Abstract
The granule-exocytosis pathway is the major mechanism via which cytotoxic lymphocytes eliminate virus-infected and tumor cells. In this pathway, cytotoxic lymphocytes release granules containing the pore-forming protein perforin and a family of serine proteases known as granzymes into the immunological synapse. Pore-formation by perforin facilitates entry of granzymes into the target cell, where they can activate various (death) pathways. Humans express five different granzymes, of which granzymes A and B have been most extensively characterized. However, much less is known about granzyme M (GrM). Recently, structural analysis and advanced proteomics approaches have determined the primary and extended specificity of GrM. GrM functions have expanded over the past few years: not only can GrM efficiently induce cell death in tumor cells, it can also inhibit cytomegalovirus replication in a noncytotoxic manner. Finally, a role for GrM in lipopolysaccharide-induced inflammatory responses has been proposed. In this review, we recapitulate the current status of GrM expression, substrate specificity, functions, and inhibitors.
Keywords: granzyme M, serine protease, apoptosis, inflammation, CMV, cytotoxic lymphocytes
Facts
Granzyme M is a serine protease expressed in multiple cytotoxic lymphocyte subsets.
Granzyme M prefers to cleave after a Met or a Leu residue and proteolyzes a restricted set of macromolecular substrates.
Human and mouse Granzyme M display species-specific substrate specificities.
Granzyme M kills human tumor cells, inhibits cytomegalovirus replication, and plays a role in LPS-induced inflammation.
Open Questions
What is the physiological relevance of granzyme M in humans?
What is the relative contribution of each substrate to the immunoregulatory functions of granzyme M?
How does granzyme M enhance LPS-induced inflammation?
To counteract virus-infected cells and tumor tissue, the immune system utilizes a variety of specialized cytotoxic lymphocytes, including natural killer (NK) cells and cytotoxic T lymphocytes. These cells constitute the effector arm of the cellular immune response and can exert their protective function by either killing potentially harmful cells or by crippling viral replication within the host cell. Upon recognition of a target cell, effector cells engage with the target cell and form an immunological synapse. Target cells are executed via two mechanisms: (1) by ligation of death receptor ligands on the surface of the effector cell with death receptors on the membrane of the target cell,1 or (2) by secretion of cytolytic granules from the effector cells into the immunological synapse.2, 3, 4 The latter mechanism is generally believed to be the principal mechanism of killing. The released cytolytic granules contain a family of structurally homologous serine proteases known as granzymes, and the pore-forming protein perforin. Perforin enables granzyme entry into the target cell,5 allowing granzymes to cleave intracellular substrates to induce cell death.2, 3
There are five human granzymes: granzyme A (GrA), GrB, GrH, GrK, and GrM. This non-sequential nomenclature in humans results from the more extensive granzyme family in mice, which express GrA–GrG and GrK–GrN.6 The five human granzymes share an amino-acid sequence homology of ∼40%,7 and are structurally related to the trypsin family of serine proteases. They all share the same catalytic triad, consisting of a His, Ser, and Asp residue. GrA and GrB are most extensively studied, but less is known about the other granzymes, which are often referred to as ‘orphan' granzymes.8 In this review, we will summarize and discuss the current insights into the substrate specificity, structure, cellular expression, functions, and inhibitors of GrM.9
Gene Characteristics
GrM (also known as Met-ase or Met-1) was first identified in a rat NK leukemia cell line as a granular protease that displayed Met-ase activity, that is, hydrolyzed peptide substrates after a methionine or leucine residue.10 Soon after this initial discovery, the human11 and mouse12 orthologs of GrM were cloned. Interestingly, the genes encoding human GrM (hGrM) and mouse GrM (mGrM) are situated on human chromosome 19p13.313, 14 and a syntenic region of mouse chromosome 10C,14, 15 respectively, which closely localize to a cluster of neutrophil elastase-like proteases.16 Moreover, the organization of the hGrM gene – which is approximately 7.5 kb long and consists of four introns and five exons – also resembles that of neutrophil elastase-like genes. Together, these findings suggest that GrM and the neutrophil elastase-like proteases may have arisen by duplication and divergence of a common ancestor gene.17
Protein Synthesis and Structure
All granzymes are synthesized as inactive precursors, or pre-pro-proteases. The pre-sequence, also known as the leader sequence, is cleaved off by signal peptidases at the endoplasmic reticulum. Removal of the propeptide sequence is subsequently catalyzed inside the secretory granules by the lysosomal cathepsin C (also known as dipeptidyl peptidase I).18 Whereas most granzymes contain a propeptide of two amino acids, the propeptide of GrM is not a dipeptide, but instead consists of six amino acids.11, 19, 20 Removal of the pre-pro-peptide reveals the N-terminal tail of GrM, containing the highly conserved Ile-Ile-Gly-Gly motif,18, 21, 22 and leads to a conformational change of the granzyme resulting in exposure of the active site. Although at this point GrM is fully processed (232 amino acids, ∼28 kDa), it is not yet enzymatically active, since granzymes are stored in granules at low pH.23 Mature GrM is maximally active at a pH between 7.5–9.24 Granzymes are highly basic (with an iso-electric point above 10) and bind to proteoglycans within the secretory granules.25
The crystal structures of active GrM and an inactive GrM-mutant complexed with a synthetic substrate have recently been solved at <2.7 Å by the group of Fan,24 and are accessible at the Protein Data Bank (http://www.rscb.org) under accession codes 2ZGC and 2ZGJ, respectively. The non-conserved L3 loop (residues 199–212), which is in close proximity to the catalytic triad, is most likely an important determinant of GrM substrate specificity, since this loop changes in conformation upon binding of a substrate.24 Although granzymes generally have three conserved disulfide bridges, GrM has four. In addition, several potential N-glycosylation sites have been identified: Asn21, Asn152, Asn158,26 and other possible N- and O-glycosylation sites,27 phosphorylation sites, and myristoylation sites7 have been proposed.
Substrate Specificity
In protease biology, nomenclature proposed by Schechter and Berger is used to define the position of amino acids in the substrate and protease.28 Amino acids surrounding the scissile bond are denoted as Pn-P2-P1↓P1'-P2'-Pn', with amine bond hydrolysis occurring after P1, and the corresponding binding sites in the protease are numbered Sn-S2-S1-S1'-S2'-Sn'. Although all granzymes share the His-Asp-Ser catalytic triad, their substrate specificities only partially overlap. GrM was initially thought to have a unique primary (P1) specificity for which it was first named Met-ase or Met-1: it was found to preferentially cleave substrates after a Met residue.11, 12, 20 Later, using a positional scanning synthetic combinatorial library of tetrapeptides, hGrM was found to indeed hydrolyze substrates after a Met, but its preferred P1 residue is actually Leu, another long hydrophobic amino acid29 (Figure 1). This is in agreement with the crystal structure of hGrM, which shows that the S1 pocket (Pro177-Cys178-Gly180-Ser182-Ser199) can only accommodate long, narrow hydrophobic amino acids due to its size and its hydrophobic properties.24
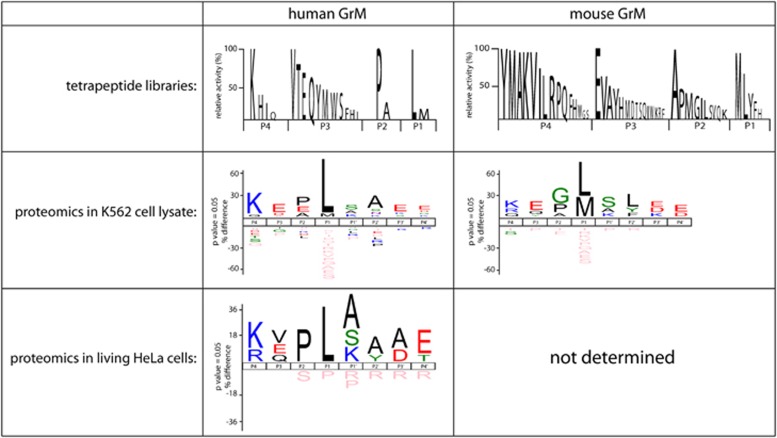
Figure 1.
The primary and extended specificities of hGrM and mGrM. A positional scanning synthetic combinatorial library of tetrapeptides has been used to determine the P4–P1 specificity of hGrM30 and mGrM.31 In addition, complementary positional proteomics have been used to determine the consensus cleavage sites of human and mGrM in K562 tumor cell lysates.31 Finally, complementary positional proteomics have been used on living HeLa tumor cells treated with the pore-forming perforin-analogue SLO and hGrM32
The P4–P1 specificity of hGrM has been further defined using a similar approach, which reveals that hGrM has a strong preference for a Lys at the P4, no clear preference for the P3, and a preference for a small amino acid such as a Pro or Ala at the P2 position24, 30 (Figure 1). These data suggest that the optimal P4–P1 tetrapeptide specificity of hGrM would be Lys-Val-Pro-Leu (KVPL).30 This tetrapeptide is highly specific for hGrM, as it cannot be hydrolyzed by other granzymes, neutrophil elastase, cathepsin G, and chymotrypsin.30 To determine the consensus cleavage site of hGrM in macromolecular substrates, we have used combined fractional diagonal chromatography (COFRADIC)-based complementary positional proteomics in human K-562 tumor cell lysates treated with recombinant hGrM31 (Figure 1). These data largely confirm the results obtained with tetrapeptide libraries: hGrM clearly prefers a Leu over a Met at the P1, a Pro or Ala at the P2, and a Lys at the P4 position.31 In addition, for the first time, these data confirm the contribution of prime residues to GrM substrate recognition.31 To determine GrM substrate specificity in a more physiological setting, we have recently used COFRADIC-based N-terminal positional proteomics coupled with a triple L-Arg SILAC (stable isotope labeling with amino acids in cell culture) labeling strategy to determine GrM substrates in living human HeLa tumor cells.32 Only 25 substrates with a Met or a Leu at the P1 are identified, and the consensus cleavage site in these substrates displays an almost perfect correlation with the previously identified substrate specificities of GrM (Figure 1). Together, these data confirm that hGrM is a highly specific serine protease with a specificity that extends beyond its preferences for P4–P1.
The substrate specificities of hGrM and mGrM have been shown to only partially overlap.31 While these two orthologs share a sequence homology of ∼69%,12 their primary and extended specificities are distinct. In combinatorial positional scanning tetrapeptide libraries, mGrM prefers a Met over a Leu at the P1, in comparison to the strong Leu-preference of hGrM. In addition, mGrM displays a broader tolerance for various amino acids at the P2 and P4 positions.31 The macromolecular substrate specificities of both orthologs are also distinct: using a fluorescence 2-dimensional digestion gel electrophoresis (2D-DIGE) proteomic approach, hGrM and mGrM specificities have been compared in lysates of human HeLa and mouse C2C12 tumor cells. The macromolecular substrate specificities of hGrM and mGrM overlap for ∼42% in the human tumor cell lysate, and for ∼34% in the mouse tumor cell lysate.31 A similar, partial overlap in hGrM and mGrM macromolecular specificities has been found using COFRADIC-based complementary positional proteomics, revealing that the orthologs share ∼67% of the cleavage events in human K562 tumor lysate.31 The species-specific differences between hGrM and mGrM extend beyond the granzymes' substrate preferences: the substrates themselves can also differ between species. This is for instance the case for known GrM substrate nucleophosmin (NPM).33 Whereas human NPM is efficiently cleaved by both hGrM and mGrM, mouse NPM cannot be cleaved by either ortholog.31 Replacement of the putative P1'–P2' residues in mouse NPM with the corresponding residues of human NPM restores cleavage by both granzymes.31 This further underlines the importance of the prime residues for GrM-mediated substrate recognition. For hGrM substrate Fas-associated protein with death domain (FADD),34 a similar species-specific difference in the cleavage site has been reported.35 Replacement of the putative P1 and P1' residues in mouse FADD with the corresponding amino acids of human FADD restores cleavage of mouse FADD by both human and mGrM.35 Collectively, these data show that hGrM and mGrM display narrow macromolecular substrate specificities that only partially overlap, and that caution is essential when extrapolating data from mouse models.
Cellular Expression
Granzymes make up ∼90% of the total amount of granule proteins in cytotoxic T lymphocytes and NK cells.13 Initially, expression of GrM was believed to be restricted to NK cells.10, 11, 12 Based on northern blots for hGrM, high mRNA expression has been identified in two NK leukemia cell lines as well as in purified CD3−CD56+ large granular NK cells. In contrast, very low levels to no expression of hGrM mRNA is detected in other leukemia/lymphoma cell lines – including Jurkat, Daudi and K-562 cells – purified CD3+CD56− peripheral blood mononuclear T cells and CD3−CD56+ small high density NK cells.11, 36 Further evidence for NK-cell specific GrM expression is based on a reporter construct consisting of the rat or mouse 5′ flanking region of GrM coupled to a chloramphenicol acetyltransferase reporter gene. Transfection of this construct into a number of mouse and rat cell lines allows transcription of chloramphenicol acetyltransferase in NK but not in T cell lines.12, 17 Another means that has been used to determine the cellular expression of hGrM is based on the proteolytic Met-ase activity of GrM. Although sorted human NK cells/NK cell lines have high Met-ase activity,11, 17 no activity is detected in a variety of leukemia/lymphoma cell lines.11 Using western blot and immunohistochemistry on FACS-purified cell populations, Sayers et al. have also found that hGrM expression is restricted to CD3−CD56+ NK cells, CD3+CD56+ NKT cells, and γδ T lymphocytes, and absent in CD4+ and CD8+ T cells, neutrophils and monocytes.37 This highly specific expression of hGrM by NK, NKT and γδ T cells suggests a role for GrM in innate immunity.
In recent years, however, GrM expression has also been detected in cells of the adaptive immune system. Murine lymphokine-activated killer (LAK) cells and murine cytotoxic T lymphocyte clones, for instance, contain similar levels of mGrM mRNA, and both display Met-ase activity.38 Bade et al. have characterized hGrM protein levels in human peripheral blood mononuclear cells using flow cytometry, and show hGrM expression in NK cells (both CD16+CD56+ and CD3−CD56bright), NKT cells, γδ T cells and in ∼35% of the CD3+CD8+ T-cell population.39 In contrast, no hGrM expression is detected in B-cells, and only a small subset of the T helper cells express hGrM.39 These data are confirmed by de Koning et al., who – by means of real-time quantitative PCR and flow cytometry – also demonstrate high expression of hGrM in human NK cells, NKT cells, γδ T cells, and in differentiated effector CD8+ CD27− CD45RO− T cells.40 hGrM expression correlates with the expression of perforin, and hGrM levels gradually increase during T-cell differentiation from naïve T cells, via effector-memory cells, to effector cells.40 Interestingly, elevated hGrM levels have recently also been detected in human cytomegalovirus (HCMV)-specific CD8+ T lymphocytes as compared with naïve CD8+ T cells.41 These findings show that hGrM is expressed by effector cells of both innate and adaptive immunity.
Transcription of certain granzymes, such as GrA, GrB, and GrK, can be upregulated in response to a variety of stimuli, such as interleukin (IL)-2, IL-12, IL-15, or interferon (IFN)α.42 None of these factors, however, have an effect on the hGrM mRNA levels in the human NK92 NK cell line, suggesting that hGrM transcription is regulated differently.42 This has later been confirmed at the protein level using flow cytometry: hGrM expression is not upregulated in response to IL-2, IL-6, IL-7, IL-12, IL-15, IL-21, IL-27, IFNα, or lipopolysaccharide (LPS).40
Functions
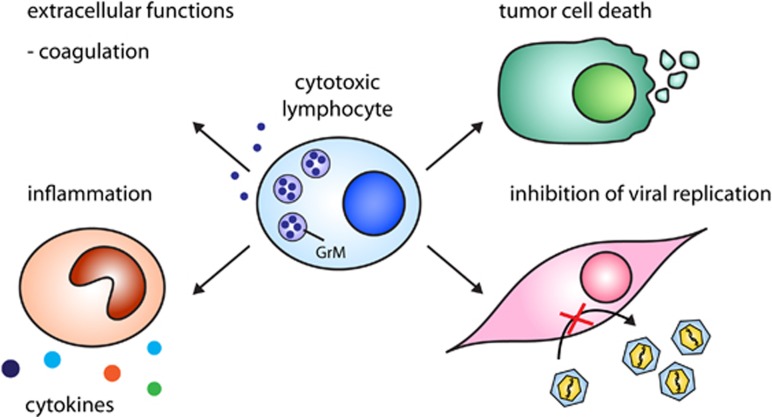
Over the years, the role of hGrM has been studied in a variety of processes, ranging from cell death induction to inflammation (Figure 2). Although its mechanism of action and the relevance of its substrates (Table 1) are not fully understood, it has become increasingly clear that hGrM is a key player in immune processes (Figure 3).
Figure 2.
Immunoregulatory functions of GrM. GrM released by cytotoxic lymphocytes (or other immune cells) functions in various processes: induction of cell death in tumor cells, inhibition of viral replication by cleavage of essential virus and host proteins, regulation of LPS-induced inflammation, and regulation of coagulation
Table 1. Validated GrM substrates.
| Substrate | Evidence | GrM cleavage site | Result of cleavage | Proposed role | Reference | Note |
|---|---|---|---|---|---|---|
| α-Tubulin | Cells, lysate, and purified protein | HFPL269↓ATYA | Increases tubulin polymerization, disturbing the cytoskeleton | Cell death | 47 | Cleavage and cleavage site confirmed by proteomics screen31 |
| Ezrin | Cells, lysate, and purified protein | N.D., HLVM467↓TAPP based on proteomics screen31 | Unknown | Unknown | 47 | Cleavage confirmed by proteomics screen31 |
| FADD | Cells, lysate, and purified protein | MSPM196↓SWNS | Self-association resulting in caspase-8 activation | Cell death | 34, 35 | |
| hnRNP K | Cells, lysate, and purified protein | Multiple, including Leu125, Leu133, and Met359 | Dissection of the functional hnRNP K domains, likely resulting in a loss of hnRNP K function | Inhibition of HCMV replication and cell death | 41, 49 | Cleavage confirmed by proteomics screen31 |
| Hsp75/TRAP | Cells, lysate, and purified protein | N.D. | Abolishes its antagonistic function to reactive oxygen species | Cell death | 45 | |
| ICAD/DFF45 | Cells, lysate, and purified protein | AWIS107↓QESF | Degrades ICAD to activate caspase-activated DNase leading to DNA laddering | Cell death | 46 | |
| NPM | Cells, lysate, and purified protein | KVKL158↓AADE | Abolishes NPM function | Cell death | 33 | Cleavage confirmed by proteomics screen31 |
| PAK2 | Cells and lysate | N.D., KDPL31↓SANH based on proteomics screen31 | Unknown | Unknown | 33 | Cleavage confirmed by proteomics screen31 |
| PARP | Cells and purified protein | N.D. | Unknown | Unknown | 46 | |
| pp71 | CMV-infected cells, CMV lysate, and purified protein | PTPL439↓SEAM | Completely abolished pp71 transactivation of the HCMV major immediate-early promoter | Inhibition of HCMV replication | 72 | |
| survivin | Cells, lysate, and purified protein | IEQL138↓AAMD | Triggers XIAP degradation, resulting in decreased caspase inhibition | Cell death | 44 | |
| topoIIα | Cells, lysate, and purified protein | QTTL1280↓AFKP | Removes the NLS from the catalytic domains of topoIIα, resulting in its nuclear exit | Cell death, cell cycle arrest | 32 | Cleavage confirmed by proteomics screen31 |
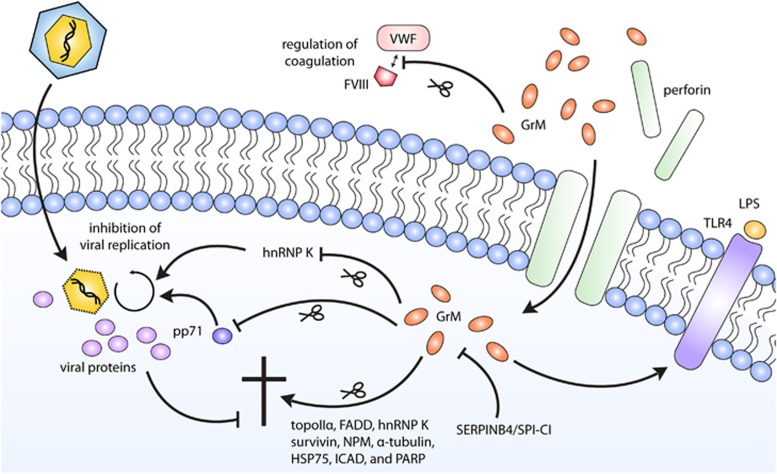
Figure 3.
The functions of GrM. After entry into a target cell via perforin-mediated pore formation in the target cell membrane, GrM can activate various signaling pathways. GrM can induce apoptosis via cleavage of a number of death substrates, among which topoIIα,32 FADD,97 hnRNP K,49 survivin,44 NPM,33 α-tubulin,47 Hsp75,45 ICAD, and PARP.46 GrM can also function in a noncytotoxic manner to inhibit HCMV replication by either cleaving the host protein hnRNP K41 or the viral protein pp71.72 In addition, GrM can stimulate LPS/TLR-4-mediated signaling via yet to be identified mechanisms.90 GrM intracellular functioning can be inhibited in tumor cells by human SERPINB492 and mouse SPI-CI.38 Next to these intracellular effects, GrM has also been found to cleave VWF, and may in that way regulate FVIII levels and coagulation91
Induction of Target Cell Death
Granzymes are best known for their pro-apoptotic role in the cellular immune response against virus-infected and tumor cells. The role of hGrM in cell death induction has therefore been studied most extensively. Even so, much remains unclear and it is difficult to find a consensus between the contradicting findings that have been published. Here, we will briefly summarize the different aspects of hGrM cell death.
The first study describing hGrM-mediated cytotoxicity was published in 2004 and shows efficient tumor cell death induction with low concentrations of hGrM and sublytic concentrations of perforin.43 hGrM-treated cells contain condensed chromatin, large vacuoles and rounded mitochondria, but lack classical apoptosis markers such as phosphatidylserine exposure in the absence of membrane permeabilization, caspase-3 activation, DNA fragmentation, and mitochondrial damage.43 Similar results are found in a later study: hGrM-treated cells die in the absence of caspase activation and phosphatidylserine exposure.33 In contrast, several other studies have shown that hGrM treatment can lead to phosphatidylserine exposure, DNA fragmentation, targeting of the mitochondria, and caspase activation.32, 34, 44, 45, 46 Some classical hallmarks of apoptosis (such as phosphatidylserine exposure, DNA fragmentation, and mitochondrial damage) can be the result of caspase activation, so whether or not caspases are activated in response to hGrM may determine whether or not these apoptotic hallmarks are observed. Differences between studies may be due to the method used to produce recombinant hGrM (in Escherichia coli or Pichia pastoris), the applied hGrM concentrations, or the means used for intracellular delivery of hGrM (purified perforin, Pro-Ject protein transfection reagent, adenovirus, or the pore-forming protein streptolysin O (SLO)).
Using different methods, including proteomic techniques such as gel-based 2D-DIGE47 and MS-based COFRADIC,31, 32 a number of hGrM substrates have been identified that may induce cell death in tumor cells. hGrM cleaves and inactivates the inhibitor of caspase-activated DNase (ICAD) and poly (ADP-ribose) polymerase (PARP).46 Both of these proteins are also caspase substrates during apoptosis.48 In addition, hGrM hydrolyzes cytoskeleton components ezrin and α-tubulin.47 Cleavage of the latter results in increased tubulin polymerization and in the destabilization of the microtubule network.47 The contribution of these substrates to hGrM-mediated cell death, however, remains unclear. A few other hGrM substrates have been studied in more detail, using siRNA-mediated knockdown or overexpression (of uncleavable mutants) to determine their physiological relevance. One such substrate is the chaperone protein TRAP1 (HSP75), cleavage of which leads to increased reactive oxygen species.45 Although RNAi-mediated depletion of TRAP1 does not induce spontaneous cell death, it does sensitize tumor cells to hGrM,45 suggesting that TRAP1 cleavage by hGrM contributes to cell death, but does not directly induce it. The nucleolar phosphoprotein NPM is also efficiently hydrolyzed by hGrM and knockdown of this protein leads to apoptosis.33 Overexpression of an uncleavable NPM mutant, however, fails to protect tumor cells from hGrM-induced cell death.33 Similarly, knockdown of heterogeneous nuclear ribonucleoprotein K (hnRNP K) – the first substrate known to be cleaved by all five human granzymes – sensitizes tumor cells to LAK-cell-mediated cytotoxicity,49 and knockdown of survivin, another hGrM substrate, sensitizes tumor cells to hGrM-induced cell death by lowering the threshold for caspase activation.44 All of these pathways are likely to contribute to hGrM-induced cell death, but none of these appear to be absolutely essential for apoptosis induction. In contrast, hGrM-mediated cleavage of FADD seems to be an important event in cell death induction. FADD-deficient Jurkat cells are significantly less sensitive to hGrM, and cells expressing an uncleavable FADD-mutant are also more resistant to hGrM-induced apoptosis.34 hGrM-mediated cleavage of FADD leads to increased self-association of the protein, resulting in the recruitment and activation of procaspase-8 and subsequent activation of the other members of the caspase cascade.34 Although FADD-deficient cells are less sensitive to hGrM, cell death is not completely impaired, suggesting that other mechanisms are still at play in these cells. Indeed, we have recently identified DNA topology enzyme topoisomerase II alpha (topoIIα) as a highly efficient hGrM substrate.32 Cleavage of topoIIα by hGrM leads to a dissection of the protein's catalytic domains from the C-terminal regulatory domain, resulting in nuclear exit of the functional domains. Knockdown of topoIIα induces G2/M cell cycle arrest and subsequently leads to apoptosis. This pathway is therefore also of importance in hGrM-mediated cytotoxicity. In agreement with the substrate specificity profiles determined for hGrM, most of the identified cleavage sites within these substrates have either a Leu (NPM, survivin, topoIIα, α-tubulin) or a Met (FADD) at the P1. Proteomics approaches have revealed more potential hGrM substrates31, 32, 47 which may also contribute to hGrM-induced cytotoxicity. Thus, hGrM targets multiple intracellular proteins that cooperate in the induction of target cell death.
Another potential interesting pro-apoptotic function of hGrM is its role as a synergistic effector that aids GrB-mediated apoptosis. GrB, which can directly cleave (and activate) both Bid and procaspase-3,50, 51 is a potent inducer of cell death. Tumor cells, however, can evade GrB-mediated cytotoxicity by overexpressing the serine protease inhibitor (serpin) proteinase inhibitor-9 (PI-9), expression of which is associated with poor clinical outcome.52, 53, 54, 55 Upon cleavage by GrB in its reactive center loop (RCL), PI-9 undergoes a conformational change after which it covalently binds GrB, rendering GrB kinetically inactive. Interestingly, hGrM can also cleave PI-9, but cannot be trapped by the serpin.29 Instead, cleavage of PI-9 by hGrM renders the serpin inactive, and hGrM may therefore bypass GrB inhibition.29 A similar synergistic mechanism has been reported for GrB and GrH: GrH can inactivate an adenovirus-encoded GrB inhibitor.56 The physiological relevance of the GrB-hGrM interplay deserves further study, since inhibition of GrB by PI-9 occurs at faster kinetics than hGrM-mediated inactivation of PI-9.29
The physiological relevance of hGrM in tumor clearance in vivo is still unclear. There is convincing evidence, however, that underlines the importance of the granule-exocytosis pathway in tumor rejection. Perforin-deficient mice are severely immunodeficient, and are significantly compromised in their ability to eradicate transplanted and injected tumor cells and viruses.57, 58 In addition, these mice spontaneously develop highly aggressive disseminated B-cell lymphomas,59 implying a role for the granule-exocytosis pathway in immune surveillance of tumors. The importance of the granule-exocytosis pathway in tumor rejection is further supported by humans with mutations in the perforin gene PRF1. These mutations are extremely rare, but recent studies suggest that carriers of PRF1 alleles with temperature-sensitive missense mutations may be more than 100 times more susceptible to the development of hematological cancers.60, 61 On the contrary, knockout mice of single or multiple granzymes rarely display a clear phenotype: knockout of GrA and GrB-F, for instance, has no effect on tumor clearance, and mice lacking these granzymes can still handle most infections.62, 63 Two studies have looked specifically at the role of mGrM in tumor clearance in vivo. In the first study, GrM knockout mice develop normally, and NK cells isolated from these mice display efficient cytotoxicity toward tumor cells ex vivo.64 GrM knockout mice can still successfully clear several mouse lymphoma cell lines, whereas perforin-deficient mice die within 50 days.64 In addition, upon intravenous inoculation of melanoma cells, no effect on the number of metastases can be observed.64 In a later study, adoptively transferred NK cells from GrM knockout mice display impaired tumor cell killing in vivo toward a chemically (MCA)-induced tumor cell line, but not in vitro.65 The absence of clear phenotypes in granzyme knockout mice is likely due to the redundancy of the granzyme system. It is of importance to note that absence of a clear role for mGrM in tumor rejection in mice may not reflect its function in humans: as mentioned previously, mGrM and hGrM display species-specific substrate specificities. mGrM is not cytotoxic toward a panel of mouse tumor cell lines and the GrM death substrates NPM and FADD cannot be cleaved in mice.31, 35 Ideally, GrM function would therefore be studied in a humanized cancer mouse model.
Noncytotoxic Inhibition of Viral Replication
A longstanding dogma in granzyme biology is that granzymes exert their functions by inducing target cell death. Recently, however, evidence is emerging that granzymes also have noncytotoxic functions. This may be particularly true for virus-infected cells, as viruses have evolved multiple mechanisms to block cell death pathways.66 Indeed, GrB can inhibit HSV-1 (herpes simplex virus 1) reactivation from neuronal latency in an apoptosis independent manner,67 GrK can inhibit influenza A viral replication via cleavage of the host cell nuclear transport complex importin α1/β,68 and GrH cleaves host cell protein La, which is important for viral RNA metabolism,69 and the HBx protein which is required for replication of hepatitis B virus.70
Interestingly, GrM knockout mice are more sensitive to murine CMV infection,64 but not to HSV-1 or influenza A infection.71 We have validated that hGrM inhibits HCMV replication in human cells in vitro.72 hGrM can inhibit HCMV replication in a noncytotoxic manner via at least two ways: via cleavage of the viral phosphoprotein pp7172 and via cleavage of the host cell protein hnRNP K.41 hGrM-mediated cleavage of HCMV protein pp71 – which is indispensible for efficient HCMV replication73– disturbs its nuclear localization and abrogates its transcriptional activity toward the HCMV major immediate-early promoter.72 hGrM can also cleave host protein hnRNP K, which is essential for the translation of HCMV immediate early 2 protein and HCMV replication.41 Lending credibility to this antiviral activity of hGrM is the fact that hGrM protein levels are elevated in HCMV-specific CD8+ T cells, supporting an active antiviral role for hGrM in HCMV infection and in the prevention of HCMV reactivation.41 In addition, HCMV reactivation in stem cell transplantation is associated with higher percentages of GrM-expressing CD4+ (total and central-memory) cells.74 Similarly, GrM levels in the plasma of these patients were elevated during HCMV reactivation.74 Interestingly, a number of other known hGrM substrates have been implicated in viral replication: α-tubulin,47 for instance, plays a crucial role in viral intracellular transport,75 the shuttling capacity of NPM can be used by viral proteins to enter the nucleus,76, 77 and topoIIα is thought to be involved in the replication of HIV-1 (human immunodeficiency virus-1),78, 79 HSV-1,80 vaccinia virus,81 and Epstein–Barr virus.82 It is therefore tempting to speculate that hGrM uses multiple pathways to attack virus infections.
Regulation of Inflammation
Although granzymes are best known for their intracellular roles – and mainly for their role in target cell killing – extracellular functions for granzymes have recently been suggested.83, 84 Granzymes have been linked to extracellular matrix degradation,85, 86 immune regulation, and inflammation.87, 88, 89
Using the GrM knockout mouse model, a role for mGrM in inflammation has been established. In comparison to wild-type mice, GrM knockout mice and perforin knockout mice are significantly more resistant to toll-like receptor (TLR)4-dependent LPS-induced endotoxicosis.90 GrM-deficient mice have decreased serum levels of proinflammatory cytokines such as IL-1α, IL-1β, TNF, and IFNγ following LPS challenge. It has been proposed that mGrM likely acts intracellular and downstream of caspase-1 in this LPS-induced model of sepsis. Interestingly, elevated plasma levels of hGrM have been detected in meningococcal sepsis patients,91 pointing to a role of hGrM in the inflammatory response to bacteria.
Other Functions
In addition to its role in inflammation, hGrM may also have an extracellular function in the regulation of blood coagulation: hGrM can cleave the pro-hemostatic multimer plasma protein Von Willebrand factor (VWF) in vitro.91 VWF normally promotes platelet aggregation and stabilizes coagulation factor VIII, but VWF cleaved by hGrM can no longer bind factor VIII in vitro.91 In this manner, hGrM may play a role in the regulation of factor VIII plasma levels and blood coagulation.
Inhibitors
Over the years, several inhibitors of GrM have been identified. The synthetic compounds z-MetP-(OPh-4-Cl)229 and Ac-KVPL-CMK24 specifically inhibit GrM. The CMK group of Ac-KVPL-CMK ensures that a covalent bond is formed between the inhibitor and the catalytic Ser182 and His41 residues, and therefore irreversibly inhibits GrM proteolytic activity.24 Pre-treatment of human Jurkat tumor cells with this inhibitor significantly inhibited hGrM-mediated cell death,24 and pre-treatment of LAK cells prior to co-culture with human HeLa tumor cells led to a significant decrease in LAK-cell-mediated cytotoxicity.34 These inhibitors are valuable tools in elucidating GrM function.
Next to these synthetic inhibitors of GrM, a number of proteinaceous inhibitors have also been identified. One of these is the bacterially expressed serine protease inhibitor ecotin, but this protein is a broad-spectrum serine protease inhibitor, and therefore not specific for GrM.29 Two extracellular human serpins in plasma, SerpinA1 (α1-antitrypsin) and SerpinA3 (α1-antichymotrypsin), inhibit hGrM activity and form SDS-stable complexes with hGrM.29 hGrM – which is undetectable in the serum of healthy controls – has been detected in the serum of sepsis patients.91 Whether or not SerpinA1 and/or SerpinA3 inhibit hGrM activity in vivo remains an open question.
Recently, two intracellular GrM-inhibiting serpins have been identified: the murine serine protease inhibitor involved in cytotoxic inhibition (SPI-CI, also known as R86)38 and human SerpinB4 (also known as squamous cell carcinoma antigen-2 or leupin).92 SPI-CI has been initially identified in mouse cytotoxic T lymphocytes93 and has later been found to inhibit purified rat GrM38 and – to a lesser extent – hGrM.94 SPI-CI forms the classical SDS-stable serpin-protease complex with rat GrM, and is expressed by purified murine CD8+ T cells and even more by murine LAK cells.38 Transfection of SPI-CI prevents both rat GrM and – with reduced efficiency – hGrM-mediated cell death in tumor cells.38, 94 No human ortholog of SPI-CI has been identified yet.
SerpinB4 is the only human intracellular B-clade serpin that contains a Leu residue at the putative P1 position in its RCL, and was therefore predicted to inhibit hGrM.92 Indeed, SerpinB4 directly inhibits hGrM proteolytic activity, and its overexpression in human HeLa tumor cells protects against both hGrM-induced and NK cell-induced cell death.92 Inhibition of hGrM by SerpinB4 occurs with kinetics that resembled SerpinB4-mediated inhibition of CathepsinG and chymase.95 More serpins that target hGrM might exist. Interestingly, SerpinB1, which also inhibits neutrophil elastase and harbors a Cys at the putative P1 and a Met at P1',96 has recently been identified as a human GrH inhibitor,97 but also forms an SDS-stable complex with hGrM.97 It is therefore tempting to speculate that this complex is formed by GrM-mediated hydrolysis of SerpinB1 at the P1' Met. Further investigation is required to determine the physiological relevance of this interaction.
Originally, granzyme-inhibiting serpins were mainly believed to act as a fail-safe mechanism for cytotoxic lymphocytes, to avoid self-injury during granule exocytosis.98 Lately, however, it is thought that tumor cells often misuse these serpins to avoid eradication by the immune system. It has been well established that some tumors evade cytotoxic lymphocyte-mediated killing by blocking the activity of GrB through expression of the intracellular serpin PI-9. Expression of PI-9 by tumors not only correlates with decreased susceptibility to GrB-mediated apoptosis, but also positively associates with unfavorable clinical outcome.52, 53, 54, 55 Expression of the hGrM inhibitor SerpinB4 may therefore constitute an analogous mechanism via which tumor cells could evade cell death. Interestingly, SerpinB4 expression has been found in numerous squamous cell carcinoma's: in the squamous cell carcinomas of the cervix,99 head and neck, and lung.100 Moreover, elevated SerpinB4 mRNA levels correlate with the progression from normal, to dysplastic, to cancerous tissue,101 and are a predictor of head and neck squamous cell carcinoma prognosis.102 Furthermore, SPI-CI is also expressed by mouse colon carcinoma cell lines, suggesting it may serve a protective role in these cells as well.94
Conclusion and Future Perspectives
Over the last five years, the arsenal of GrM functions has expanded enormously. Although it was initially believed to function only in the induction of apoptosis in target cells, evidence is emerging that GrM functions in many other biological processes, among which non-cytotoxic inhibition of viral replication and the regulation of inflammation. With the discovery of new GrM substrates, the mechanisms via which GrM exerts its function will become increasingly clear. Proteomics-based profiling of protease substrates (degradomics) has been proven a valuable tool in this process.103
A common theme in GrM functioning is its substrate redundancy. In the induction of apoptosis, GrM targets multiple death substrates that all contribute to the execution of cell death. Cleavage of FADD and subsequent caspase activation appears to be one of the key pathways via which GrM induces apoptosis.34 In addition, we have recently shown that GrM cleaves topoIIα, depletion of which results in G2/M cell cycle arrest and caspase-dependent apoptosis.32 To inhibit CMV replication, GrM not only targets important viral proteins such as pp71,72 but also essential host cell proteins such as hnRNP K.41 This high level of redundancy has likely evolved in an arms-race of the immune system with viruses and tumor cells, which have also developed multiple strategies to counteract granzyme attacks.
In the coming years, studies should concentrate on the importance of GrM in each of its immunoregulatory functions (Figure 2). Mouse models with reconstituted human immune systems will play an essential role in this. Knockdown of hGrM in these mouse models should reveal the relevance of GrM in various aspects of immune regulation in tumor immunology and viral elimination. In addition, the role of GrM in LPS-induced inflammation requires further attention. Here, important GrM substrates remain to be identified.
Increased knowledge of the immunoregulatory functions of GrM will help in the discovery of novel therapies or the improvement thereof, by tipping the balance in favor of GrM activity.
Acknowledgments
Authors are supported by the Dutch Cancer Society (KWF) (grant number UU-2009–4302 (to NB)) and the Netherlands Organization for Scientific Research (NWO) (grant number 916.66.044 (to NB)).
Glossary
- 2D-DIGE
2-dimensional digestion gel electrophoresis
- CMV
cytomegalovirus
- COFRADIC
combined fractional diagonal chromatography
- FADD
Fas-associated protein with death domain
- Gr
granzyme
- GrM
granzyme M
- HCMV
human CMV
- hGrM
human GrM
- hnRNP K
heterogeneous nuclear ribonucleoprotein K
- HIV
human immunodeficiency virus
- HSV-1
herpes simplex virus 1
- ICAD
inhibitor of caspase-activated DNase
- IFN
interferon
- IL
interleukin
- i.v.
intravenous
- LAK cells
lymphokine-activated killer cells
- LPS
lipopolysaccharide
- MCMV
mouse CMV
- mGrM
mouse GrM
- NK cells
natural killer cells
- NPM
nucleophosmin
- PARP
Poly (ADP-ribose) polymerase
- PBMCs
peripheral blood mononuclear cells
- PI-9
proteinase inhibitor 9
- pp71
phosphoprotein 71
- RCL
reactive center loop
- SILAC
stable isotope labeling with amino acids in cell culture
- SLO
streptolysin O
- SPI-CI
serine protease inhibitor involved in cytotoxicity inhibition
- TLR
toll-like receptor
- topoIIα
topoisomerase II alpha
- VWF
Von Willebrand factor
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Walczak H, Krammer PH. The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp Cell Res. 2000;256:58–66. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- Cullen SP, Brunet M, Martin SJ. Granzymes in cancer and immunity. Cell Death Differ. 2010;17:616–623. doi: 10.1038/cdd.2009.206. [DOI] [PubMed] [Google Scholar]
- Lieberman J. Anatomy of a murder: how cytotoxic T cells and NK cells are activated, develop, and eliminate their targets. Immunol Rev. 2010;235:5–9. doi: 10.1111/j.0105-2896.2010.00914.x. [DOI] [PubMed] [Google Scholar]
- Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- Voskoboinik I, Dunstone MA, Baran K, Whisstock JC, Trapani JA. Perforin: structure, function, and role in human immunopathology. Immunol Rev. 2010;235:35–54. doi: 10.1111/j.0105-2896.2010.00896.x. [DOI] [PubMed] [Google Scholar]
- Grossman WJ, Revell PA, Lu ZH, Johnson H, Bredemeyer AJ, Ley TJ. The orphan granzymes of humans and mice. Curr Opin Immunol. 2003;15:544–552. doi: 10.1016/s0952-7915(03)00099-2. [DOI] [PubMed] [Google Scholar]
- Sattar R, Ali SA, Abbasi A. Bioinformatics of granzymes: sequence comparison and structural studies on granzyme family by homology modeling. Biochem Biophys Res Commun. 2003;308:726–735. doi: 10.1016/s0006-291x(03)01458-x. [DOI] [PubMed] [Google Scholar]
- Bovenschen N, Kummer JA. Orphan granzymes find a home. Immunol Rev. 2010;235:117–127. doi: 10.1111/j.0105-2896.2010.00889.x. [DOI] [PubMed] [Google Scholar]
- de Koning PJ, Kummer JA, Bovenschen N. Biology of granzyme M: a serine protease with unique features. Crit Rev Immunol. 2009;29:307–315. doi: 10.1615/critrevimmunol.v29.i4.20. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Wiltrout T, Trapani JA, Ottaway KS, Sowder R, Henderson LE, et al. Purification and cloning of a novel serine protease, RNK-Met-1, from the granules of a rat natural killer cell leukemia. J Biol Chem. 1992;267:24418–24425. [PubMed] [Google Scholar]
- Smyth MJ, Sayers TJ, Wiltrout T, Powers JC, Trapani JA. Met-ase: cloning and distinct chromosomal location of a serine protease preferentially expressed in human natural killer cells. J Immunol. 1993;151:6195–6205. [PubMed] [Google Scholar]
- Kelly JM, O'Connor MD, Hulett MD, Thia KY, Smyth MJ. Cloning and expression of the recombinant mouse natural killer cell granzyme Met-ase-1. Immunogenetics. 1996;44:340–350. doi: 10.1007/BF02602778. [DOI] [PubMed] [Google Scholar]
- Baker E, Sutherland GR, Smyth MJ. The gene encoding a human natural killer cell granule serine protease, Met-ase 1, maps to chromosome 19p13.3. Immunogenetics. 1994;39:294–295. doi: 10.1007/BF00188796. [DOI] [PubMed] [Google Scholar]
- Pilat D, Fink T, Obermaier-Skrobanek B, Zimmer M, Wekerle H, Lichter P, et al. The human Met-ase gene (GZMM): structure, sequence, and close physical linkage to the serine protease gene cluster on 19p13.3. Genomics. 1994;24:445–450. doi: 10.1006/geno.1994.1651. [DOI] [PubMed] [Google Scholar]
- Thia KY, Jenkins NA, Gilbert DJ, Copeland NG, Smyth MJ. The natural killer cell serine protease gene Lmet1 maps to mouse chromosome 10. Immunogenetics. 1995;41:47–49. doi: 10.1007/BF00188433. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Medcalf RL, Fink TM, Mattmann C, Lichter P, Jenne DE. Three human elastase-like genes coordinately expressed in the myelomonocyte lineage are organized as a single genetic locus on 19pter. Proc Natl Acad Sci. 1992;89:8215–8219. doi: 10.1073/pnas.89.17.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Hulett MD, Thia KY, Young HA, Sayers TJ, Carter CR, et al. Cloning and characterization of a novel NK cell-specific serine protease gene and its functional 5'-flanking sequences. Immunogenetics. 1995;42:101–111. doi: 10.1007/BF00178584. [DOI] [PubMed] [Google Scholar]
- McGuire MJ, Lipsky PE, Thiele DL. Generation of active myeloid and lymphoid granule serine proteases requires processing by the granule thiol protease dipeptidyl peptidase I. J Biol Chem. 1993;268:2458–2467. [PubMed] [Google Scholar]
- Smyth MJ, O'Connor MD, Trapani JA. Granzymes: a variety of serine protease specificities encoded by genetically distinct subfamilies. J Leukoc Biol. 1996;60:555–562. doi: 10.1002/jlb.60.5.555. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, O'Connor MD, Trapani JA, Kershaw MH, Brinkworth RI. A novel substrate-binding pocket interaction restricts the specificity of the human NK cell-specific serine protease, Met-ase-1. J Immunol. 1996;156:4174–4181. [PubMed] [Google Scholar]
- Pham CT, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA. 1999;96:8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer JA, Kamp AM, Citarella F, Horrevoets AJ, Hack CE. Expression of human recombinant granzyme A zymogen and its activation by the cysteine proteinase cathepsin C. J Biol Chem. 1996;271:9281–9286. doi: 10.1074/jbc.271.16.9281. [DOI] [PubMed] [Google Scholar]
- Rukamp BJ, Kam CM, Natarajan S, Bolton BW, Smyth MJ, Kelly JM, et al. Subsite specificities of granzyme M: a study of inhibitors and newly synthesized thiobenzyl ester substrates. Arch Biochem Biophys. 2004;422:9–22. doi: 10.1016/j.abb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Wu L, Wang L, Hua G, Liu K, Yang X, Zhai Y, et al. Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M. J Immunol. 2009;183:421–429. doi: 10.4049/jimmunol.0803088. [DOI] [PubMed] [Google Scholar]
- Galvin JP, Spaeny-Dekking LH, Wang B, Seth P, Hack CE, Froelich CJ. Apoptosis induced by granzyme B-glycosaminoglycan complexes: implications for granule-mediated apoptosis in vivo. J Immunol. 1999;162:5345–5350. [PubMed] [Google Scholar]
- Smyth MJ, O'Connor MD, Kelly JM, Ganesvaran P, Thia KY, Trapani JA. Expression of recombinant human Met-ase-1: a NK cell-specific granzyme. Biochem Biophys Res Commun. 1995;217:675–683. doi: 10.1006/bbrc.1995.2827. [DOI] [PubMed] [Google Scholar]
- Khurshid R, Saleem M, Akhtar MS, Salim A. Granzyme M: characterization with sites of post-translational modification and specific sites of interaction with substrates and inhibitors. Mol Biol Rep. 2011;38:2953–2960. doi: 10.1007/s11033-010-9959-7. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Kisiel W, Craik CS. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J Biol Chem. 2004;279:54275–54282. doi: 10.1074/jbc.M411482200. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Craik CS. Selective chemical functional probes of granzymes A and B reveal granzyme B is a major effector of natural killer cell-mediated lysis of target cells. Chem Biol. 2005;12:567–577. doi: 10.1016/j.chembiol.2005.03.006. [DOI] [PubMed] [Google Scholar]
- de Poot SA, Westgeest M, Hostetter DR, Van Damme P, Plasman K, Demeyer K, et al. Human and mouse granzyme M display divergent and species-specific substrate specificities. Biochem J. 2011;437:431–442. doi: 10.1042/BJ20110210. [DOI] [PubMed] [Google Scholar]
- de Poot SA, Lai KW, van der Wal L, Plasman K, Van Damme P, Porter AC, et al. Granzyme M targets topoisomerase II alpha to trigger cell cycle arrest and caspase-dependent apoptosis Cell Death Differ 2013. e-pub ahead print 1 November 2013doi: 10.1038/cdd.2013.155 [DOI] [PMC free article] [PubMed]
- Cullen SP, Afonina IS, Donadini R, Luthi AU, Medema JP, Bird PI, et al. Nucleophosmin is cleaved and inactivated by the cytotoxic granule protease granzyme M during natural killer cell-mediated killing. J Biol Chem. 2009;284:5137–5147. doi: 10.1074/jbc.M807913200. [DOI] [PubMed] [Google Scholar]
- Wang S, Xia P, Shi L, Fan Z. FADD cleavage by NK cell granzyme M enhances its self-association to facilitate procaspase-8 recruitment for auto-processing leading to caspase cascade. Cell Death Differ. 2012;19:605–615. doi: 10.1038/cdd.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Poot SA, Lai KW, Hovingh ES, Bovenschen N. Granzyme M cannot induce cell death via cleavage of mouse FADD. Apoptosis. 2013;18:533–534. doi: 10.1007/s10495-013-0810-1. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Browne KA, Kinnear BF, Trapani JA, Warren HS. Distinct granzyme expression in human CD3- CD56+ large granular- and CD3- CD56+ small high density-lymphocytes displaying non-MHC-restricted cytolytic activity. J Leukoc Biol. 1995;57:88–93. doi: 10.1002/jlb.57.1.88. [DOI] [PubMed] [Google Scholar]
- Sayers TJ, Brooks AD, Ward JM, Hoshino T, Bere WE, Wiegand GW, et al. The restricted expression of granzyme M in human lymphocytes. J Immunol. 2001;166:765–771. doi: 10.4049/jimmunol.166.2.765. [DOI] [PubMed] [Google Scholar]
- Bots M, Kolfschoten IG, Bres SA, Rademaker MT, de Roo GM, Kruse M, et al. SPI-CI and SPI-6 cooperate in the protection from effector cell-mediated cytotoxicity. Blood. 2005;105:1153–1161. doi: 10.1182/blood-2004-03-0791. [DOI] [PubMed] [Google Scholar]
- Bade B, Boettcher HE, Lohrmann J, Hink-Schauer C, Bratke K, Jenne DE, et al. Differential expression of the granzymes A, K and M and perforin in human peripheral blood lymphocytes. Int Immunol. 2005;17:1419–1428. doi: 10.1093/intimm/dxh320. [DOI] [PubMed] [Google Scholar]
- de Koning PJ, Tesselaar K, Bovenschen N, Colak S, Quadir R, Volman TJ, et al. The cytotoxic protease granzyme M is expressed by lymphocytes of both the innate and adaptive immune system. Mol Immunol. 2010;47:903–911. doi: 10.1016/j.molimm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- van Domselaar R, de Poot SA, Remmerswaal EB, Lai KW, Ten Berge IJ, Bovenschen N. Granzyme M targets host cell hnRNP K that is essential for human cytomegalovirus replication. Cell Death Differ. 2013;20:419–429. doi: 10.1038/cdd.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhang J, Tian Z. Comparison in the effects of IL-2, IL-12, IL-15 and IFNalpha on gene regulation of granzymes of human NK cell line NK-92. Int Immunopharmacol. 2008;8:989–996. doi: 10.1016/j.intimp.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Kelly JM, Waterhouse NJ, Cretney E, Browne KA, Ellis S, Trapani JA, et al. Granzyme M mediates a novel form of perforin-dependent cell death. J Biol Chem. 2004;279:22236–22242. doi: 10.1074/jbc.M401670200. [DOI] [PubMed] [Google Scholar]
- Hu D, Liu S, Shi L, Li C, Wu L, Fan Z. Cleavage of survivin by Granzyme M triggers degradation of the survivin-X-linked inhibitor of apoptosis protein (XIAP) complex to free caspase activity leading to cytolysis of target tumor cells. J Biol Chem. 2010;285:18326–18335. doi: 10.1074/jbc.M109.083170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua G, Zhang Q, Fan Z. Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J Biol Chem. 2007;282:20553–20560. doi: 10.1074/jbc.M703196200. [DOI] [PubMed] [Google Scholar]
- Lu H, Hou Q, Zhao T, Zhang H, Zhang Q, Wu L, et al. Granzyme M directly cleaves inhibitor of caspase-activated DNase (CAD) to unleash CAD leading to DNA fragmentation. J Immunol. 2006;177:1171–1178. doi: 10.4049/jimmunol.177.2.1171. [DOI] [PubMed] [Google Scholar]
- Bovenschen N, de Koning PJ, Quadir R, Broekhuizen R, Damen JM, Froelich CJ, et al. NK cell protease granzyme M targets alpha-tubulin and disorganizes the microtubule network. J Immunol. 2008;180:8184–8191. doi: 10.4049/jimmunol.180.12.8184. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- van Domselaar R, Quadir R, van der Made AM, Broekhuizen R, Bovenschen N. All human granzymes target hnRNP K that is essential for tumor cell viability. J Biol Chem. 2012;287:22854–22864. doi: 10.1074/jbc.M112.365692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord SJ, Rajotte RV, Korbutt GS, Bleackley RC. Granzyme B: a natural born killer. Immunol Rev. 2003;193:31–38. doi: 10.1034/j.1600-065x.2003.00044.x. [DOI] [PubMed] [Google Scholar]
- Trapani JA, Sutton VR. Granzyme B: pro-apoptotic, antiviral and antitumor functions. Curr Opin Immunol. 2003;15:533–543. doi: 10.1016/s0952-7915(03)00107-9. [DOI] [PubMed] [Google Scholar]
- Bird CH, Sutton VR, Sun J, Hirst CE, Novak A, Kumar S, et al. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol Cell Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema JP, de Jong J, Peltenburg LT, Verdegaal EM, Gorter A, Bres SA, et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc Natl Acad Sci USA. 2001;98:11515–11520. doi: 10.1073/pnas.201398198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladergroen BA, Meijer CJ, ten Berge RL, Hack CE, Muris JJ, Dukers DF, et al. Expression of the granzyme B inhibitor, protease inhibitor 9, by tumor cells in patients with non-Hodgkin and Hodgkin lymphoma: a novel protective mechanism for tumor cells to circumvent the immune system. Blood. 2002;99:232–237. doi: 10.1182/blood.v99.1.232. [DOI] [PubMed] [Google Scholar]
- van Houdt IS, Oudejans JJ, van den Eertwegh AJ, Baars A, Vos W, Bladergroen BA, et al. Expression of the apoptosis inhibitor protease inhibitor 9 predicts clinical outcome in vaccinated patients with stage III and IV melanoma. Clin Cancer Res. 2005;11:6400–6407. doi: 10.1158/1078-0432.CCR-05-0306. [DOI] [PubMed] [Google Scholar]
- Andrade F, Fellows E, Jenne DE, Rosen A, Young CS. Granzyme H destroys the function of critical adenoviral proteins required for viral DNA replication and granzyme B inhibition. Embo J. 2007;26:2148–2157. doi: 10.1038/sj.emboj.7601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- Lowin B, Beermann F, Schmidt A, Tschopp J. A null mutation in the perforin gene impairs cytolytic T lymphocyte- and natural killer cell-mediated cytotoxicity. Proc Natl Acad Sci USA. 1994;91:11571–11575. doi: 10.1073/pnas.91.24.11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Thia KY, Street SE, MacGregor D, Godfrey DI, Trapani JA. Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. J Exp Med. 2000;192:755–760. doi: 10.1084/jem.192.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J, Yeo KP, Whisstock JC, Dunstone MA, Trapani JA, Voskoboinik I. Temperature sensitivity of human perforin mutants unmasks subtotal loss of cytotoxicity, delayed FHL, and a predisposition to cancer. Proc Natl Acad Sci USA. 2009;106:9809–9814. doi: 10.1073/pnas.0903815106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan AJ, Chia J, Trapani JA, Voskoboinik I. Perforin deficiency and susceptibility to cancer. Cell Death Differ. 2010;17:607–615. doi: 10.1038/cdd.2009.212. [DOI] [PubMed] [Google Scholar]
- Davis JE, Smyth MJ, Trapani JA. Granzyme A and B-deficient killer lymphocytes are defective in eliciting DNA fragmentation but retain potent in vivo anti-tumor capacity. Eur J Immunol. 2001;31:39–47. doi: 10.1002/1521-4141(200101)31:1<39::aid-immu39>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Street SE, Trapani JA. Cutting edge: granzymes A and B are not essential for perforin-mediated tumor rejection. J Immunol. 2003;171:515–518. doi: 10.4049/jimmunol.171.2.515. [DOI] [PubMed] [Google Scholar]
- Pao LI, Sumaria N, Kelly JM, van Dommelen S, Cretney E, Wallace ME, et al. Functional analysis of granzyme M and its role in immunity to infection. J Immunol. 2005;175:3235–3243. doi: 10.4049/jimmunol.175.5.3235. [DOI] [PubMed] [Google Scholar]
- Pegram HJ, Haynes NM, Smyth MJ, Kershaw MH, Darcy PK. Characterizing the anti-tumor function of adoptively transferred NK cells in vivo. Cancer Immunol Immunother. 2010;59:1235–1246. doi: 10.1007/s00262-010-0848-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Domselaar R, Bovenschen N. Cell death-independent functions of granzymes: hit viruses where it hurts. Rev Med Virol. 2011;21:301–314. doi: 10.1002/rmv.697. [DOI] [PubMed] [Google Scholar]
- Knickelbein JE, Khanna KM, Yee MB, Baty CJ, Kinchington PR, Hendricks RL. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science. 2008;322:268–271. doi: 10.1126/science.1164164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Li C, Wang X, Toyoda T, Gao G, Fan Z. Granzyme K inhibits replication of influenza virus through cleaving the nuclear transport complex importin alpha1/beta dimer of infected host cells. Cell Death Differ. 2012;19:882–890. doi: 10.1038/cdd.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero V, Fellows E, Jenne DE, Andrade F. Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death Differ. 2009;16:340–348. doi: 10.1038/cdd.2008.165. [DOI] [PubMed] [Google Scholar]
- Tang H, Li C, Wang L, Zhang H, Fan Z. Granzyme H of cytotoxic lymphocytes is required for clearance of the hepatitis B virus through cleavage of the hepatitis B virus X protein. J Immunol. 2012;188:824–831. doi: 10.4049/jimmunol.1102205. [DOI] [PubMed] [Google Scholar]
- Anthony DA, Andrews DM, Watt SV, Trapani JA, Smyth MJ. Functional dissection of the granzyme family: cell death and inflammation. Immunol Rev. 2010;235:73–92. doi: 10.1111/j.0105-2896.2010.00907.x. [DOI] [PubMed] [Google Scholar]
- van Domselaar R, Philippen LE, Quadir R, Wiertz EJ, Kummer JA, Bovenschen N. Noncytotoxic inhibition of cytomegalovirus replication through NK cell protease granzyme M-mediated cleavage of viral phosphoprotein 71. J Immunol. 2010;185:7605–7613. doi: 10.4049/jimmunol.1001503. [DOI] [PubMed] [Google Scholar]
- Bresnahan WA, Shenk TE. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci USA. 2000;97:14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenschen N, Spijkers SN, Wensink AC, Schellens IM, van Domselaar R, van Baarle D. Elevated granzyme M-expressing lymphocytes during cytomegalovirus latency and reactivation after allogeneic stem cell transplantation. Clin Immunol. 2013;150:1–11. doi: 10.1016/j.clim.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Dohner K, Nagel CH, Sodeik B. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 2005;13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Mai RT, Yeh TS, Kao CF, Sun SK, Huang HH, Wu Lee YH. Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene. 2006;25:448–462. doi: 10.1038/sj.onc.1209052. [DOI] [PubMed] [Google Scholar]
- Huang WH, Yung BY, Syu WJ, Lee YH. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J Biol Chem. 2001;276:25166–25175. doi: 10.1074/jbc.M010087200. [DOI] [PubMed] [Google Scholar]
- Kondapi AK, Satyanarayana N, Saikrishna AD. A study of the topoisomerase II activity in HIV-1 replication using the ferrocene derivatives as probes. Arch Biochem Biophys. 2006;450:123–132. doi: 10.1016/j.abb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Lokeswara Balakrishna S, Satyanarayana N, Kondapi AK. Involvement of human topoisomerase II isoforms in HIV-1 reverse transcription. Arch Biochem Biophys. 2013;532:91–102. doi: 10.1016/j.abb.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Advani SJ, Weichselbaum RR, Roizman B. Herpes simplex virus 1 activates cdc2 to recruit topoisomerase II alpha for post-DNA synthesis expression of late genes. Proc Natl Acad Sci USA. 2003;100:4825–4830. doi: 10.1073/pnas.0730735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Li J, Irwin CR, Jenkins H, DeLange L, Evans DH. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J Virol. 2008;82:5922–5932. doi: 10.1128/JVI.02723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Chen JY, Wang JT, Kimura K, Takemoto A, Lu CC, et al. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J Virol. 2007;81:5166–5180. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich CJ, Pardo J, Simon MM. Granule-associated serine proteases: granzymes might not just be killer proteases. Trends Immunol. 2009;30:117–123. doi: 10.1016/j.it.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Romero V, Andrade F. Non-apoptotic functions of granzymes. Tissue Antigens. 2008;71:409–416. doi: 10.1111/j.1399-0039.2008.01013.x. [DOI] [PubMed] [Google Scholar]
- Buzza MS, Zamurs L, Sun J, Bird CH, Smith AI, Trapani JA, et al. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280:23549–23558. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- Hiebert PR, Boivin WA, Abraham T, Pazooki S, Zhao H, Granville DJ. Granzyme B contributes to extracellular matrix remodeling and skin aging in apolipoprotein E knockout mice. Exp Gerontol. 2011;46:489–499. doi: 10.1016/j.exger.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, Freudenberg M, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29:720–733. doi: 10.1016/j.immuni.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Irmler M, Hertig S, MacDonald HR, Sadoul R, Becherer JD, Proudfoot A, et al. Granzyme A is an interleukin 1 beta-converting enzyme. J Exp Med. 1995;181:1917–1922. doi: 10.1084/jem.181.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeckel LT, Wallich R, Martin P, Sanchez-Martinez D, Weber FC, Martin SF, et al. Mouse granzyme K has pro-inflammatory potential. Cell Death Differ. 2011;18:1112–1119. doi: 10.1038/cdd.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony DA, Andrews DM, Chow M, Watt SV, House C, Akira S, et al. A role for granzyme M in TLR4-driven inflammation and endotoxicosis. J Immunol. 2010;185:1794–1803. doi: 10.4049/jimmunol.1000430. [DOI] [PubMed] [Google Scholar]
- Hollestelle MJ, Lai KW, van Deuren M, Lenting PJ, de Groot PG, Sprong T, et al. Cleavage of von Willebrand factor by granzyme M destroys its factor VIII binding capacity. PLoS One. 2011;6:e24216. doi: 10.1371/journal.pone.0024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning PJ, Kummer JA, de Poot SA, Quadir R, Broekhuizen R, McGettrick AF, et al. Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death. PLoS One. 2011;6:e22645. doi: 10.1371/journal.pone.0022645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ooms L, Bird CH, Sutton VR, Trapani JA, Bird PI. A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9) J Biol Chem. 1997;272:15434–15441. doi: 10.1074/jbc.272.24.15434. [DOI] [PubMed] [Google Scholar]
- Bots M, VANB L, Rademaker MT, Offringa R, Medema JP. Serpins prevent granzyme-induced death in a species-specific manner. Immunol Cell Biol. 2006;84:79–86. doi: 10.1111/j.1440-1711.2005.01417.x. [DOI] [PubMed] [Google Scholar]
- Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, et al. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J Biol Chem. 1997;272:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, et al. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci. 2004;61:301–325. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Q, Wu L, Liu S, Zhang Y, Yang X, et al. Identification of SERPINB1 as a physiological inhibitor of human granzyme H. J Immunol. 2013;190:1319–1330. doi: 10.4049/jimmunol.1202542. [DOI] [PubMed] [Google Scholar]
- Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, De Jong TA, et al. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- de Bruijn HW, Duk JM, van der Zee AG, Pras E, Willemse PH, Boonstra H, et al. The clinical value of squamous cell carcinoma antigen in cancer of the uterine cervix. Tumour Biol. 1998;19:505–516. doi: 10.1159/000030044. [DOI] [PubMed] [Google Scholar]
- Cataltepe S, Gornstein ER, Schick C, Kamachi Y, Chatson K, Fries J, et al. Co-expression of the squamous cell carcinoma antigens 1 and 2 in normal adult human tissues and squamous cell carcinomas. J Histochem Cytochem. 2000;48:113–122. doi: 10.1177/002215540004800112. [DOI] [PubMed] [Google Scholar]
- Hsu KF, Huang SC, Shiau AL, Cheng YM, Shen MR, Chen YF, et al. Increased expression level of squamous cell carcinoma antigen 2 and 1 ratio is associated with poor prognosis in early-stage uterine cervical cancer. Int J Gynecol Cancer. 2007;17:174–181. doi: 10.1111/j.1525-1438.2006.00663.x. [DOI] [PubMed] [Google Scholar]
- Deng Z, Hasegawa M, Yamashita Y, Matayoshi S, Kiyuna A, Agena S, et al. Prognostic value of human papillomavirus and squamous cell carcinoma antigen in head and neck squamous cell carcinoma. Cancer Sci. 2012;103:2127–2134. doi: 10.1111/cas.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Domselaar R, de Poot SA, Bovenschen N. Proteomic profiling of proteases: tools for granzyme degradomics. Expert Rev Proteomics. 2010;7:347–359. doi: 10.1586/epr.10.24. [DOI] [PubMed] [Google Scholar]