Abstract
Psoriasis is a chronic and common human skin disorder currently with no cure. Psoriatic skin displays inflammatory, raised, and scaly lesions with widely aberrant gene expression. Recent studies have revealed critical roles that microRNAs play as a class of posttranscriptional gene regulator in skin development and skin diseases. A substantial number of novel microRNAs have been identified in skin, and much has been learned about the dysregulated expression and functional roles of microRNAs in psoriasis, as well as the robustness and plasticity of microRNA-mediated gene expression regulation. Here we review recent progresses in discovery, profiling, and characterization of microRNAs in human psoriatic skin, discuss insights to their biological functions, and share our view on remaining challenges to be addressed.
Keywords: psoriasis, skin development, microRNA, endogenous siRNA
human skin is the largest organ of the body; it regenerates throughout the life of every individual, serves as the outermost barrier to prevent inner organs from dehydration, is able to repair when injured through a complex healing process, and responds to a variety of environmental stress and hazards (25, 54). Skin has developed a system of regulatory mechanisms to maintain these functions, orchestrated by various mediators that have local or systemic effects (79–81).
As a common skin disease, psoriasis is a chronic, autoimmune, and complex genetic disorder that affects 2∼3% of the European population. Psoriatic skin has symptoms of inflammation and raised and scaly lesions (8). Three different types of cellular alteration occur in psoriatic involved skin: abnormal differentiation of keratinocyte, hyperproliferation of keratinocyte, and infiltration of immune cells (70). Much has been learned about the molecular components and cellular pathways of inflammation that contribute to the pathogenesis of psoriasis (30, 55, 107), as well as genetic and environmental factors that influence the onset and progression of psoriasis (35, 71).
Results from recent studies have indicated that microRNAs (miRNAs), as a novel regulator of gene expression, play important roles in psoriasis. miRNAs form a class of short, nonprotein-coding, regulatory RNAs that play critical roles in nearly all biological processes, including cell differentiation, development, and metabolism, as well as complex diseases (6, 12). The maturation of miRNAs involves multiple steps, during which two intermediate forms of miRNAs, primary (pri-) and precursor (pre-)miRNAs, are produced sequentially. In this process, RNase III enzyme Drosha and partner double-stranded RNA (dsRNA) binding protein Dgcr8 cleave pri-miRNAs to produce hairpin-shaped pre-miRNAs that are recognized by Exportin5 and are subsequently transported from nucleus to cytoplasm. There, another RNase III enzyme Dicer cleaves the pre-miRNAs to release ∼22 nucleotide (nt) dsRNA duplexes, namely miRNA/miRNA* duplexes, with ∼2 nt 3′ overhangs. One strand of a RNA duplex, termed mature miRNA, is then loaded into an Argonaute protein in the RNA-induced silencing complex to exert its regulatory function by binding to target transcripts (6, 12).
Next-generation sequencing (NGS) is an enabling, high-throughput sequencing technology that is capable of profiling the expression of various RNA species in a genome-wide scale with single-nucleotide resolution (41, 53). Compared with hybridization-based techniques, such as quantitative PCR and microarray, which are more suitable for profiling known genes, NGS directly sequences RNA transcripts and thus is able to facilitate de novo discovery of novel miRNA genes and accurate quantification of miRNA expression (9, 32). Recent studies, particularly those based on NGS, have discovered a pool of miRNAs, miRNA-like RNAs, and their variants that are more diverse than we previously anticipated. These diverse miRNAs include canonical and noncanonical miRNAs (7, 13, 57, 65), miRNA-like RNAs (4, 22, 96, 97), and miRNA isoforms (58, 62) (Fig. 1). Canonical miRNAs are generated from a biogenesis pathway that requires Drosha and Dicer, as discussed earlier. Noncanonical miRNAs are produced from alternative biogenesis pathways where the essential player of the canonical miRNA biogenesis pathway, Drosha, is typically not involved. The first example of noncanonical miRNA is the class of mirtrons, which arise from short, ∼60 to 100 nt long, debranched intron lariats that form stem-loop structures that serve as substrates for Dicer cleavage, thus bypassing the Drosha/dgcr8 processing (7, 16, 65, 73). Dicer-dependent but Dgcr8-independent miRNA-like RNAs can also arise from local hairpin formation within larger noncoding RNA (ncRNA) species, such as small nucleolar RNAs (snoRNAs) (4, 10, 22). miRNAs are typically defined as the most abundant small RNAs on pre-miRNA hairpins. Nevertheless, other less abundant but cognate small RNAs from the same pre-miRNAs, which differ by a few bases from miRNAs, have been reported (46, 72). These miRNA variants have been named miRNA isoforms, or isomiRs (59), and observed to exist in animals, plants, and viruses (21, 23, 68). isomiRs can function as regular miRNAs (31, 95, 98); they often share common mRNA targets with their companion miRNAs but may also have their own exclusive target genes (98). The emergence of such diverse miRNAs as additional gene expression regulators with potential functions complementary to that of canonical miRNAs reflects the robustness and plasticity of miRNA-mediated gene expression regulation.
Fig. 1.
Biogenesis pathways of canonical and noncanonical microRNAs (miRNAs) and miRNA-like RNAs, with miRNA isoforms (isomiRs) included. A: RNA polymerase II transcribes primary miRNAs (pri-miRNAs) into long single-stranded primary RNA transcripts in the nucleus, where pri-miRNAs form double-stranded hairpin structures. Pri-miRNAs are then processed to ∼60 to 70 nt long precursor miRNAs (pre-miRNAs) by the Microprocessor (Drosha-DGCR8/Pasha complex). After being exported to the cytoplasm by Exportin-5, pre-miRNAs are further processed to ∼22 nt miRNA/miRNA* duplexes with 2-nt 3′ overhangs by the Dicer1-TRBP/PACT/Loqs complex. During the process, variants of miRNAs, termed isomiRs, can be produced due to inaccurate Drosha and/or Dicer processing. Finally, mature miRNAs are loaded into Argonaute (Ago) protein complexes, while miRNA* are discarded. b: mirtron-derived pathway. Mirtrons are debranched short introns forming pre-miRNA hairpin mimics, thus bypassing the Drosha process. C: snoRNA-derived pathway. A number of box H/ACA snoRNAs can give rise to Ago-associated, miRNA-like species in a Drosha/DGCR8 independent, Dicer-dependent manner. d: short-hairpin RNAs (shRNAs) are directly transcribed as short RNAs folding to pre-miRNA-like hairpins, thus bypassing the Drosha process. E: in the case of tRNA-Ile/miR-1983, a pre-tRNA folds into an alternative structure consisting of a pre-miRNA-like hairpin. F: a number of tRNA and miRNA polycistrons in murine gamma herpesvirus are processed to pre-miRNA-like hairpins by tRNase Z instead of Drosha. G: miRNAs derived from a terminal hairpin of an endogenous siRNA (endo-siRNA) precursor. Endo-siRNA precursors are processed by the Dicer2 complex to yield pre-miRNA-like terminal hairpins, which are then processed by the Dicer1 complex. H: Dicer-independent pathway. Drosha cleavage of pri-miR-451 proceeds normally, and the pre-miR-451 is directly incorporated into Ago2 without Dicer cleavage. I: Dicer-independent and tRNase Z-dependent pathway. A 3′-trailer fragment of a precursor tRNA is released by tRNase Z cleavage. Organisms that represent each pathway (B–I) are indicated at the bottom of the panels. D.m., Drosophila melanogaster, G. lamblia, Giardia lamblia. Adapted and modified from Fig. 1 of (57) with permission.
It has been estimated that miRNAs regulate over one-third of protein-encoding mRNAs in humans (24, 50). In mammalian skin development, recent studies have revealed that the interaction between miRNAs and their target mRNAs is critical in regulating distinct signaling pathways during cell differentiation (48, 104). Noncanonical miRNAs, miRNA-like RNAs, and isomiRs in normal and psoriatic human skin are also of interest and have recently been reported (38, 96). Dysregulation of miRNAs and their regulated targets has been implicated in the pathogenesis of psoriasis (75, 82, 108) as well as other forms of disorder of the skin, including malignant melanoma (11, 60, 76, 77). Such knowledge of the aberrantly expressed small ncRNAs (sncRNAs) in psoriatic skin has suggested functional roles of sncRNAs in psoriasis. An interesting and emerging theme from these studies is that miRNAs that take important parts in early skin development can also function in critical ways in psoriasis. This theme of “what goes wrong early in life can go wrong again later in life” echoes a similar scenario that dysregulation of some miRNAs critically important during early cardiovascular muscle development contributes to cardiovascular diseases (51). This emerging theme on miRNAs suggests that these miRNAs may potentially be excellent candidates for therapeutic treatment of psoriasis. We discuss in this short review the latest development in identification and characterization of miRNAs and recent findings about their regulatory functions during skin development and in normal and psoriatic skin. We finish by sharing our view on challenges to be addressed for future development.
miRNAs DURING SKIN DEVELOPMENT
Significant progress has been made in identifying miRNAs and characterizing their specific functions in skin morphogenesis and homeostasis. Several miRNAs with such functions have been studied (Table 1). Initial efforts have been devoted to discovering novel miRNAs (45), investigating phenotypes of Dicer and Dgcr8 skin conditional knockouts (1, 102), and determining their spatiotemporal patterns of expression in mammalian skin by using cloning and sequencing methods (102).
Table 1.
miRNAs that function in skin development
| miRNA | Function | Ref. List No. |
|---|---|---|
| miR-203 | induced in suprabasal layer to inhibit cell proliferation by repressing p63; regulate the transition from basal to suprabasal layer in epidermis | 48, 104 |
| miR-34a/c | repressed by p63 in epidermal cells to maintain cell cycle progression and expression of cyclin D1 and Cdk4 | 2 |
| mir-125b | expressed in skin stem cells to balance self-renewal and early lineage commitment | 106 |
| miR-200/miR-205 | restricted to basal layer. 1) maintains proliferation of progenitor cells | 91 |
| 2) maintains epithelial-mesenchymal transition | 28, 42, 67 |
miR, miRNA: microRNA.
By investigating phenotypes of mutants knocking out either Dicer or Dgcr8 in murine embryonic skin, the importance of miRNAs in skin development has been revealed (1, 102). Overall, mutants of Dicer and Dgcr8 conditional knockouts present similar phenotypes of severe defects in murine embryonic skin development (103). Specifically, both mutants have been characterized by rough skin, failure to gain weight, defects in hair follicle down-growth, abnormal apoptosis, and no more than 6-day survival after birth (103). Hyperproliferation has been observed in the Dicer conditional knockout epidermis, showing the importance of miRNAs in regulating epidermal proliferation (1).
Profiling miRNA expression at epidermal skin and hair follicle at embryonic day 17.5 (E17.5) has revealed a set of differentially expressed murine miRNAs between the two places. (102). For example, the highly expressed microRNA (miR)-199 family is only found in the hair follicle but not from the epidermis at E17.5, indicating a potential regulatory function that relates to hair morphogenesis (102). Further investigation by profiling miRNA expression in murine skin at embryonic day 13.5 (E13.5) and day 15.5 (E15.5) has revealed that miR-203 is barely detectable in E13.5 skin but emerges as one of the most abundant miRNAs after E15.5. Phenotypic difference between E13.5 and E15.5 skin is also prominent. When only single-layered multipotent embryonic skin stem cells exist at E13.5, the stratification of epidermis begins after E15.5, suggesting that miR-203 has a critical function in skin differentiation and stratification (104).
To appreciate the spatiotemporal pattern of miR-203 expression, in situ hybridization (ISH) has been carried out to localize the expression of miR-203 in great detail. It has been shown that the expression of miR-203 depends on the stages of skin development and is cell specific; it is highly expressed only in differentiated cells of mature skin such as suprabasal epidermis and inner root sheath of the hair follicle, but not in progenitor or stem cells such as basal epidermis and outer root sheath (48, 104). Furthermore, analysis of zebrafish skin (94) has demonstrated that the expression of miR-203 is restricted to the outermost layer of skin, indicating a conserved function of miR-203 contributing to skin differentiation across species.
To comprehend the physiological mechanism of miR-203 functions in differentiated skin cells, targets of miR-203 have been examined. Among these targets, the transcription factor p63, which plays an essential role in maintaining “stemness” in the skin, has been well examined (78). Highly expressed miR-203 in suprabasal layer has been shown to repress the expression of p63, thus reducing the proliferative potential of terminally differentiating keratinocytes (48, 104).
Besides miR-203, recent studies have also recognized several other miRNAs involved in skin development and homeostasis (Table 1 and Fig. 2). For example, repression of miR-34 transcription by p63 is important to maintain cell cycle progression in epidermal cells (2). miR-125b has been shown to be highly expressed in skin stem cells, in contrast to dramatically lower expression in their early progeny (106). In an inducible mouse system, miR-125b has been implicated as a repressor of stem cell differentiation (106). In addition, miR-205 has been found restricted to basal progenitor cells and had roles in maintaining the expansion of skin stem cells by antagonizing negative regulators of PI(3)K signaling (91). miR-200 family and miR-205 are both highly expressed in normal skin. These miRNAs have been shown to target ZEB1 and ZEB2, which are both transcriptional repressors of E-cadherin (17, 28, 42, 67). Downregulation of the miR-200 family and miR-205 induces an epithelial-to-mesenchymal transition in conjunction with upregulation of ZEB1 and ZEB2 (28).
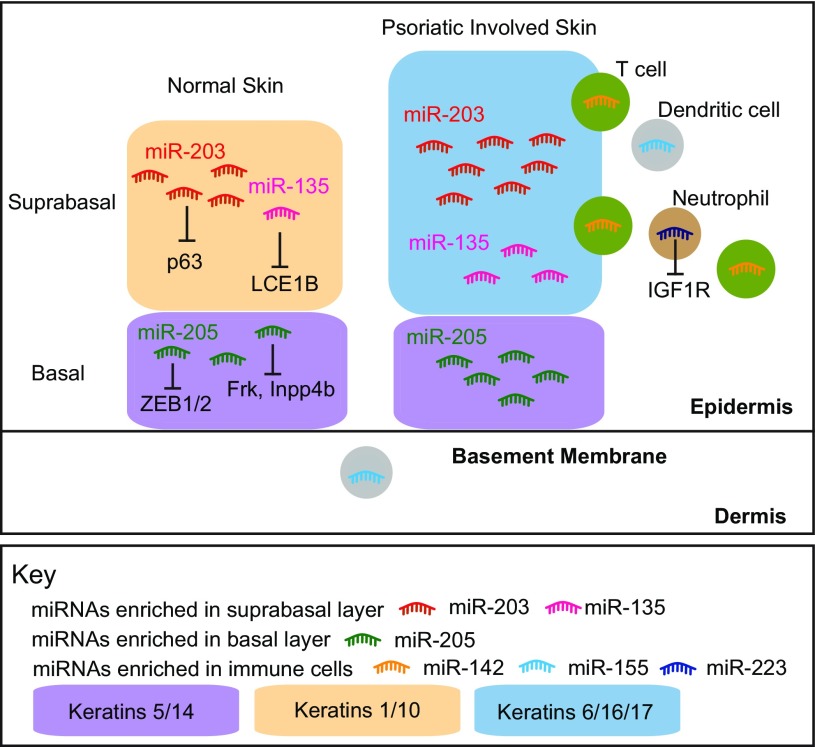
Fig. 2.
Localization of miRNAs in normal and psoriatic-involved skin. Normal epidermis contains the basal and suprabasal layers. The basal layer contains progenitor cells in which marker proteins such as keratin-5 and keratin-14 are highly expressed, and the suprabasal layer contains mostly differentiated cells characterized by keratin-1 and keratin-10. miR-203 and miR-135 are highly expressed in the suprabasal layer and miR-205 is restricted to basal progenitor cells. In psoriasis lesions, the suprabasal layer is thicker and disorganized, where keratins 6/16/17 are induced in large amounts in response to trauma. miR-203 and miR-205 are moderately upregulated (1.6-fold), and miR-135 shows higher upregulation (5-fold) in psoriatic-involved vs. normal epidermis. With infiltrated immune cells, psoriatic-involved epidermis also contains miRNAs specific to immune cells such as miR-142 and miR-223.
miRNAs IN PSORIATIC SKIN
Novel miRNAs and miRNA Variations
A recent study has adopted NGS to comprehensively profile microRNAome in human psoriatic and normal skin (38). Compared with the results from profiling murine skin (102), the miRNAs that are most abundantly expressed in human and murine skin largely overlap. Furthermore, the depth of the collected data has not only allowed detection of low abundant miRNAs but also enabled discovery of novel human miRNAs at an unprecedented rate (38). Overall, >200 loci in the human genome have been identified to host novel miRNAs and miRNA candidates, and 38 novel miRNAs from this profiling study have been registered in miRbase (38). Many of these miRNAs and candidates have been reaffirmed as miRNAs in other studies. Over one-third of the novel miRNAs are intronic, and some are of particular interest in skin. The genomic loci of the intronic miRNAs suggest their coexpression and functional relationship with their host genes. For example, miR-4632, a newly identified mirtron, is located within TNFRSF1B, which has been implicated in psoriatic arthritis (69). Intronic miR-6510 is encoded in KRT15, which shows downregulation in psoriatic skin (30). A known human-specific miRNA, miR-944, is located in an intron of p63, a well-characterized transcription factor with essential function in maintaining stemness in skin (78).
Several novel miRNAs align uniquely to the antisense strands of known miRNAs (38). For example, miR-203-AS has been identified as a distinct miRNA on the DNA strand at the locus antisense to miR-203 (38). A few known miRNA loci, such as miR-103, encode similar but distinct miRNAs on both sense and antisense strands. As a note, distinct function of antisense miRNA has been described in detail at the miR-iab-4 locus in Drosophila melanogaster, where sense and antisense miRNAs coordinately regulate Hox genes during larval development (84).
A variety of noncanonical miRNAs and miRNA-like RNAs have been discovered in human psoriatic and normal skin with NGS-based profiling (38, 96). The largest group of noncanonical miRNAs discovered is the class of mirtrons; >100 mirtron candidates have been identified in human skin (38, 96), which have important implications for biogenesis and function. Genomic loci of many ncRNAs, such as snoRNAs and tRNAs, may also host Dicer-dependent but Dgcr8-independent miRNA-like RNAs. For example, miRNA-like RNAs have been identified from small cajal body-associated RNAs, ACA45, in human (22) and mouse (4), as well as psoriatic and normal skin (38). Although there have been several snoRNA-derived miRNAs in eukaryotes, only one tRNA-derived miRNA has been reported so far (4). In this case, a murine tRNA, IleTAT, folds into an alternative secondary structure consisting of a stem-loop hairpin from which mmu-miR-1983 is derived (4). hsa-miR-1983, a homolog of mmu-miR-1983 located within human tRNA-IleTAT, has been detected to express in psoriatic and normal human skin at a lower level than mmu-miR-1983 in murine stem cells (96).
A systematic analysis has revealed that miRNA isoforms, i.e., isomiRs, can consistently originate from a majority of miRNA hairpins in diverse tissues and across species. Of particular interest is the class of so-called 5′-isomiRs, whose 5′-ends shift by a few bases from the 5′-ends of the cognate miRNAs (98). A number of miRNA loci that are essential for skin development and homeostasis produce substantial amounts of 5′-isomiRs. Particularly, the 5′-isomiR from hsa-miR-203 hairpin accumulates to the same level of abundance as the major miR-203 in human psoriatic and normal skin (Table 2) (98). Murine mmu-miR-203 hairpin also carries an abundant 5′-isomiR in murine skin (98), suggesting that the mechanism of generating 5′-isomiRs is conserved in mammals. In addition, miR-142 and miR-223, two hematopoietic-specific miRNAs, have high 5′-heterogeneities in human psoriatic lesions (98). miR-142 is highly expressed in dendritic cells (86) and miR-223 in neutrophils in humans and mice (5, 47). The abundant expression of miR-142 and miR-223 can be anticipated, given that immune cells including dendritic cells and neutrophils are known to infiltrate in affected skin lesions (38, 44).
Table 2.
miRNAs that express aberrantly in human psoriatic skin and relative amount of their companion 5′-isomiRs
| miRNA | Description | Fold Change (PP/NN) | IsomiRs, % |
|---|---|---|---|
| miR-31 | regulates cytokine/chemokine expression via targeting serine/threonine kinase 40 (STK40) | 42.9 | 0.4 |
| miR-977 | mirtron; upregulated in psoriatic skin | 6.4 | 1.3 |
| miR-135b/miR-7 | targets late cornified envelope-1B (LCE1B); impairs barrier development and response to environmental stimuli | 5.6/3.1 | 0.4/1.0 |
| miR-1983 | tRNA-derived human homolog of murine miR-1983 | 4.9 | 0.2 |
| miR-21 | contributes to T cell-derived psoriatic skin inflammation | 4.0 | 0.6 |
| miR-203-AS | antisense to miR-203 | 2.7 | 7 |
| miR-155/miR-142/miR-146a | cell fate decisions in hematopoietic development | 2.7/2.5/2.3 | 1.1/46/0.8 |
| miR-223 | suppresses cell proliferation by targeting IGF-1R | 2.5 | 9.8 |
| miR-203 | regulates the transition from basal to suprabasal layer in epidermis | 1.6 | 43 |
| miR-205 | primarily in the basal layer expressed in normal skin and regulates the transcriptional repressor of E-cadherin to maintain epithelial-mesenchymal transition | 1.6 | 0.5 |
| miR-99a/miR-100 | inhibits angiogenesis by repressing the mammalian target of rapamycin (mTOR) in endothelial cells | −2.1/−2.3 | 3.1/0.7 |
| mir-10a | regulates the behavior of endothelial cells during angiogenesis | −2.3 | 22 |
| miR-124 | downregulation in skin cancer, cutaneous squamous cells | −5.9 | 23 |
| miR-4490 | novel miRNA from intergenic region | −8.1 | 1.2 |
Fold change is a value of miRNA expression in psoriatic skin (PP) divided by that in normal skin (NN). IsomiRs, the percentage of reads mapping to miRNA hairpins identified as 5′-isomiRs.
With a large pool of novel miRNA candidates, experimental validation further supports endogenous expression of novel miRNAs in human skin. For example, stem-loop quantitative real-time PCR (qRT-PCR) has showed that novel miRNAs, including miR-203-AS, miR-3613, and miR-4490, have visible bands of the same length of miR-203 in normal skin (38). Stem-loop qRT-PCR uses primers specific to sequences in the loop regions of miRNAs so that they have higher RT efficiency than the conventional qRT-PCR and are able to discriminate among paralogous miRNAs (15, 43).
Ectopic expression in HEK293 cells has indicated that miR-203-AS undergoes miRNA biogenesis processing and incorporates into the Argonaute 2 (AGO2) protein (38). qRT-PCR has also confirmed the expression of miRNA-like RNAs from noncanonical miRNAs, including miR-6499 (mirtron) and miRNAs from tRNA-pseudo-TTA and snoRNA-U60, in normal human skin (96). Ectopic expression of these miRNA-like RNAs has shown their enrichment in AGO2 immunoprecipitates, suggesting that they can associate with AGO2 protein (96).
Aberrant Expression of miRNAs in Psoriatic Skin
In addition to playing critical roles in skin development, miRNAs also regulate gene expression in skin disorders, including psoriasis. Earlier studies using microarray-based miRNA profiling have implicated the involvement of a number of miRNAs in pathogenesis of psoriasis (83, 108). Profiling using NGS, which is sensitive to low abundant transcripts, has identified 125 miRNAs from a diverse origins of canonical and noncanonical miRNAs as well as 5′-isomiRs that exhibit more than twofold differential expression in psoriatic skin (Supplemental Table S1) (38, 96, 98).1 The majority of the differentially expressed miRNAs are upregulated (90 upregulated vs. 35 downregulated) in psoriatic-involved (PP) skin vs. normal (NN) skin. This result is consistent with the overall mRNA expression changes in psoriatic skin, where there are more upregulated than downregulated mRNA transcripts and substantially fewer differentially expressed miRNAs between psoriatic-uninvolved (PN) skin vs. NN skin (30, 107).
A subset of differentially expressed (DE) miRNAs has been validated by stem-loop qRT-PCR, including 12 significantly DE miRNAs (8 upregulated and 4 downregulated) in PP skin compared with PN or NN skin (38). The most highly upregulated miRNAs in psoriatic skin include the abundantly expressed miR-21 and miR-31 (4- and 43-fold, respectively; Table 2), which are potential proangiogenic miRNAs (92). Upregulation of miR-21 and miR-31 has been observed in various cancers including skin cancer (74). miR-155, miR-142-3p, and miR-146a, which regulate various cell fate decisions in hematopoietic development (27), show ∼2.5-fold upregulation in PP vs. NN skin (Table 2). miR-203-AS, discovered in human skin, has also been validated by qRT-PCR to have a 2.7-fold upregulation in psoriatic skin (38). Interestingly, miR-203, which plays a critical role in skin development, shows a moderate 1.6-fold upregulation in psoriatic vs. normal skin (38).
The most significantly downregulated miRNAs in psoriatic skin include miR-124 and miR-4490 (5.8- and 8.0-fold downregulated, respectively; Table 2). miR-124 also exhibits downregulation in the cutaneous squamous cell, one of the most common skin cancers (100). Several highly expressed miRNAs including miR-10a, miR-99a, and miR-100 (Table 2) in human skin also exhibit significant downregulation (2- to 2.3-fold) in psoriatic skin.
Although the overall abundance of noncanonical miRNAs is low in human skin, 12 of them, including hsa-miR-1983 (Supplemental Table S1), show differential expression in PP and/or PN skin (96). The differential expression of hsa-miR-937 (Table 2), which is recently reclassified as a mirtron, has also been validated by qRT-PCR (96). Furthermore, 18 5′-isomiRs also exhibit aberrant expression in psoriatic skin (98). One prominent example is the 5′-isomiR from miR-203; the 5′-isomiR is more than twofold upregulated in psoriasis. In addition, 5′-isomiRs from miR-142 and miR-223 are upregulated by 2.3- and 2.5-fold in psoriatic vs. normal skin. Given the high abundance and upregulation of many 5′-isomiRs in psoriasis, DE miRNAs and 5′-isomiRs may be important for pathogenesis of psoriasis.
Overall, the dramatic alteration in miRNA expression in psoriatic skin reflects the pathogenic features of psoriasis such as defects in skin cells, infiltration of immune cells, and impaired vasculature.
Functions of DE miRNAs in Psoriasis
An early study exploring the function of miRNAs in psoriasis has revealed an upregulation of miR-203 in psoriatic skin; miR-203 has been implicated in targeting suppressor of cytokine signaling 3 (SOCS3) (83). However, subsequent studies have challenged the conclusion that SOCS3 is a genuine target of miR-203 (48, 104). Furthermore, miR-203 has been characterized as an inhibitor of cell proliferation by targeting the transcription factor p63 in normal skin (Table 1 and Fig. 2) (48, 104). Thus, it is arguable for miR-203 to be downregulated in psoriatic skin, potentially accounting for the hyperproliferative skin involving a proinflammatory response. miR-203, however, has been observed to be slightly upregulated in PP skin in several studies (38, 83, 108), suggesting a minor effect of miR-203 to suppress the pathogenic hyperproliferation in psoriatic skin. Although further study is needed to reconcile the disparity over miR-203 targets and to determine the function of miR-203 in psoriatic skin, it has been speculated that the newly identified miR-203-AS as well as its cognate miRNA isoform (see below) may indicate their role in psoriasis (38, 98).
miR-21 and miR-31 are among the most abundant miRNAs with dramatic upregulation (4- and 40-fold, respectively; Table 2) in psoriatic lesions (38). The expression of miR-21 in epidermal cells and dermal T cells between psoriatic and healthy skin has also been examined, showing an elevated expression in both cell types (56). Inhibition of miR-21 increases the rate of apoptosis in activated T cells, suggesting that miR-21 suppresses apoptosis in activated T cells. Therefore, overexpression of miR-21 in psoriatic lesions may reflect the infiltration of activated T cells in psoriatic skin, contributing to T cell-derived psoriatic skin inflammation (56). Overexpression of miR-31 has been shown to contribute to skin inflammation in psoriasis lesions by regulating the production of inflammatory mediators and leukocyte chemotaxis to the skin (99). Specifically, miR-31 regulates cytokine/chemokine expression via targeting serine/threonine kinase 40 (STK40), a negative regulator of NF-κB signaling, in keratinocytes (99). Besides, transforming growth factor-β1, a cytokine highly expressed in psoriasis epidermis, has been shown to induce upregulation of miR-31 in keratinocytes in vitro and in vivo (99).
miR-135b and miR-7, which are upregulated in psoriatic skin (Table 2), have been reported to regulate late cornified envelope-1B (LCE1B) through direct interactions with the 3′-untranslated region of LCE1B (39). The LCE gene family is known to have dozens of members that are clustered within the epidermal differentiation complex region on human chromosome 1. LCE genes are transcribed synergistically in response to environmental stimuli such as calcium levels and ultraviolet light (36). In skin, LCE1B is normally expressed in a skin barrier but shows a 2.1-fold downregulation in psoriatic skin (30). Therefore, downregulation of gene LCE1B targeted by upregulated miR-135b and miR-7 may result in defects in barrier development, contributing to the pathogenesis of psoriasis.
Downregulated miRNAs are of the same significance as upregulated miRNAs. miR-10a regulates the behavior of endothelial cells during angiogenesis by positively titrating proangiogenic signaling in zebrafish and human (33). Downregulation of miR-10a may contribute to abnormal behavior of endothelial cells in psoriasis. miR-100 is an antiangiomiR that has been shown to inhibit angiogenesis by repressing the mammalian target of rapamycin (mTOR) in endothelial cells (29). miR-99a has the same seed (ACCCGUA) as miR-100 and thus is anticipated to have a similar target specificity (49). Indeed, miR-99a and miR-100 have been shown to target mTOR in human skin (37). Downregulation of miR-99a and miR-100 has been observed in diverse cancers such as esophageal squamous cell carcinoma and endometrioid endometrial carcinoma (85, 90). Given the function of miR-99a and miR-100 as inhibitors of angiogenesis, downregulation of the two miRNAs in psoriatic skin may have an implication in pathogenic characteristics of psoriasis, particularly increased angiogenesis.
A system-wide target analysis has revealed that DE miRNAs show anticorrelated expression with their target mRNAs (38, 96). In particular, 287 DE mRNAs express anticorrelated with 55 DE canonical miRNAs (43 upregulated and 12 downregulated), and 59 DE mRNAs exhibit expression anticorrelated with nine noncanonical miRNAs (7 upregulated and 2 downregulated). A gene ontology analysis has shown that the anticorrelated targets of canonical DE miRNAs are significantly enriched in biological processes such as response to cytokine stimulus, organic substance, and estrogen stimulus. The genes related to cytokine stimulus are of particular interest because overexpression of proinflammatory cytokines, including IFN-alpha and TNF-alpha, has been implicated in the pathophysiology of psoriasis (44). The gene ontology analysis also shows that the enriched molecular functions involve transcription factor activity and cytoskeletal protein binding. These functional terms are consistent with the early results from mRNA transcriptome analyses in Refs. 30, 87, suggesting that miRNAs may respond to the dysregulated transcriptome of psoriasis.
Target analyses show that more than half of the targets between miRNAs and their 5′-isomiRs are different, implying their potentially distinct functions (98). For example, hsa-miR-203 and its most abundant 5′-isomiR share about half (53.7%) of their common targets. The 5′-isomiR of miR-203 may have additional and complementary functions with miR-203 by putatively regulating p63 to affect multiple pathways during skin development. Overall, DE 5′-isomiRs have a total of 148 anticorrelated pairs with 110 distinct target genes (21 upregulated and 89 downregulated) in psoriatic and normal skin (98).
While 5′-isomiRs are highly expressed in diverse tissues including skin, their impact on mRNA expression remains to be studied. Recent studies investigating a few examples of 5′-isomiRs (3, 16, 34) have shown that 5′-isomiRs of miR-223 have exclusive targets that are significantly de-repressed after the miR-223 gene was knocked out in neutrophils, indicating a direct impact of 5′-isomiRs on target gene expression (16, 98). Existing experimental evidence from in vitro target cleavage assays supports the notion that miR-142-5p and its 5′-isomiR have different target specificities (3). In light of the fact that 5′-isomiRs are functional, the dysregulated 5′-isomiRs, including those from the loci of abundantly expressed hsa-miR-203, hsa-miR-142, and hsa-miR-223, are of particular interest in psoriatic skin.
Cell Type-specific miRNA Expression in Psoriatic Skin
RNA ISH has been adopted to localize miRNA expression to determine cell-specific expression patterns (40, 94). It has helped reveal the spatial pattern of miR-203 expression in the suprabasal layer of the epidermis. Adaption of ISH of miRNAs in psoriatic skin has also revealed that miR-135b, one of the upregulated miRNAs, is expressed in the suprabasal epidermis of psoriatic epidermis but is excluded from the basal layer (Fig. 4 of Ref. 38). In contrast, miR-205 is expressed primarily in the basal layer of psoriatic uninvolved skin, but not in the basal and suprabasal layers of PP skin (38). Although miR-205 does not show more than twofold differential expression in psoriasis, its modest upregulation in psoriatic skin coupled with a previously described function in the establishment of epithelial cell fate (28) points toward its potential role in psoriasis pathogenesis. The spatial patterns of miR-135b and miR-205 in psoriatic skin suggest a role in keratinocyte differentiation. ISH of DE miRNAs has also helped recognize the epidermal infiltration of miR-142-3p, a hematopoietic-specific miRNA, in psoriatic lesions (Fig. 4 of Ref. 38), consistent with the infiltration of dendritic cells, where miR-142 is highly expressed, in affected skin lesions (38, 44).
OTHER SMALL RNAs IN NORMAL AND PSORIATIC SKIN
Endogenous siRNAs (endo-siRNAs) arise from long dsRNA transcripts from repetitive or transposable elements (66, 88, 93), whose biogenesis requires Dicer but not Drosha/Dgcr8 (3, 10). Endo-siRNAs have been described in plants (12, 14), Caenorhabditis elegans (72), D. melanogaster (19, 26, 63, 64), and murine embryonic stem cells and oocytes (4, 88, 93). It has also been indicated that endo-siRNAs are expressed in murine embryonic skin (103). Endo-siRNAs have functions to repress mobile elements and to regulate downstream targets (88).
In human skin, specific sncRNAs from repeat regions are endo-siRNA candidates. In particular, an siRNA candidate derived from an Alu-short interspersed element exhibits a 17-fold upregulation in PP skin (96). This endo-siRNA resides within an intron of the gon-4-like (GON4L) gene on chromosome 1, where three Alu-SINEs appear in tandem. In sharp contrast to the huge upregulation of the endo-siRNA, the expression of the host gene GON4L shows little variation between PP and NN skin. This discrepancy implies that the endo-siRNA and the host gene may be transcribed or processed independently. The cluster of the siRNAs appears only in the first two Alu SINEs, suggesting that the repetitive elements may harbor the novel endo-siRNAs instead of the intron. A few targets, such as MDM4 (mouse double minute 4 homolog), are anticorrelated with the Alu-derived endo-siRNAs (96). This gene is an inhibitor of p53 that is frequently upregulated in cancers (20, 89) but modestly downregulated in psoriasis (61). Thus, the downregulation of MDM4 in psoriasis, which may be mediated by the novel Alu-SINE-derived siRNA, might be one factor that distinguishes the benign hyperplasia in PP skin from cancerous tumors. MDM4 is also a putative target of canonical miRNA miR-19b that is upregulated by 2.1-fold in psoriasis. These results indicate MDM4 is tightly regulated by miRNAs and endo-siRNAs in psoriatic skin.
SUMMARY AND FUTURE DIRECTIONS
Although still in its infancy, the study of miRNAs and endo-siRNAs in mammalian skin and skin disorders has already provided deep insights into this novel layer of gene regulation. Knowledge about the aberrantly expressed sncRNAs in psoriatic skin has laid a foundation for understanding the roles of miRNAs and endo-siRNAs in psoriasis. We now have a good collection of miRNAs as well as their validated and putative targets, many of which are discussed here, that provide not only new knowledge of psoriasis mechanisms, but also novel threads for future research.
An interesting and converging theme has emerged from the results and findings obtained so far, i.e., two key miRNAs, miR-203 and miR-205 (Tables 1 and 2 and Fig. 2), which play essential roles in early skin development also function in critical ways in psoriasis. This theme of “what goes wrong early in life can go wrong again later in life” echoes a similar observation that miR-1 and miR-133 are critically important during early cardiovascular muscle development as well as in cardiovascular diseases (51). This emerging theme on miRNAs suggests that these miRNAs may potentially be excellent candidates for therapeutic treatment of psoriasis.
Despite significant progress that has been made so far, recent results and findings require more research to be done to unravel the complex genetic networks involved in the etiology of autoimmune skin disorders such as psoriasis. First, it is important, albeit challenging, to build a high-quality regulatory network underlying psoriasis. Such a network can be used to identify the major genetic components of the disease. Building such a network, nevertheless, requires a comprehensive profile of the total transcriptome, including mRNA genes, miRNAs, and other sncRNA species. A good collection of such transcriptome data can also be used to address the second challenge, i.e., identification and characterization of potential long noncoding RNAs (lncRNAs) in psoriasis. Although little results on lncRNA have been reported yet for psoriasis, it is possible to hypothesize the involvement of novel genetic elements, particularly lncRNAs, since extensive research has not brought us close to the genetic culprits of the disease. The third challenge is to integrate the information of genotypic variations, e.g., single nucleotide polymorphisms and copy number variations, with the data of transcriptome variations in hunting for causal genetic variations that ultimately lead to disease phenotypes. The results from biological assays analyzing individual genes and genome-wide profiling have shown that both techniques are effective and complementary to each other. It will be more effective, although challenging, to integrate the results from genome-wide profiling with experimental functional analyses of key elements. For example, the current results have pointed to a few miRNAs, e.g., miR-203, miR-205, and miR-125, to be critical players in skin development and psoriasis. Their regulatory functions need to be further scrutinized in animal models or cell lines with, e.g., loss- and/or gain-of-function mutants of these miRNAs. The recent progress on transgenic technologies, such as the latest TALEN (105) and CRISPR/Cas (18, 52, 101) techniques, can be adopted to meet the challenge of such functional analysis. They can be used to manipulate individual genes for their functions in psoriasis and also to develop animal models for psoriasis research.
Finally, we are optimistic with the prospects of this field. Because of the easy accessibility of the skin and the collective effort of the research community, we anticipate that skin-related diseases will be among the first to be treated with small RNA-based therapies.
GRANTS
The research was supported by National Institutes of Health Grants R01GM-100364 and RC1AR-058681, National Science Foundation Grant DBI-0743797, and an internal grant of Jianghan University, Wuhan, Hubei, China.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.X. performed experiments; J.X. and W.Z. analyzed data; J.X. and W.Z. interpreted results of experiments; J.X. prepared figures; J.X. and W.Z. drafted manuscript; J.X. and W.Z. edited and revised manuscript; W.Z. conception and design of research; W.Z. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
Thanks to Kevin Zhang for a critical reading of the manuscript.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 16: 1041–1049, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 130: 1249–1257, 2010. [DOI] [PubMed] [Google Scholar]
- 3.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA 105: 7964–7969, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev 22: 2773–2785, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 455: 64–71, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell 28: 328–336, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhalerao J, Bowcock AM. The genetics of psoriasis: a complex disorder of the skin and immune system. Hum Mol Genet 7: 1537–1545, 1998. [DOI] [PubMed] [Google Scholar]
- 9.Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A. Absolute quantification of microRNAs by using a universal reference. RNA 15: 2375–2384, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 39: 675–86, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol 130: 2062–2070, 2010. [DOI] [PubMed] [Google Scholar]
- 12.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castellano L, Stebbing J. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res 41: 3339–3351, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAsμ: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christoffersen NR, Silahtaroglu A, Ørom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA 13: 1172–1178, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel a J, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danovi D, Meulmeester E, Pasini D, Capra M, Frenk R, de Graaf P, Gasparini P, Gobbi A, Helin K, Pelicci PG, Jochemsen AG, Migliorini D, Francoz S. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor. Mol Cell Biol 24: 5835–5843, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res 37: 2461–2470, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528, 2008. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA 16: 1881–1888, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman R, Farh K, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs E. Scratching the surface of skin development. Nature 445: 834–842, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gracias D, Katsikis P. MicroRNAs: key components of immune regulation. In: Crossroads between Innate and Adaptive Immunity III, edited by Pulendran B, Katsikis PD, Schoenberger SP. New York: Springer, p. 15–26. [Google Scholar]
- 28.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601, 2008. [DOI] [PubMed] [Google Scholar]
- 29.Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, Sluijter JPG, Hoefer I, Pasterkamp G, Bode C, Moser M. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 123: 999–1009, 2011. [DOI] [PubMed] [Google Scholar]
- 30.Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR, Elder JT. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol 130: 1829–1840, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L, Yang Q, Lu J, Li H, Ge Q, Gu W, Bai Y, Lu Z. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PLoS One 6: e21072, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassel D, Cheng P, White MP, Ivey KN, Kroll J, Augustin HG, Katus HA, Stainier DYR, Srivastava D. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ Res 111: 1421–1433, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphreys DT, Hynes CJ, Patel HR, Wei GH, Cannon L, Fatkin D, Suter CM, Clancy JL, Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One 7: e30933, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwu WL, Yang CF, Fann CSJ, Chen CL, Tsai TF, Chien YH, Chiang SC, Chen CH, Hung SI, Wu JY, Chen YT. Mapping of psoriasis to 17q terminus. J Med Genet 42: 152–158, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson B, Tilli CM, Hardman MJ, Avilion AA, MacLeod MC, Ashcroft GS, Byrne C. Late cornified envelope family in differentiating epithelia–response to calcium and ultraviolet irradiation. J Invest Dermatol 124: 1062–1070, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y, Dragas D, Dai Y, Marucha PT, Zhou X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS One 8: e64434, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, Zhang W, Bowcock AM. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 20: 4025–4040, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joyce CE. Expression and Function of Small Noncoding RNAs in Psoriatic Skin (thesis). St. Louis, MO: Washington University in St. Louis, 2012, p. 227. [Google Scholar]
- 40.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RHA. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Meth 3: 27–29, 2006. [DOI] [PubMed] [Google Scholar]
- 41.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell 155: 27–38, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer MF. Stem-loop RT-qPCR for miRNAs. In: Current Protocols in Molecular Biology. John Wiley & Sons, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krueger JG, Bowcock A. Psoriasis pathophysiology: current concepts of pathogenesis. Ann Rheum Dis 64: ii30–ii36, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858, 2001. [DOI] [PubMed] [Google Scholar]
- 46.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen MT, Hother C, Häger M, Pedersen CC, Theilgaard-Mönch K, Borregaard N, Cowland JB. MicroRNA profiling in human neutrophils during bone marrow granulopoiesis and in vivo exudation. PLoS One 8: e58454, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses “stemness” by repressing DeltaNp63. Cell Death Differ 15: 1187–1195, 2008. [DOI] [PubMed] [Google Scholar]
- 49.Lerman G, Avivi C, Mardoukh C, Barzilai A, Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S, Orenstein A, Hornstein E, Sidi Y, Avni D. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS One 6: e20916, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20, 2005. [DOI] [PubMed] [Google Scholar]
- 51.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell 18: 510–525, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 339: 823–826, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9: 387–402, 2008. [DOI] [PubMed] [Google Scholar]
- 54.Martin P. Wound healing–aiming for perfect skin regeneration. Science 276: 75–81, 1997. [DOI] [PubMed] [Google Scholar]
- 55.Mee JB, Johnson CM, Morar N, Burslem F, Groves RW. The psoriatic transcriptome closely resembles that induced by interleukin-1 in cultured keratinocytes: dominance of innate immune responses in psoriasis. Am J Pathol 171: 32–42, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meisgen F, Xu N, Wei T, Janson PC, Obad S, Broom O, Nagy N, Kauppinen S, Kemény L, Ståhle M, Pivarcsi A, Sonkoly E. MiR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Exp Dermatol 21: 312–314, 2012. [DOI] [PubMed] [Google Scholar]
- 57.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics 284: 95–103, 2010. [DOI] [PubMed] [Google Scholar]
- 58.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra M. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mueller D, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol 129: 1740–1751, 2009. [DOI] [PubMed] [Google Scholar]
- 61.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, Schreiber S, Weichenthal M, Gladman D, Rahman P, Schrodi SJ, Prahalad S, Guthery SL, Fischer J, Liao W, Kwok PY, Menter A, Lathrop GM, Wise a C, Begovich AB, Voorhees JJ, Elder JT, Krueger GG, Bowcock AM, Abecasis GR. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet 41: 199–204, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neilsen CT, Goodall GJ, Bracken CP. IsomiRs the overlooked repertoire in the dynamic microRNAome. Trends Genet 28: 544–549, 2012. [DOI] [PubMed] [Google Scholar]
- 63.Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15: 581–590, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol 84: 10344–10353, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reich K, Hüffmeier U, König IR, Lascorz J, Lohmann J, Wendler J, Traupe H, Mössner R, Reis A, Burkhardt H. TNF polymorphisms in psoriasis: association of psoriatic arthritis with the promoter polymorphism TNF*-857 independent of the PSORS1 risk allele. Arthritis Rheum 56: 2056–2064, 2007. [DOI] [PubMed] [Google Scholar]
- 70.Roberson EDO, Bowcock AM. Psoriasis genetics: breaking the barrier. Trends Genet 26: 415–423, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rongloetti F, Fiorucci C, Parodi A. Psoriasis induced or aggravated by drugs. J Rheumatol 83: 59–61, 2009. [DOI] [PubMed] [Google Scholar]
- 72.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207, 2006. [DOI] [PubMed] [Google Scholar]
- 73.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, Kapp A, Gutzmer R. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp Dermatol 21: 509–514, 2012. [DOI] [PubMed] [Google Scholar]
- 75.Schneider MR. MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol 166: 22–28, 2012. [DOI] [PubMed] [Google Scholar]
- 76.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18: 549–557, 2008. [DOI] [PubMed] [Google Scholar]
- 77.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA 106: 1814–1819, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536, 2007. [DOI] [PubMed] [Google Scholar]
- 79.Slominski A, Zbytek B, Nikolakis G, Manna PR, Skobowiat C, Zmijewski M, Li W, Janjetovic Z, Postlethwaite A, Zouboulis CC, Tuckey RC. Steroidogenesis in the skin: implications for local immune functions. J Steroid Biochem Mol Biol 137: 107–123, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slominski AT, Zmijewski MA, Skobowiat C, Zbytek B, Slominski RM, Steketee JD. Sensing the environment: regulation of local and global homeostasis by the skin's neuroendocrine system. Adv Anat Embryol Cell Biol 212: 1–115, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slominski AT, Zmijewski MA, Zbytek B, Tobin DJ, Theoharides TC, Rivier J. Key role of CRF in the skin stress response system. Endocr Rev 34: 827–884, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonkoly E, Lovén J, Xu N, Meisgen F, Wei T, Brodin P, Jaks V, Kasper M, Shimokawa T, Harada M, Heilborn J, Hedblad M, Hippe a Grandér D, Homey B, Zaphiropoulos PG, Arsenian-Henriksson M, Ståhle M, Pivarcsi A. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis 1: e3, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sonkoly E, Wei T, Janson PCJ, Sääf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Ståhle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2: e610, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev 22: 8–13, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao Y, Sun N, Xu X, Shao K, He J. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med Oncol 30: 1–9, 2013. [DOI] [PubMed] [Google Scholar]
- 86.Sun Y, Varambally S, Maher CA, Cao Q, Chockley P, Toubai T, Malter C, Nieves E, Tawara I, Wang Y, Ward PA, Chinnaiyan A, Reddy P. Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood 117: 6172–6183, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swindell WR, Johnston A, Carbajal S, Han G, Wohn C, Lu J, Xing X, Nair RP, Voorhees JJ, Elder JT, Wang XJ, Sano S, Prens EP, DiGiovanni J, Pittelkow MR, Ward NL, Gudjonsson JE. Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis. PLoS One 6: e18266, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Terzian T, Torchia EC, Dai D, Robinson SE, Murao K, Stiegmann RA, Gonzalez V, Boyle GM, Powell MB, Pamela M, Lozano G, Robinson WA, Roop DR, Box NF. p53 prevents progression of nevi to melanoma predominantly through cell cycle regulation. Pigment Cell Melanoma Res 23: 781–794, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, Zamboni G, Maciejewski R. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 12: 369, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang D, Zhang Z, O'Loughlin E, Wang L, Fan X, Lai EC, Yi R. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol 15: 1153–1163, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S, Olson EN. AngiomiRs—key regulators of angiogenesis. Curr Opin Genet Dev 19: 205–211, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543, 2008. [DOI] [PubMed] [Google Scholar]
- 94.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311, 2005. [DOI] [PubMed] [Google Scholar]
- 95.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21: 1450–1461, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xia J, Joyce CE, Bowcock AM, Zhang W. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Hum Mol Genet 22: 737–748, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia J, Zhang W. Noncanonical microRNAs and endogenous siRNAs in lytic infection of murine gammaherpesvirus. PLoS One 7: e47863, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia J, Zhang W. A meta-analysis revealed insights into the sources, conservation and impact of microRNA 5′-isoforms in four model species. Nucleic Acids Res [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu N, Meisgen F, Butler LM, Han G, Wang X, Söderberg-Nauclér C, Ståhle M, Pivarcsi A, Sonkoly E. MicroRNA-31 is overexpressed in psoriasis and modulates inflammatory cytokine and chemokine production in keratinocytes via targeting serine/threonine kinase 40. J Immunol 190: 678–688, 2013. [DOI] [PubMed] [Google Scholar]
- 100.Yamane K, Jinnin M, Etoh T, Kobayashi Y, Shimozono N, Fukushima S, Masuguchi S, Maruo K, Inoue Y, Ishihara T, Aoi J, Oike Y, Ihn H. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J Mol Med 91: 69–81, 2013. [DOI] [PubMed] [Google Scholar]
- 101.Yang H, Wang H, Shivalila C, Cheng A, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yi R, O′Carroll D, Pasolli a H, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362, 2006. [DOI] [PubMed] [Google Scholar]
- 103.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA 106: 498–502, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing “stemness”. Nature 452: 225–229, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotech 29: 149–153, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang L, Stokes N, Polak L, Fuchs E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 8: 294–308, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou X, Krueger JG, Kao MJ, Lee E, Du F, Menter A, Wong WH, Bowcock AM. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics 13: 69–78, 2003. [DOI] [PubMed] [Google Scholar]
- 108.Zibert JR, Løvendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci 58: 177–185, 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.