Abstract
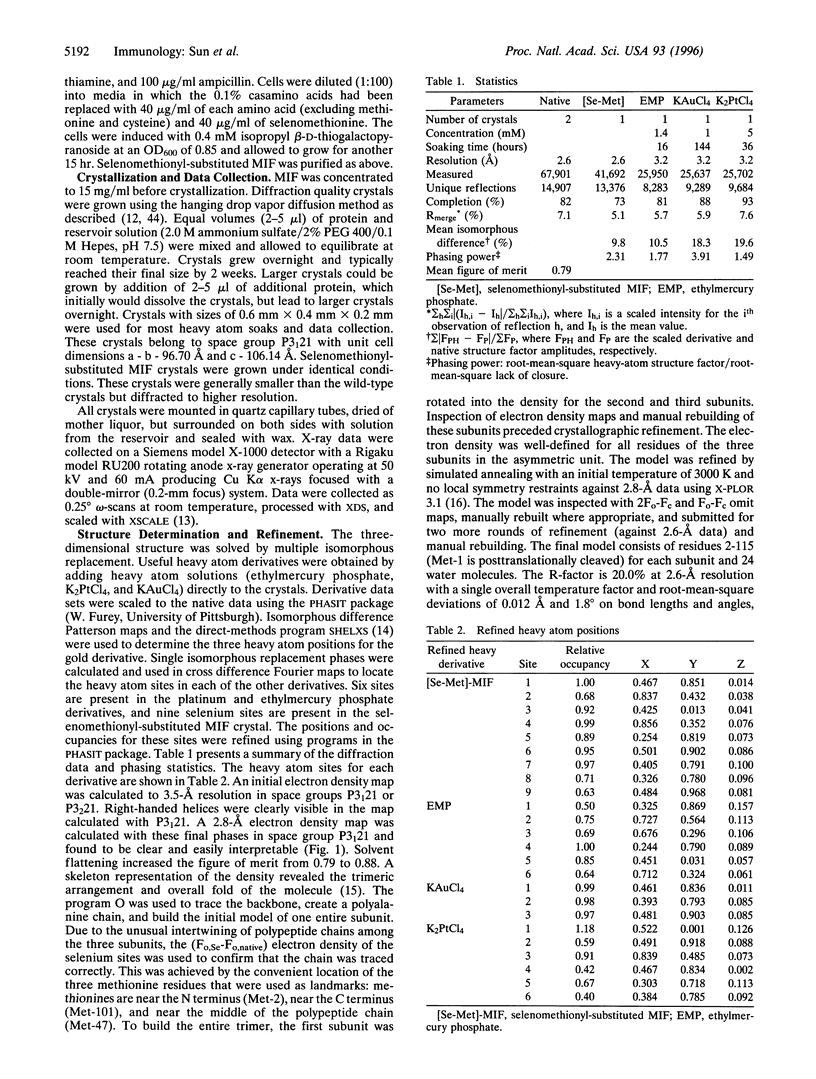
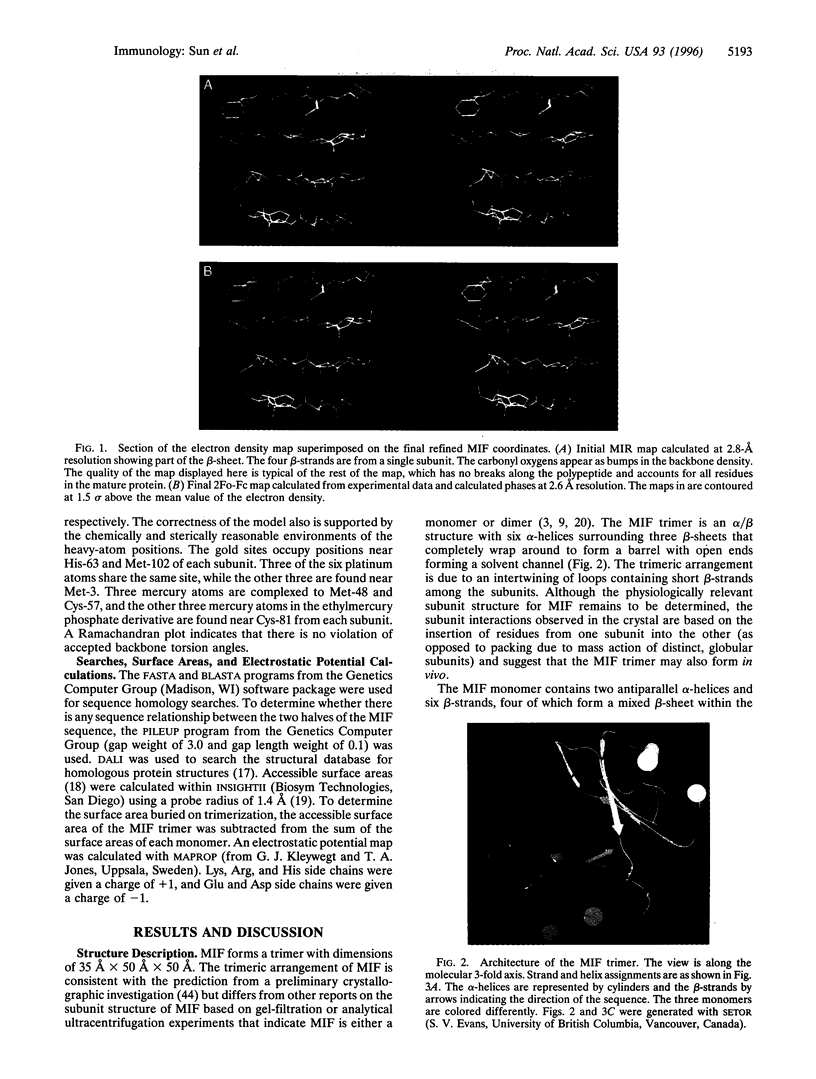
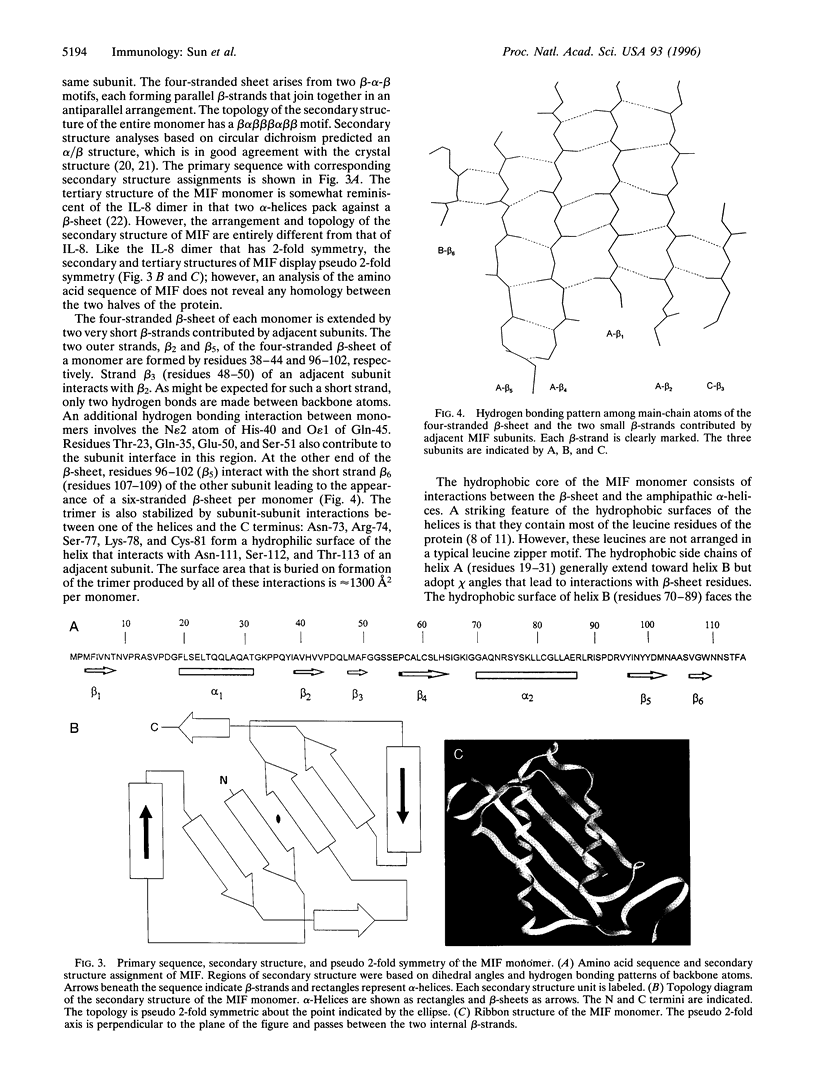
Macrophage migration inhibitory factor (MIF) was the first cytokine to be described, but for 30 years its role in the immune response remained enigmatic. In recent studies, MIF has been found to be a novel pituitary hormone and the first protein identified to be released from immune cells on glucocorticoid stimulation. Once secreted, MIF counterregulates the immunosuppressive effects of steroids and thus acts as a critical component of the immune system to control both local and systemic immune responses. We report herein the x-ray crystal structure of human MIF to 2.6 angstrom resolution. The protein is a trimer of identical subunits. Each monomer contains two antiparallel alpha-helices that pack against a four-stranded beta-sheet. The monomer has an additional two beta-strands that interact with the beta-sheets of adjacent subunits to form the interface between monomers. The three beta-sheets are arranged to form a barrel containing a solvent-accessible channel that runs through the center of the protein along a molecular 3-fold axis. Electrostatic potential maps reveal that the channel has a positive potential, suggesting that it binds negatively charged molecules. The elucidated structure for MIF is unique among cytokines or hormonal mediators, and suggests that this counterregulator of glucocorticoid action participates in novel ligand-receptor interactions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin E. T., Weber I. T., St Charles R., Xuan J. C., Appella E., Yamada M., Matsushima K., Edwards B. F., Clore G. M., Gronenborn A. M. Crystal structure of interleukin 8: symbiosis of NMR and crystallography. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):502–506. doi: 10.1073/pnas.88.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhagen J., Calandra T., Mitchell R. A., Martin S. B., Tracey K. J., Voelter W., Manogue K. R., Cerami A., Bucala R. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993 Oct 21;365(6448):756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- Bernhagen J., Mitchell R. A., Calandra T., Voelter W., Cerami A., Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF). Biochemistry. 1994 Nov 29;33(47):14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- Blocki F. A., Ellis L. B., Wackett L. P. MIF protein are theta-class glutathione S-transferase homologs. Protein Sci. 1993 Dec;2(12):2095–2102. doi: 10.1002/pro.5560021210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocki F. A., Schlievert P. M., Wackett L. P. Rat liver protein linking chemical and immunological detoxification systems. Nature. 1992 Nov 19;360(6401):269–270. doi: 10.1038/360269a0. [DOI] [PubMed] [Google Scholar]
- Bloom B. R., Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966 Jul 1;153(3731):80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- Braig K., Otwinowski Z., Hegde R., Boisvert D. C., Joachimiak A., Horwich A. L., Sigler P. B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994 Oct 13;371(6498):578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Calandra T., Bernhagen J., Metz C. N., Spiegel L. A., Bacher M., Donnelly T., Cerami A., Bucala R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995 Sep 7;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- Calandra T., Bernhagen J., Mitchell R. A., Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994 Jun 1;179(6):1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- David J. R. Delayed hypersensitivity in vitro: its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci U S A. 1966 Jul;56(1):72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A., Rivière S., Bouet F., Ménez A. A diversified family of 12-kDa proteins with a high amino acid sequence similarity to macrophage migration-inhibitory factor (MIF). Eur J Biochem. 1994 Sep 1;224(2):417–421. doi: 10.1111/j.1432-1033.1994.00417.x. [DOI] [PubMed] [Google Scholar]
- Gulick A. M., Fahl W. E. Mammalian glutathione S-transferase: regulation of an enzyme system to achieve chemotherapeutic efficacy. Pharmacol Ther. 1995 May;66(2):237–257. doi: 10.1016/0163-7258(94)00079-i. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hubbard S. J., Gross K. H., Argos P. Intramolecular cavities in globular proteins. Protein Eng. 1994 May;7(5):613–626. doi: 10.1093/protein/7.5.613. [DOI] [PubMed] [Google Scholar]
- Ji X., Zhang P., Armstrong R. N., Gilliland G. L. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 1992 Oct 27;31(42):10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- Ji X., von Rosenvinge E. C., Johnson W. W., Tomarev S. I., Piatigorsky J., Armstrong R. N., Gilliland G. L. Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods. Biochemistry. 1995 Apr 25;34(16):5317–5328. doi: 10.1021/bi00016a003. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kisljuk O. S., Kachalova G. S., Lanina N. P. An algorithm to find channels and cavities within protein crystals. J Mol Graph. 1994 Dec;12(4):305-7, 296. doi: 10.1016/0263-7855(94)80110-x. [DOI] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Lasters I., Wodak S. J., Alard P., van Cutsem E. Structural principles of parallel beta-barrels in proteins. Proc Natl Acad Sci U S A. 1988 May;85(10):3338–3342. doi: 10.1073/pnas.85.10.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. Y., David J. R., Remold H. G. Glycolipid affinity purification of migration inhibitory factor. Nature. 1982 Mar 4;296(5852):78–80. doi: 10.1038/296078a0. [DOI] [PubMed] [Google Scholar]
- Nishihira J., Kuriyama T., Sakai M., Nishi S., Ohki S., Hikichi K. The structure and physicochemical properties of rat liver macrophage migration inhibitory factor. Biochim Biophys Acta. 1995 Feb 22;1247(1):159–162. doi: 10.1016/0167-4838(94)00215-3. [DOI] [PubMed] [Google Scholar]
- Nishino T., Bernhagen J., Shiiki H., Calandra T., Dohi K., Bucala R. Localization of macrophage migration inhibitory factor (MIF) to secretory granules within the corticotrophic and thyrotrophic cells of the pituitary gland. Mol Med. 1995 Nov;1(7):781–788. [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. MIF proteins are not glutathione transferase homologs. Protein Sci. 1994 Mar;3(3):525–527. doi: 10.1002/pro.5560030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Huber R., Lo Bello M., Federici G., Parker M. W. Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution. J Mol Biol. 1992 Sep 5;227(1):214–226. doi: 10.1016/0022-2836(92)90692-d. [DOI] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Schäffer J., Gallay O., Huber R. The three-dimensional structure of class pi glutathione S-transferase in complex with glutathione sulfonate at 2.3 A resolution. EMBO J. 1991 Aug;10(8):1997–2005. doi: 10.1002/j.1460-2075.1991.tb07729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F. M. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974 Jan 5;82(1):1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- Rosengren E., Bucala R., Aman P., Jacobsson L., Odh G., Metz C. N., Rorsman H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996 Jan;2(1):143–149. [PMC free article] [PubMed] [Google Scholar]
- Sakai M., Nishihira J., Hibiya Y., Koyama Y., Nishi S. Glutathione binding rat liver 13k protein is the homologue of the macrophage migration inhibitory factor. Biochem Mol Biol Int. 1994 Jun;33(3):439–446. [PubMed] [Google Scholar]
- Schirmer T., Keller T. A., Wang Y. F., Rosenbusch J. P. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995 Jan 27;267(5197):512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- Sinning I., Kleywegt G. J., Cowan S. W., Reinemer P., Dirr H. W., Huber R., Gilliland G. L., Armstrong R. N., Ji X., Board P. G. Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes. J Mol Biol. 1993 Jul 5;232(1):192–212. doi: 10.1006/jmbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Murata E., Tanaka I., Nishihira J., Sakai M. Crystallization and a preliminary X-ray diffraction study of macrophage migration inhibitory factor from human lymphocytes. J Mol Biol. 1994 Jan 21;235(3):1141–1143. doi: 10.1006/jmbi.1994.1063. [DOI] [PubMed] [Google Scholar]
- Weiss M. S., Schulz G. E. Structure of porin refined at 1.8 A resolution. J Mol Biol. 1992 Sep 20;227(2):493–509. doi: 10.1016/0022-2836(92)90903-w. [DOI] [PubMed] [Google Scholar]
- Wistow G. J., Shaughnessy M. P., Lee D. C., Hodin J., Zelenka P. S. A macrophage migration inhibitory factor is expressed in the differentiating cells of the eye lens. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1272–1275. doi: 10.1073/pnas.90.4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Smerdon S. J., Jones D. H., Dodson G. G., Soneji Y., Aitken A., Gamblin S. J. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995 Jul 13;376(6536):188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- Zhang M., Aman P., Grubb A., Panagopoulos I., Hindemith A., Rosengren E., Rorsman H. Cloning and sequencing of a cDNA encoding rat D-dopachrome tautomerase. FEBS Lett. 1995 Oct 16;373(3):203–206. doi: 10.1016/0014-5793(95)01041-c. [DOI] [PubMed] [Google Scholar]







