforms, are currently dominating the NGS market and widely used in RNA-seq, including the 454 pyrosequencing system (a subsidiary of Roche), the AB SOLiD system (Life Technologies), and the Illummina Genome Analyzer (Illumina) (Liu et al., 2012a; Marguerat and Bähler, 2010). All these three NGS platforms rely on an in vitro cloning step (clonal amplification) to amplify each fragmented cDNA molecule in a cell-free system, because their sensitivities are not high enough for the single molecule sequencing (Metzker, 2009). Specifically, both the 454 and the SOLiD systems employ an innovative emulsion PCR. In the emulsion PCR, the cDNA fragments from a library are attached to beads and subsequently compartmentalized in the aqueous droplets of a water-in-oil emulsion such that each droplet contains a single DNA molecule; the segregated template fragments are then amplified in the tiny aqueous droplets of the emulsion (Dressman et al., 2003). Different from the 454 and the SOLiD systems, the Illummina Genome Analyzer performs a so-called bridge PCR amplification, in which the adapter-linked, single-stranded cDNA fragments are first immobilized on a glass slide by oligonucleotide hybridization in a bridging way, followed by clonal PCR amplification (Adessi et al., 2000; Fedurco et al., 2006). Clonal amplification results in a population of identical templates, each of which is subjected to the following sequencing reaction. Due to PCR artifacts, clonal amplication may introduce bias in the RNA-seq results as well. One way to discriminate PCR artifacts is to perform different biological replicates and determine whether same short reads are concurrently present in different replicates (Wang et al., 2009).
NGS platforms use different sequencing strategies (Metzker, 2009). The sequencing mechanism employed by Roche 454 is pyrosequencing, which is a nonelectrophoretic, bioluminescence-based method (Metzker, 2009; Ronaghi et al., 1998). In brief, every bead, containing the clonally amplified template originating from a single cDNA molecule, is transferred to a ∼29 μm well on a Picotiter Plate (a fiberoptic chip). A mixture of enzymes, such as DNA polymerase, ATP sulfurylase, and luciferase, are also packed into each well. The four DNA nucleotides are added sequentially in a fixed order across the Picotiter Plate during a sequencing run. When one or more complementary nucleotides are added, it generates a light signal that is recorded by the camera in the instrument. The signal strength is proportional to the number of nucleotide added. For the SOLiD system, a mechanism, known as sequencing by ligation (SBL), is used (Landegren et al., 1988; Metzker, 2009; Shendure and Ji, 2008). In its simplest form, after clonal amplification, a sequencing primer is hybridized to template with 3′ at position ‘n’. A set of four fluorescently labeled di-base-encoded probes (8 bp oligonucleotides) then compete for ligation to the sequencing primer. The best-matching probe is linked to the primer by DNA ligase, followed by removal of nonligated probes by wash. Fluorescence imaging is then done to determine the identity of the ligated probe. Following a series of ligation cycles, the extension product is removed and the template is reset with the second sequencing primer with 3′ at the ‘n-1′ position for a second round of ligation cycles. After five rounds of primer reset, each base is examined twice by two different primers, although SOLiD is hence not running as fast as other methods. Last, the method of cyclic reversible termination (CRT) is used by IIIumina (Bentley et al., 2008). CRT is a cyclic sequencing-by-synthesis method that differs from SBL in its use of nucleotide monomer and DNA polymerase. In particular, four types of reversible terminator bases are added in the presence of DNA polymerase and template, and after incubation non-incorporated nucleotides are washed away. Imaging is then performed to determine the identity of the incorporated nucleotide, and, then the dye and the terminal 3′ blocker are chemically removed, allowing for the next cycle to begin. In addition to CRT, ion semiconductor sequencing (Life Technologies) uses a method of “sequencing by synthesis” as well, which is based on the detection of hydrogen ions released during the polymerization of DNA. The major benefits of this method are rapid sequencing speed and low operating costs (Rothberg et al., 2011). Several reviews describing the mechanisms and comparing the advantages and disadvantages of these NGS technologies were published elsewhere (Ansorge, 2009; Liu et al., 2012a; Metzker, 2009; Shendure and Ji, 2008).
Despite the popularization of the NGS technologies, the so-called third generation sequencing methods, also known as single-molecule sequencing methods, are on their way. These methods, such as Heliscope sequencing and single-molecule real-time (SMRT) (Pacific Biosciences), are featured by omission of template clonal amplification and real-time signal capture (Liu et al., 2012a). Heliscope sequencing has already been used for a published RNA-seq study (Ozsolak et al., 2010). Additionally, the single-molecule direct RNA sequencing (DRS) technology, developed by Heliscope, is emerging. DRS sequences RNA molecules directly in a massively-parallel manner without biasing sample manipulations such as RNA reverse transcription, ligation, and clonal amplification (Ozsolak and Milos, 2011a). However, these technologies are at varying stages of development and suffer from many drawbacks, such as low single-pass accuracy (81%∼83%), low sequencing efficiency, and low throughput (Niedringhaus et al., 2011; Schadt et al., 2010).
Bioinformatic analysis
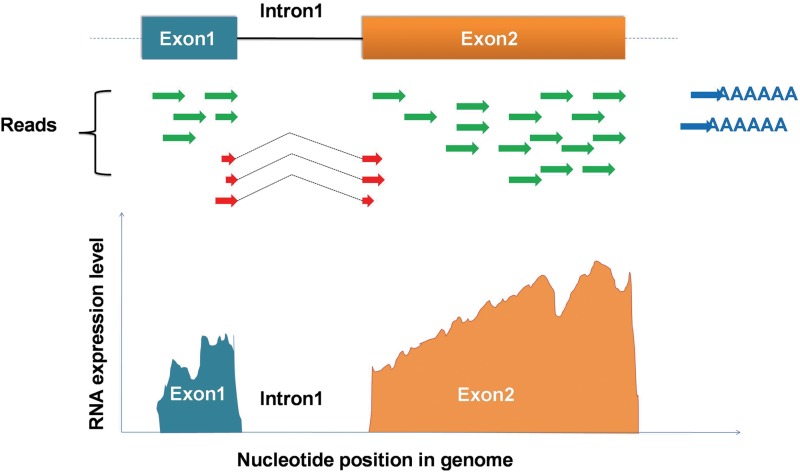
After the signal processing, NGS platforms generate millions of short sequences, termed reads, associated with their base-call quality scores that indicate the reliability of each base call. The lengths of these short reads are within a range of 25–450 bp, depending on the type of NGS platform. The resulting reads are categorized into three types: exonic reads, exon–intron junction reads, and poly(A) reads (Fig. 2) (Wang et al., 2009). Although the application of NGS technologies in RNA-seq makes transcriptomic sequences handy, the analysis of the huge amount of RNA-seq data are still the bottleneck for understanding the transcriptome. Fortunately, during the past few years, various bioinformatic tools (software) for RNA-seq data analysis have been developed. Especially, the significance of Bioconductor should be emphasized here. Bioconductor is a free, open source, and open development software project, mainly based on the R programming language, for the analysis and annotation of RNA-seq data and other types of high-throughput genomic data (Gentleman et al., 2004). Most Bioconductor components are organized as R packages. These Bioconductor packages and other software for RNA-seq data analysis have been well reviewed elsewhere (Chen et al., 2011; Febrer et al., 2011; Oshlack et al., 2010). Nowadays, researchers can routinely combine these tools to form the best analysis pipeline per their research interests.
FIG. 2.
Short reads mapping and quantification of the digital signal (modified from Wang et al., 2009). The resulting reads are aligned against a reference genome and fall into three groups, including exonic reads (green), junction reads (red-brown), and poly(A) end reads (blue) (Wang et al., 2009). These reads are used to generate an expression profile with a single-base resolution.
A typical analysis pipeline of RNA-seq data is outline in Figure 1. Quality assessment is the first step for the RNA-seq data analysis. To ensure a coherent final result, low-quality sequences, over-represented sequences, and adapter sequences have to be filtered out, and this step can be accomplished with Bioconductor packages, such as ShortRead (Morgan et al., 2009) and Biostrings (Pages et al., 2009). Once high-quality reads have been obtained, these short reads are subsequently aligned to a reference genome or transcriptome. Alignment is usually done with software outside Bioconductor. In contrast to conventional alignment algorithms, these software are based on the indexing strategies that are able to align millions of short reads in a reasonable period of time (Langmead et al., 2009). Basically, these aligners fall into two categories: one is based on the Burrows-Wheeler transform algorithm, such as Bowtie (Langmead et al., 2009) and BWA (Li and Durbin, 2010), and the other based on Needleman-Wunsch or Smith-Waterman algorithm, such as GNUMAP (Clement et al., 2010), BFAST (Homer et al., 2009), and SHRiMP (Rumble et al., 2009). The aligners in the first category is much faster, the aligners in the second category, however, despite more time needed, are usually more sensitive and generate more reads correctly aligned (Garber et al., 2011). Since many reads span exon–exon junctions and therefore cannot be directly aligned, specialized aligners (spliced aligners), such as Erange (Trapnell et al., 2009) and IsoformEx (Kim et al., 2011), are developed to split the junction reads and then independently align the split read fragments. The methods available to study RNA splicing from short RNA-Seq data have been well reviewed elsewhere (Alamancos et al., 2013). When the reference genome or transcriptome is unavailable, de novo transcriptome assembly can be performed (Robertson et al., 2010). De novo assembly is used for most fish studies using RNA-seq because only limited fish species have the whole genome information. De novo assembly software, for instance, ABySS (Simpson et al., 2009), Velvet (Zerbino and Birney, 2008), and Trinity (Grabherr et al., 2011), use a de Bruijn graph approach, which aligns the user-defined sequence overlap (referred as k-mer) between two reads to create contigs (Grabherr et al., 2011). Transcriptome de novo assembly is impeded by the repeats within huge amounts of short reads, alternatively spliced transcripts, as well as biased transcriptome sequencing coverage (Liu et al., 2012b). Therefore, different improvements for the de Bruijn graph approach, such as using various k-mer lengths instead of a single one, have been made to optimize transcriptome assembly (Surget-Groba and Montoya-Burgos, 2010). Besides, a reference proteome or genome from an evolutionarily linked species can be used to aid de novo assembly.
Once all reads have been appropriately filtered and mapped or assembled, they can then be counted, and gene expression levels can thus be inferred from the total counts of reads belonging to the exons of a particular gene. In this way, an expression score can be assigned to every base, leading to a genome-scale transcriptome map in terms of quality and quantity (Fig. 2). The single-base (digital) resolution of RNA-seq allows for detection of gene expression at the isoformic and allelic levels and discovery of previously unannotated genes (Jiang and Wong, 2009; Levin et al., 2009; Oshlack et al., 2010; Trapnell et al., 2010). RNA-seq analysis allows not only quantifying gene expression levels within a single RNA sample but also detecting differential expression (DE) across treatments or conditions (Kvam et al., 2012; Oshlack et al., 2010); the latter is usually the real interest of most studies. However, for the DE analysis of RNA-seq data, normalization has to be performed to adjust for between-sample differences such as library size and within-sample gene-specific features regarding GC content and gene length (Dillies et al., 2012; Kvam et al., 2012). There is no standard method to detect DE due to the short history of RNA-seq technology. The currently popular tools for DE analysis in Bioconductor include edger (Robinson et al., 2010), DESeq (Anders and Huber, 2010), baySeq (Hardcastle and Kelly, 2010), and tweeDEseq (Esnaola et al., 2013), and methods, not included in Bioconductor, also exist, such as ShrinkBayes (Van De Wiel et al., 2013) and TSPM (Auer and Doerge, 2011). These DE analysis tools have already been reviewed in detail and well compared in their gene ranking performances (Dillies et al., 2012; Kvam et al., 2012; Soneson et al., 2013). In addition, RNA-Seq analysis can also enable us to detect SNPs, fusion genes, and post-transcriptional gene regulation, such as RNA editing, RNA degradation, and RNA translation (Marguerat and Bähler, 2010).
New Biological Insights Gained in Fish Studies Using RNA-seq
The advent of RNA-seq technology has begun to revolutionize fish transcriptomic studies. In the past several years, RNA-seq has been applied to a number of studies involved with various fish species, including not only model species, such as zebrafish (Collins et al., 2012), but also commercially important and eco-environmentally relevant fish species, such as channel catfish (Ictalurus punctatus) (Liu et al., 2012b), European sea bass (Dicentrarchus labrax) (Sarropoulou et al., 2012), and rainbow trout (Oncorhynchus mykiss) (Palstra et al., 2013). Although the application of RNA-seq in fish transcriptomics is at the nascent stage, the results of a Pubmed literature search (with keywords, “fish” and “RNA-seq”) indicated that the number of publications in this field has increased considerably in the last 3 years. And, to our knowledge, no comprehensive review paper has appeared on this subject. Therefore, the aim of this section is to review the literature on fish transcriptomic studies using RNA-seq.
Transcriptome mapping and genome annotation
The high-throughput RNA-seq technology, independent of prior knowledge, allows efficient transcriptome annotation, including, for instance, transcript start and end sites and the identification of novel transcripts (Wang et al., 2009). For example, using RNA-seq, Liu and colleagues sequenced and de novo assembled the doubled haploid channel catfish transcriptome and generated 370,798 non-redundant transcript-derived contigs (Liu et al., 2012b). Functional annotation of these contigs revealed 25,144 unique protein-encoding transcripts. Of these 225,144 unique transcripts, over 14,000 transcripts were identified as full-length transcripts with complete open reading frame, and about 90% of these full-length transcripts were identified with the complete 5′ and 3′ ends. The lengths of 5′ UTRs (∼254 bp) were found to be much shorter than those of 3′ UTRs (∼1,096 bp). Similarly, reference transcriptomes have been constructed for other fish species, including zebrafish (Collins et al., 2012), guppy (Poecilia reticulata) (Fraser et al., 2011), and turquoise killifish (Nothobranchius furzeri) (Petzold et al., 2013)’
Creating a comprehensive reference transcriptome using RNA-seq provides invaluable information for genome annotation, such as the gene and exon boundaries, as well as the identification of novel transcribed regions. For example, to improve zebrafish genome annotation, RNA-seq data, based on an optimized analysis pipeline, was used to adjust intron/exon boundaries of the defined genes, confirm their expression, and improve the coverage of 3′ untranslated regions of genes (Collins et al., 2012). This optimized pipeline can potentially be applied to improve genome annotation for other organisms.
Novel transcript discovery
RNA-seq has been used to discover novel transcribed regions in the genome. The results from RNA-seq suggest the existence of a large number of unknown transcribed regions in every fish species surveyed, including zebrafish (Pauli et al., 2012), rohu carp (Labeo rohita, Hamilton) (Robinson et al., 2012), and rainbow trout (Palstra et al., 2013). Among these unknown transcripts, many nc RNAs other than rRNA and tRNA are of great interest due to their essential roles in many cellular processes, including translation, RNA splicing, gene regulation, and genome defense (Mattick and Makunin, 2006). These ncRNAs can be broadly classified as either long ncRNAs (lncRNAs; >200 nucleotides) or small RNAs (sRNAs; <200 nucleotides). More than 550 distinct long intervening noncoding RNAs (lincRNAs, one type of lncRNAs) were identified in zebrafish embryo (Ulitsky et al., 2011); 224 unique mature microRNAs (miRNAs, one type of sRNAs) were identified in Atlantic salmon (Salmo salar) (Kure et al., 2013). These newly discovered transcripts will facilitate the annotation of sequenced genome and lay the ground for future functional studies.
Interrogation of post-transcriptional modification (RNA splicing)
The sequence of the mature mRNA molecule can differ substantially from the corresponding genome sequence. This is because the precursor mRNA undergoes three main post-transcriptional modifications, including 5′ capping, 3′ polyadenylation, and RNA splicing, in the cell nucleus before the translation process begins. RNA splicing has been studied in fish using RNA-seq (Aanes et al., 2011; Pauli et al., 2012). Analysis of zebrafish transcriptome dynamics during maternal to zygotic transition by RNA-seq revealed the frequency of alternative splicing in zebrafish embryo to be in a range between 50% and 60%, which is considerably high, but much lower when compared to the splicing frequency estimated for human, ranging from 92% to 95% (Aanes et al., 2011). Another RNA-seq study in zebrafish embryo identified 3532 transcripts as the variants of known RefSeq genes (novel isoforms and partial transcripts), supplementing the existing exon–intron structures of many genes in zebrafish (Pauli et al., 2012). These extensive splicing variants, in conjunction with many novel transcribed regions, give strong evidence of previously unappreciated transcriptome complexity.
In addition to identification of splicing isoforms, RNA-seq was used to study the process of alternative splicing regulation in zebrafish. A genome-wide and target-specific role of U1C protein in 5′ splice-site recognition and selection was characterized, adding this protein to the growing list of splicing regulators (Rösel et al., 2011).
SNP discovery
SNP is a single nucleotide variation at a given position in the genome between members of a biological species. SNPs are widely distributed throughout the genome and have been extensively used as the markers for many applications in genomics and genetics. With its superior sensitivity and single-base resolution, RNA-seq has been proved to be a very effective tool for the identification of gene-associated SNPs at a genome-wide scale. Several SNP identification-directed RNA-seq studies in fishes have already been reported. To developing SNP arrays in catfish, Liu and colleagues conducted RNA-seq in multiple individuals of both channel catfish and blue catfish (Ictalurus furcatus) (Liu et al., 2011). With the help of the SNP calling module in CLC Genomics Workbench (CLC bio, Aarhus, Denmark), as well as a SNP quality screening procedure, they identified 342,104 intra-specific SNPs for channel catfish, 366,269 intra-specific SNPs for blue catfish, and 420,727 inter-specific SNPs between channel catfish and blue catfish; these SNPs were found to be distributed within 16,562 unique genes in channel catfish and 17,423 unique genes in blue catfish. In another study, RNA-seq was performed to discover gene-associated SNPs in four strains of common carp (Cyprinus carpio) (Xu et al., 2012). BWA and SAMtools software were applied to align the reads to reference transcriptome and call SNPs, and, in total, 712,042 intra-strain SNPs and 53,893 inter-SNPs were identified. These identified SNPs provide a solid base for the future genetic studies in these fish species and will contribute to the development of a high throughput SNP genotyping platform.
Since 90% of the genetic difference between individuals is explained by SNPs, SNPs serve as invaluable markers for selection of important traits in breeding (Collins et al., 1998). In one RNA-seq study regarding identification of SNP markers for growth traits, 22 SNP markers and one mitochondrial haplotype were found to be significantly associated with growth traits in rainbow trout (Salem et al., 2012). In addition, SNPs are widely used as markers for distinguishing allelic transcripts when studying allele-specific expression that is essential for normal development and many cellular processes (Bell and Beck, 2009). RNA-seq provides a perfect tool for allele-specific expression studies by identifying SNPs and quantifying transcripts at the same time. For example, RNA-seq was successfully performed to assess the allele-specific expression in a F1 interspecies hybridized from southern platyfish (Xiphophorus maculates) and monterrey platyfish (Xiphophorus couchianus) (Shen et al., 2012).
Quantifying transcript level
One superior advantage of RNA-seq is its quantitative nature, enabling researchers to capture the transcriptome dynamic changes in response to environment or to intrinsic programs. Almost every RNA-seq studies published so far has included this analysis, and they all suggest that RNA-seq data gives reliable measurements of transcript levels within or between samples. In the last few years, RNA-seq, with a striking speed, has been widely used to detect differential gene expression in fish studies relating to developmental biology, immunology, evolutionary biology, physiology, toxicology, and diseases.
Developmental biology
Zebrafish is used as a model system, with the most prominent application in developmental biology. So far, the application of RNA-seq in the studies of fish developmental biology has been confined to zebrafish. RNA-Seq was employed to compare the transcriptome profiles of four early developmental stages (1-, 16-, 512-cell stage, and 50% epiboly) in zebrafish on a global scale (Vesterlund et al., 2011). In this study, only 177 genes were detected as developmentally regulated, while a majority of gene transcripts were present at a steady level, and a major transition in gene regulation and transcriptional activity took place between the 512-cell and 50% epiboly stages. To determine the role of vitamin D receptor (VDR) in zebrafish embryogenesis, transcriptome dynamics were assessed using RNA-seq in zebrafish embryos/larvae treated with 1α,25(OH)2D3 (active metabolite of vitamin D3) or vehicle for various periods of time (Craig et al., 2012). The expression levels of genes for transcription factors, peptide hormones, receptor-activator of NFκB ligand (RANKL) and of genes encoding proteins that plays key roles in fatty acid, amino acid, and xenobiotic metabolism pathways were significantly affected, demonstrating that 1α,25(OH)2D3 regulates multiple pathways in zebrafish embryogenesis. RNA-seq was also used to study zebrafish retinogenesis, and Id2a protein was identified as an intrinsic regulator of retinogenesis that balances between proliferation and differentiation during retinogenesis by modulating Notch pathway gene expression (Uribe et al., 2012).
Immunology
Comparison of the fish transcriptomes before and after immune challenges leads to the identification and subsequent characterization of immune-related genes and specific pathways involved in the immune responses, helping to create immune-based therapy for fish diseases, select disease-resistant fish brood stocks, and understand the origin and evolution of immune system. RNA-seq has been used in this kind of study for live fish, fish embryo, and fish primary cells.
The RNA-seq studies regarding fish immune responses to pathogens are mainly performed in economically important species. For instance, the transcriptomic response of channel catfish gill to Flavobacterium columnare, a Gram-negative bacterium implicated in fish disease outbreaks worldwide, was investigated using Illumina sequencing (Sun et al., 2012). Using a 1.5-fold change cut-off, 2605 uniquely annotated genes, with critical roles in pathogen recognition, cytoskeletal dynamics, cell junction integrity, oxidative stress responses, apoptosis, lysosomal processes, and pro- and anti-inflammatory pathways, were found to exhibit significant differential expression patterns. Fifteen differentially expressed genes detected by RNA-seq were confirmed by quantitative PCR (qPCR). In this study, a rhamnose-binding lectin (RBL) gene was highlighted, with a 105-fold increase in expression level. This discovery led to a subsequent study from the same group, in which RBL ligands, L-rhamnose and D-galactose, strongly protected channel catfish against columnaris disease in a dose-dependent manner (Beck et al., 2012). The same group also investigated the transcriptomic change in the intestinal epithelium of channel catfish following Edwardsiella ictaluri challenge (Li et al., 2012). In this study, 1633 differentially expressed genes, implicated in actin cytoskeletal polymerization/remodeling and junctional regulation in pathogen entry and subsequent inflammatory responses, were identified. Similar studies were carried out in Asian seabass (Lates calcarifer) (Xia et al., 2013) and Japanese sea bass (Lateolabrax japonicus) (Xiang et al., 2010), both of which were challenged with Vibrio harveyi.
To clarify the host immune mechanisms underlying the protective effects of vaccines and improve its immunogenicity in the future efforts, RNA-seq was used to investigate the immunization-related gene expression patterns of zebrafish and European sea bass immunized with vaccines against Edwardsiella tarda and Vibrio anguillarum, respectively. In the study with zebrafish, 4565 genes were expressed differentially in liver transcriptome samples before and after immunization (2186 up-egulated and 2379 down-egulated) (Yang et al., 2012). Further qPCR analysis confirmed that the genes encoding the factors involved in major histocompatibility complex (MHC)-I processing pathway were upregulated, while those involved in the MHC-II pathway were downregulated. In the RNA-seq study with European sea bass, differential expression was detected for 496 transcripts in head kidney and for 336 in gut (Sarropoulou et al., 2012).
Aside from in vivo studies, the fish immune response was also investigated in fish embryo and primary cells using RNA-seq. The innate host immune response to inflammatory bacterial infection was probed with zebrafish embryos infected with Salmonella typhimurium using both RNA-seq and tag-based sequencing (Ordas et al., 2011). In this study, two sequencing methods showed a strong correlation of sequence read counts per transcript and an overlap of 241 transcripts differentially expressed in response to infection. These transcripts were found to encode transcription factors, signal transduction proteins, cytokines and chemokines, complement factors, proteins involved in apoptosis and proteolysis, proteins with anti-microbial activities, as well as many known or novel proteins not previously linked to the immune response. Additionally, RNA-Seq analysis of Poly (I:C) (polyinosinic:polycytidylic acid)-challenged rainbow trout erythrocytes revealed diverse groups of differentially expressed mRNA transcripts related to multiple physiological systems including the endocrine, reproductive, and immune systems (Morera et al., 2011).
Aquatic toxicology
Aquatic toxicology aims to elucidate the effects of toxic chemicals on aquatic organisms at multiple levels, from subcellular level through individual organisms to communities and even ecosystems. Fish is one of the most significant research subjects in aquatic toxicology. RNA-seq enables researchers, from the transcriptomic perspective, to deduce how organisms respond to environmental pollutants. On the basis of such fundamental knowledge, the ultimate goal in aquatic toxicology is to predict and thus diminish or prevent the harmful effects of aquatic pollutants on the environment.
Oleksiak and colleagues (2011) utilized RNA-seq, in conjunction with a previous microarray data, to probe the transcriptomic response to polychlorinated biphenyl (PCB) exposure in embryos and larvae from a PCB-sensitive population and a PCB-resistant population of Atlantic killifish (Fundulus heteroclitus). Differential expression analysis revealed a sizeable set of PCB-responsive genes in the sensitive population, a much smaller set of PCB-responsive genes in the resistant fish, and few similarities in PCB-responsive genes between the two populations. The RNA-seq data corroborated most of the microarray results and detected novel transcripts that were not captured by microarray. This discrepancy in results between microarray and RNA-seq was also observed in other researches and might be improved by statistical methods with proper assessment the statistical significance of the observed changes (Esnaola et al., 2013). In another study, the toxicological effects of perfluorooctane sulfonate (PFOS), a widely-distributed persistent organic pollutant, on Oryzias melastigma embryos were examined using RNA-seq (Huang et al., 2012). Of the 145,394 genes detected by RNA-seq, 325 genes were significantly upregulated, while 349 genes were significantly downregulated, and these differentially expressed genes were found to be implicated in neurobehavioral defects, mitochondrial dysfunction and the metabolisms of proteins and fats. In a most recent study, RNA-seq was used to identify miRNAs in Atlantic salmon muscle tissue as potential biomarkers of toxicological stress (Kure et al., 2013). A total of 18 miRNAs were significantly differentially expressed in response to acidic aluminum-rich water, 4 downregulated and 14 upregulated. These identified differential expressed miRNAs may have the potential as biomarkers for other fish species as well. Other similar studies using RNA-seq were carried out in brown trout (Salmo trutta) (Uren Webster et al., 2013), Gulf killifish (Fundulus grandis) (Garcia et al., 2012), and yellow perch (Perca flavescens) (Pierron et al., 2011) stressed by a mixture copper and zinc, deepwater horizon oil, and a mixture cadmium and copper, respectively. In general, these studies have successfully shown that RNA-seq is a powerful approach to study the ecotoxicological response of fish to the polluted environments.
Physiological processes
Stress triggers certain physiological responses and plays an essential role in the natural selection. Due to the volatile aquatic environment in the wild, fish are exposed to different kinds of stressors, including lack of food, extreme water salinity, extreme temperature, pathogens, toxicants, and many others. Physiological responses to stresses are manifested by the changes in gene expression. So, identifying differentially expressed genes and pathways under stress conditions is important for the understanding of stress responses in fish. Stress responses to pathogens and toxicants have been discussed in the foregoing sections, and we thus focus on other stressors here.
The transcriptomic response of the liver of crimson-spotted rainbowfish (Melanotaenia duboulayi) to elevated temperature (21°C versus 33°C) was studied using RNA-seq (Smith et al., 2013). In this study, of the 107,749 assembled transcripts, 4251 transcripts were differentially expressed, and over 1000 of these differentially expressed transcripts were annotated; in addition to the well-characterized temperature-responsive genes relating to protection against apoptosis or maintenance of protein structure, gene ontology analysis revealed many novel temperature-responsive genes involved in catabolism, lipid metabolism, and oxidoreductase activity, indicating increased metabolism to cope with the increased temperature and the resulting hypoxic conditions. Analogously, RNA-seq was applied to determine the heat stress-induced gene expression profile in channel catfish (25°C versus 36°C), revealing similar results as crimsonspotted rainbowfish (Liu et al., 2013). These studies suggest that fish possess great plasticity in dealing with environmental temperature variations and assist the development of heat-tolerant fish strains for aquaculture. In addition, RNA-seq was performed to study the effects of the temperature variations during embryonic development on the adult thermal adaption ability in zebrafish (Scott and Johnston, 2012). Aside from heat stress, transcriptomic responses to salinity and fasting have been studied in Asian seabass (Xia et al., 2013).
Furthermore, RNA-seq studies regarding other aspects of fish physiology, such as swimming (Palstra et al., 2013) and circadian clock (Tovin et al., 2012), have been reported as well.
Evolution biology
Elucidation of the molecular basis of population divergence and speciation is one of the major challenges in evolutionary biology. Gene expression divergence is thought to be one of the mechanisms underlying phenotypic divergence. During the past few years, RNA-seq has become an invaluable tool for the study of phenotypic divergence by allowing whole transcriptome sequencing. For instance, RNA-seq analysis was conducted to study the adaptive transcriptomic divergence between dwarf and normal lake whitefish (Coregonus clupeaformis spp., Salmonidae) (Jeukens et al., 2010). In this study, normal whitefish was found to overexpress the genes related to protein synthesis, while dwarf fish overexpress the genes associated with immunity, DNA replication and repair, and energy metabolism, and the correlation between RNA-seq results and a previous microarray data was positive. Transcriptomic analyses using RNA-seq were similarly employed to study the genetic bases for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush) (Goetz et al., 2010), for the divergent pigment patterns in marine and freshwater sticklebacks (Gasterosteus aculeatus) (Greenwood et al., 2012), for the adaptation of Mexican tetra (Astyanax mexicanus) to the cave environment (Gross et al., 2013), and for the homologous relationship between zebrafish swimbladder and mammalian lung (Zheng et al., 2011). In these transcriptomic studies, many differentially expressed genes were detected, laying the ground for future functional genomic studies investigating the heritable genetic changes governing the phenotypic divergence in the evolutionary process.
Conclusion
RNA-seq technology is still in its infancy stage, however, it has superior advantages over other transcriptomic approaches, such as microarray, tilling array, and tag-based sequencing approaches. In the past few years, RNA-seq has made substantial contributions to our understanding of the fish transcriptome in terms of transcriptomes annotation, determination of the transcriptional structure of genes, and quantitative analysis of transcriptome dynamics during different biological processes. As RNA-seq technology continuously evolve and its cost keeps decreasing, within the next few years RNA-seq will without doubt be exploited to a larger extent and lead to many more exciting discoveries regarding the transciptomics of fish and other organisms.
Abbreviations Used
- CRT
cyclic reversible termination
- DE
differential expression
- DRS
direct RNA sequencing
- lincRNA
long intervening non-coding RNA
- lncRNA
long non-coding RNA
- miRNA
microRNA
- MPSS
massively parallel signature sequencing
- mRNA
messenger RNA
- ncRNA
non-coding RNA
- NGS
next-generation sequencing
- PMAGE
polony multiplex analysis of gene expression
- RBL
rhamnose-binding lectin
- RNA-seq
RNA sequencing
- rRNA
ribosomal RNA
- SAGE
serial analysis of gene expression
- SBL
sequencing by ligation
- SMRT
single molecule real time
- SNP
single nucleotide polymorphism
- sRNA
small RNA
- tRNA
transfer RNA
- WTSS
whole transcriptome shotgun sequencing
Acknowledgments
Preparation of this review was supported by Shanghai University Knowledge Service Platform Project (ZF1206).
Author Disclosure Statement
The authors declare that no conflicts of interest exist.
References
- Aanes H, Winata CL, Lin CH, et al. (2011). Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res 21, 1328–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adessi C, Matton G, Ayala G, et al. (2000). Solid phase DNA amplification: Xharacterisation of primer attachment and amplification mechanisms. Nucleic Acids Res 28, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamancos GP, Agirre E, and Eyras E. (2013). Methods to study splicing from high-throughput RNA Sequencing data . eprint arXiv:1304.5952. [DOI] [PubMed]
- Anders S, and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. (2009). Next-generation DNA sequencing techniques. N Biotechnol 25, 195–203 [DOI] [PubMed] [Google Scholar]
- Auer PL, and Doerge RW. (2011). A two-stage Poisson model for testing RNA-seq data. Stat Appl Genet Mol Biol 10, 1–26 [Google Scholar]
- Beck BH, Farmer BD, Straus DL, Li C, and Peatman E. (2012). Putative roles for a rhamnose binding lectin in Flavobacterium columnare pathogenesis in channel catfish Ictalurus punctatus. Fish Shellfish Immunol 33, 1008–1015 [DOI] [PubMed] [Google Scholar]
- Bell CG, and Beck S. (2009). Advances in the identification and analysis of allele-specific expression. Genome Med 1, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, et al. (2008). Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, and Miller CJ. (2010). A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics 11, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18, 630–634 [DOI] [PubMed] [Google Scholar]
- Cambier S, Gonzalez P, Durrieu G, Maury-Brachet R, Boudou A, and Bourdineaud JP. (2009). Serial analysis of gene expression in the skeletal muscles of zebrafish fed with a methylmercury-contaminated diet. Environ Sci Technol 44, 469–475 [DOI] [PubMed] [Google Scholar]
- Chen G, Wang C, and Shi T. (2011). Overview of available methods for diverse RNA-Seq data analyses. Sci China Life Sci 54, 1121–1128 [DOI] [PubMed] [Google Scholar]
- Chen R, Mias GI, Li-Pook-Than J, et al. (2012). Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, et al. (2005). Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 [DOI] [PubMed] [Google Scholar]
- Clement NL, Snell Q, Clement MJ, et al. (2010). The GNUMAP algorithm: Unbiased probabilistic mapping of oligonucleotides from next-generation sequencing. Bioinformatics 26, 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, and Chakravarti A. (1998). A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8, 1229–1231 [DOI] [PubMed] [Google Scholar]
- Collins JE, White S, Searle SM, and Stemple DL. (2012). Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res 22, 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V, Angelini C, De Feis I, and Ciccodicola A. (2010). Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol 2010, 853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig TA, Zhang Y, McNulty MS, et al. (2012). Research resource: Whole transcriptome RNA sequencing detects multiple 1α,25-dihydroxyvitamin D(3)-sensitive metabolic pathways in developing zebrafish. Mol Endocrinol 26, 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, et al. (2011). Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29, 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, et al. (2006). A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103, 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies MA, Rau A, Aubert J, et al. (2012). A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 14, 671–683 [DOI] [PubMed] [Google Scholar]
- Douglas SE. (2006). Microarray studies of gene expression in fish. Omics 10, 474–489 [DOI] [PubMed] [Google Scholar]
- Dressman D, Yan H, Traverso G, Kinzler KW, and Vogelstein B. (2003). Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA 100, 8817–8822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnaola M, Puig P, Gonzalez D, Castelo R, and Gonzalez JR. (2013). A flexible count data model to fit the wide diversity of expression profiles arising from extensively replicated RNA-seq experiments. BMC Bioinformatics 14, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulhammer D, Lipton RJ, and Landweber LF. (2000). Fidelity of enzymatic ligation for DNA computing. J Comput Biol 7, 839–848 [DOI] [PubMed] [Google Scholar]
- Febrer M, Mclay K, Caccamo M, Twomey KB, and Ryan RP. (2011). Advances in bacterial transcriptome and transposon insertion-site profiling using second-generation sequencing. Trends Biotechnol 29, 586–594 [DOI] [PubMed] [Google Scholar]
- Fedurco M, Romieu A, Williams S, Lawrence I, and Turcatti G. (2006). BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res 34, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser BA, Weadick CJ, Janowitz I, Rodd FH, and Hughes KA. (2011). Sequencing and characterization of the guppy (Poecilia reticulata) transcriptome. BMC Genomics 12, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Fu N, Guo S, et al. (2009). Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Wei CL, Liu ET, and Ruan Y. (2009). Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res 19, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Grabherr MG, Guttman M, and Trapnell C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8, 469–477 [DOI] [PubMed] [Google Scholar]
- Garcia TI, Shen Y, Crawford D, Oleksiak MF, Whitehead A, and Walter RB. (2012). RNA-Seq reveals complex genetic response to deepwater horizon oil release in Fundulus grandis. BMC Genomics 13, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz F, Rosauer D, Sitar S, et al. (2010). A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush). Mol Ecol 19, 176–196 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AK, Cech JN, and Peichel CL. (2012). Molecular and developmental contributions to divergent pigment patterns in marine and freshwater sticklebacks. Evol Dev 14, 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Furterer A, Carlson BM, Stahl BA. (2013). An integrated transcriptome-wide analysis of cave and surface dwelling Astyanax mexicanus. PloSOne 8, e55659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbers M, Carninci P. aand. (2005). Tag-based approaches for transcriptome research and genome annotation. Nat Methods 2, 495–502 [DOI] [PubMed] [Google Scholar]
- Hardcastle TJ, and Kelly KA. (2010). baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wurtzel O, Singh K, et al. (2010). Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Methods 7, 807–812 [DOI] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, and Kinzler KW. (2008). The antisense transcriptomes of human cells. Science 322, 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N, Merriman B, and Nelson SF. (2009). BFAST: An alignment tool for large scale genome resequencing. PLoS One 4, e7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook S. (2010). Promise and progress in environmental genomics: A status report on the applications of gene expression-based microarray studies in ecologically relevant fish species. J Fish Biol 77, 1999–2022 [DOI] [PubMed] [Google Scholar]
- Huang Q, Dong S, Fang C, Wu X, Ye T, and Lin Y. (2012). Deep sequencing-based transcriptome profiling analysis of Oryzias melastigma exposed to PFOS. Aquat Toxicol 120, 54–58 [DOI] [PubMed] [Google Scholar]
- Islam S, Kjällquist U, Moliner A, et al. (2011). Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21, 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A. (2009). The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10, 833–844 [DOI] [PubMed] [Google Scholar]
- Jeukens J, Renaut S, St-Cyr J, Nolte AW, and Bernatchez L. (2010). The transcriptomics of sympatric dwarf and normal lake whitefish (Coregonus clupeaformis spp., Salmonidae) divergence as revealed by next-generation sequencing. Mol Ecol 19, 5389–5403 [DOI] [PubMed] [Google Scholar]
- Jiang H, and Wong WH. (2009). Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25, 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, et al. (2004). Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res 14, 331– [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Bi Y, Pal S, Gupta R, and DavulurI RV. (2011). IsoformEx: Isoform level gene expression estimation using weighted non-negative least squares from mRNA-Seq data. BMC Bioinformatics 12, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Porreca GJ, Song L, et al. (2007). Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science 316, 1481–1484 [DOI] [PubMed] [Google Scholar]
- Knoll-Gellida A, André M, Gattegno T, Forgue J, Admon A, and Babin P. (2006). Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals. BMC Genomics 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kure EH, Sæbø M, Stangeland AM, et al. (2013). Molecular responses to toxicological stressors: Profiling microRNAs in wild Atlantic salmon (Salmo salar) exposed to acidic aluminum-rich water. Aquat Toxicol 138–139, 98–104 [DOI] [PubMed] [Google Scholar]
- Kvam VM, Liu P, and Si Y. (2012). A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am J Bot 99, 248–256 [DOI] [PubMed] [Google Scholar]
- Landegren U, Kaiser R, Sanders J, and Hood L. (1988). A ligase-mediated gene detection technique. Science 241, 1077–1080 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, Berger MF, Adiconis X, et al. (2009). Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol 10, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang Y, Wang R, et al. (2012). RNA-seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish Shellfish Immunol 32, 816–827 [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg J, and Lundeberg J. (2010). The plasticity of the mammalian transcriptome. Genomics 95, 1–6 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, et al. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Y, Li S, et al. (2012a). Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012, 251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang X, Sun F, et al. (2013). RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol Genomics 45, 462–476 [DOI] [PubMed] [Google Scholar]
- Liu S, Zhang Y, Zhou Z, et al. (2012b). Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote. BMC Genomics 13, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhou Z, Lu J, et al. (2011). Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics 12, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, and Bähler J. (2010). RNA-seq: From technology to biology. Cell Mol Life Sci 67, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, and Gilad Y. (2008). RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18, 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Ito A, Saitoh H, et al. (2005). SuperSAGE. Cell Microbiol 7, 11–18 [DOI] [PubMed] [Google Scholar]
- Mattick JS, and Makunin IV. (2006). Non-coding RNA. Hum Mol Genet 15, R17–R29 [DOI] [PubMed] [Google Scholar]
- Metzker ML. (2009). Sequencing technologies — The next generation. Nat Rev Genet 11, 31–46 [DOI] [PubMed] [Google Scholar]
- Morera D, Roher N, Ribas L, et al. (2011). RNA-seq reveals an integrated immune response in nucleated erythrocytes. PLoS One 6, e26998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, and Gentleman R. (2009). ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25, 2607–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova O, Hirst M, and Marra MA. (2009). Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet 10, 135–151 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, and Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, et al. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, and Barron AE. (2011). Landscape of next-generation sequencing technologies. Anal Chem 83, 4327–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JL, and Pavey SA. (2010). Perspectives: Gene expression in fisheries management. Curr Zool 56, 157–174 [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, et al. (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573 [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Karchner SI, Jenny MJ, Franks DG, Welch DBM, and Hahn ME. (2011). Transcriptomic assessment of resistance to effects of an aryl hydrocarbon receptor (AHR) agonist in embryos of Atlantic killifish (Fundulus heteroclitus) from a marine Superfund site. BMC Genomics 12, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordas A, Hegedus Z, Henkel CV, et al. (2011). Deep sequencing of the innate immune transcriptomic response of zebrafish embryos to Salmonella infection. Fish Shellfish Immunol 31, 716–724 [DOI] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, and Young MD. (2010). From RNA-seq reads to differential expression results. Genome Biol 11, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Goren A., Gymrek M., Guttman M., Regev A., Bernstein BE, and Milos PM. (2010). Digital transcriptome profiling from attomole-level RNA samples. Genome Res 20, 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, and Milos PM. (2011a). Transcriptome profiling using single-molecule direct RNA sequencing. Methods Mol Biol 733, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, and Milos PM. (2011b). Single-molecule direct RNA sequencing without cDNA synthesis. Wiley Interdiscip Rev RNA 2, 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages H, Aboyoun P, Gentleman R, and DebRoy S. (2009). Biostrings: String objects representing biological sequences, and matching algorithms. R package version 2.28.0 [Google Scholar]
- Palstra AP, Beltran S, Burgerhout E, et al. (2013). Deep RNA sequencing of the skeletal muscle transcriptome in swimming fish. PloS one 8, e53171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, and Blencowe BJ. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, et al. (2012). Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22, 577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold A, Reichwald K, Groth M, et al. (2013). The transcript catalogue of the short-lived fish Nothobranchius furzeri provides insights into age-dependent changes of mRNA levels. BMC Genomics 14, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron F, Normandeau E, Defo MA, Campbell PG, Bernatchez L, and Couture P. (2011). Effects of chronic metal exposure on wild fish populations revealed by high-throughput cDNA sequencing. Ecotoxicology 20, 1388–1399 [DOI] [PubMed] [Google Scholar]
- Rösel TD, Hung LH, Medenbach J, et al. (2011). RNA-Seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J 30, 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, and Pachter L. (2011). Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, et al. (2010). De novo assembly and analysis of RNA-seq data. Nat Methods 7, 909–912 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, and Smyth GK. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N, Sahoo PK, Baranski M, et al. (2012). Expressed sequences and polymorphisms in rohu carp (Labeo rohita, Hamilton) revealed by mRNA-seq. Mar Biotechnol 14, 620–633 [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlén M, and Nyrén P. (1998). A sequencing method based on real-time pyrophosphate. Science 281, 363–365 [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hinz W, Rearick TM, et al. (2011). An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, et al. (2011). Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 30, 111ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Le Ber P, Ng HH, and Liu ET. (2004). Interrogating the transcriptome. Trends Biotechnol 22, 23–30 [DOI] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, and Brudno M. (2009). SHRiMP: accurate mapping of short color-space reads. PLoS Computat Biol 5, e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M, Vallejo RL, Leeds TD, et al. (2012). RNA-Seq identifies SNP markers for growth traits in rainbow trout. PloS One 7, e36264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarropoulou E, Galindo-Villegas J, García-Alcázar A, Kasapidis P, and Mulero V. (2012). Characterization of European sea bass transcripts by RNA seq after oral vaccine against V. anguillarum. Mar Biotechnol 14, 634–642 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Turner S, and Kasarskis A. (2010). A window into third-generation sequencing. Hum Mol Genet 19, R227–R240 [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, and Brown PO. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 [DOI] [PubMed] [Google Scholar]
- Scott GR, and Johnston IA. (2012). Temperature during embryonic development has persistent effects on thermal acclimation capacity in zebrafish. Proc Natl Acad Sci USA 109, 14247–14252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, and Linnarsson S. (2013). Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14, 618–630 [DOI] [PubMed] [Google Scholar]
- Shen Y, Catchen J, Garcia T, et al. (2012). Identification of transcriptome SNPs between Xiphophorus lines and species for assessing allele specific gene expression within F1 interspecies hybrids. Comp Biochem Physiol C Toxicol Pharmacol 155, 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, and Ji H. (2008). Next-generation DNA sequencing. Nat Biotechnol 26, 1135–1145 [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, and Birol I. (2009). ABySS: A parallel assembler for short read sequence data. Genome Res 19, 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Bernatchez L, and Beheregaray LB. (2013). RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genomics 14, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, and Delorenzi M. (2013). A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Peatman E, Li C, et al. (2012). Transcriptomic signatures of attachment, NF-κB suppression and IFN stimulation in the catfish gill following columnaris bacterial infection. Dev Comp Immunol 38, 169–180 [DOI] [PubMed] [Google Scholar]
- Surget-Groba Y, and Montoya-Burgos JI. (2010). Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res 20, 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6, 377–382 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, et al. (2010). Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6, 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ’t Hoen PAC, Ariyurek Y, Thygesen HH, et al. (2008). Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovin A, Alon S, Ben-Moshe Z, et al. (2012). Systematic identification of rhythmic genes reveals camk1gb as a new element in the circadian clockwork. PLoS Genet 8, e1003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, and Bartel DP. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren Webster TM, Bury NR, van Aerle R, and Santos EM. (2013). Global transcriptome profiling reveals molecular mechanisms of metal tolerance in a chronically exposed wild population of brown trout. Environ Sci Technol 47, 8869–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe RA, Kwon T, Marcotte EM, and Gross JM. (2012). Id2a functions to limit Notch pathway activity and thereby influence the transition from proliferation to differentiation of retinoblasts during zebrafish retinogenesis. Dev Biol 371, 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Wiel MA, Leday GGR, Pardo L, et al. (2013). Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors. Biostatistics 14, 113–128 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, and Kinzler KW. (1995). Serial analysis of gene expression. Science 270, 484–487 [DOI] [PubMed] [Google Scholar]
- Vesterlund L, Jiao H, Unneberg P, Hovatta O, and Kere J. (2011). The zebrafish transcriptome during early development. BMC Dev Biol 11, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, and Snyder M. (2009). RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, and Landry JR. (2009). RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48, 249–257 [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, et al. (2008). Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243 [DOI] [PubMed] [Google Scholar]
- Xia JH, Liu P, Liu F, et al. (2013). Analysis of stress-responsive transcriptome in the intestine of Asian seabass (Lates calcarifer) using RNA-Seq. DNA Res 20, 449—460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang LX, He D, Dong WR, Zhang YW, and Shao JZ. (2010). Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics 11, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ji P, Zhao Z, et al. (2012). Genome-wide SNP discovery from transcriptome of four common carp strains. PloS One 7, e48140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Liu Q, Yang M, et al. (2012). RNA-seq liver transcriptome analysis reveals an activated MHC-I pathway and an inhibited MHC-II pathway at the early stage of vaccine immunization in zebrafish. BMC Genomics 13, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, and Birney E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chu W, and Fu G. (2009). DNA microarray technology and its application in fish biology and aquaculture. Front Biol China 4, 305–313 [Google Scholar]
- Zheng W, Wang Z, Collins JE, Andrews RM, Stemple D, and Gong Z. (2011). Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. PloS One 6, e24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Xu H, Lam SH, Luo H, Karuturi RKM, and Gong Z. (2013). Transcriptomic analyses of sexual dimorphism of the zebrafish liver and the effect of sex hormones. PloS One 8, e53562. [DOI] [PMC free article] [PubMed] [Google Scholar]