Abstract
Stat1 is a pivotal transcription factor for generation of the interferon (IFN)-dependent antiviral response. Two Stat1 knockout mouse lines have been previously generated, one deleted the N-terminal domain (DNTD) and one in the DNA-binding domain (DDBD). These widely-used strains are assumed interchangeable, and both are highly susceptible to various pathogens. In this study, primary cells derived from DNTD mice were shown to be significantly more responsive to IFN, and established an antiviral state with greater efficiency than cells derived from DDBD mice, following infection with vesicular stomatitis virus and herpes simplex virus type-1. Also, while mice from both strains succumbed rapidly and equally to virus infection, DDBD mice supported significantly higher replication in brains and spleens than DNTD mice. Endpoint-type experimental comparisons of these mouse strains are therefore misleading in failing to indicate important differences in virus replication and innate response.
Keywords: Innate immunity, Stat1, vesicular stomatitis virus, herpes simplex virus
INTRODUCTION
Stat1 is a pivotal component of the signaling pathway for both type I and type II interferons (IFNβ) (O’Shea et al., 2011; Stark and Darnell, 2012). For type I IFNβ, IFNα and IFNβ engagement with their cognate receptor leads to phosphorylation of Stat1 and the formation of a heterotrimeric complex of Stat1, Stat2 and ISGF3 which translocates to the nucleus to stimulate expression of genes that contain IFN-stimulated response elements or ISREs (Schindler et al., 2007). Engagement of type II IFN (IFNγ) with its cognate receptor leads to formation of phosporylated Stat1 homodimers that stimulate expression of genes downstream of gamma-activated sequence (GAS) motifs (Lew et al., 1989). Collectively, stimulation of ISRE- and GAS-containing genes leads to initiation of the innate antiviral response and development of adaptive immunity. In addition to its role in establishing the antiviral state (Durbin et al., 1996; Meraz et al., 1996), Stat1 additionally plays a role in growth arrest and apoptosis (Kumar et al., 1997), and cancer (Chan et al., 2012; Hix et al., 2013). Most importantly, Stat1 is critical for the control of virus infection in humans (Boisson-Dupuis et al., 2012).
Two domains that are especially important for the function of Stat1 are the N-terminal domain (NTD) which is necessary for homo- and hetero-dimerization of the protein (Vinkemeier et al., 1996), and the DNA-binding domain (DBD) that is required for its function as a site-specific transcription factor (Schindler et al., 1995). Both of these domains have been deleted in the context of knockout mouse strains (Durbin et al., 1996; Meraz et al., 1996) and these two lines have been used extensively in literally hundreds of studies of signal transduction in innate immunity, innate immunity to bacterial and viral pathogens, and tumorigenesis (see for example (Bente et al., 2010; Hofer et al., 2012; Khodarev et al., 2004; Klover et al., 2010). These mouse lines have been used and interpreted more or less interchangeably since their separate development in 1996, and in their respective initial characterizations both lines were highly susceptible to vesicular stomatitis virus (VSV) infection. In 2005, a study using both Stat1-deficiant mouse lines concluded that both lines were relatively resistant to dengue virus infection relative to IFNαβγR−/− mice (Shresta et al., 2005). That study reported that mice lacking the NTD were capable of making IFNα in response to virus infection and concluded that Stat1-independent mechanisms were responsible. A subsequent study showed that while both Stat1-deficient mice were very susceptible to lethal infection, HSV-1 was neurotropic in the NTD-deficient line (DNTD), but viscerotropic in the DBD-deficient line (DDBD), with significant involvement of the liver (Pasieka et al., 2011). This and another study has led to the suggestion that the DNTD may have a small amount of residual Stat1-activity (Bowick et al., 2012). An important caveat of the previous HSV study, however, was that HSV is resistant to IFN, encoding multiple genes that inhibit IFN responses and Stat1 signaling (for example ICP0, ICP34.5 and vhs (Halford et al., 2006; Leib et al., 2000; Pasieka et al., 2008). This complicates the overall interpretation of the data with HSV since these genes may function with differential efficacy against residual Stat1 activity. Moreover, the direct comparison of DNTD and DDBD mice was limited to a bioluminescence imaging analysis of increased viral hepato-tropism in DDBD mice, with no analyses of IFN responses, comparative replication, or mortality performed.
In the present study, we therefore directly compared the replication of HSV in parallel with VSV, an IFN-sensitive virus, in primary fibroblasts derived from DNTD and DDBD mice. This approach also allows us to directly compare the susceptibility of DNTD and DDBD mice to both RNA and DNA viruses. We examined the response of these fibroblasts to IFN and also assessed HSV and VSV pathogenesis in both mouse strains. Our study revealed that despite significant differences in the abilities of DNTD and DDBD fibroblasts to respond to IFN and control VSV and HSV replication in vitro, and clear differences in viral replication in vivo, both strains of mice are highly and equally susceptible to VSV and HSV as judged by endpoint analyses. These data show that despite the ability of primary cells derived from DNTD mice to respond to IFN and partially control virus replication, these partial IFN responses are insufficient to substantially alter VSV- and HSV-induced mortality. The comparability of these two Stat1-deficient strains is therefore dependent upon experimental system and virus, and future and previous virological data from these two mouse strains must be carefully interpreted and compared.
RESULTS
VSV and HSV-1 replicate with significantly reduced efficiency in IFN-treated primary MEFs derived from DNTD relative to DDBD mice
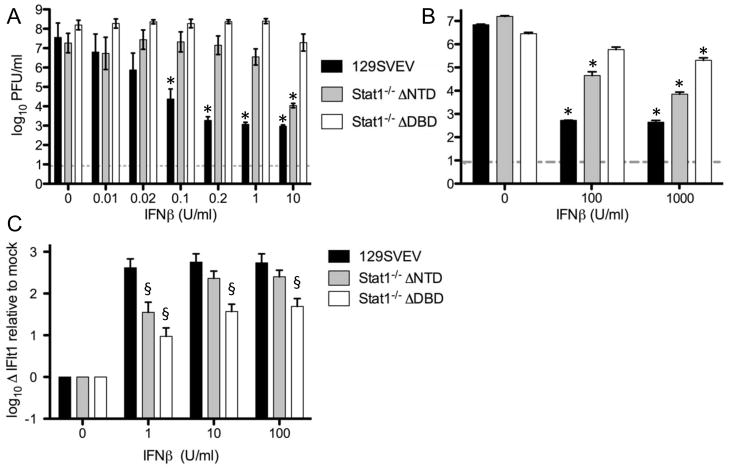
Monolayers of primary MEFs derived from control, DNTD and DDBD mice were treated overnight with varying doses of IFNβ, and subsequently infected with VSV at an MOI 0.1, and monolayers harvested and titered at 12 hours post-infection (Fig. 1A). In the absence of IFN treatment, all three lines yielded equivalent levels of VSV at 12 hours, suggesting that at this timepoint endogenous IFN synthesis was insufficient to control VSV. Pre-treatment with as little as 0.02units/ml of IFNβ, however, led to a >50-fold (p<0.05) lower yield of VSV from control and DNTD relative to DDBD MEFs. Yields from control MEFs decreased significantly in a dose-dependent fashion as IFN levels were increased, reaching a nadir at 0.2U/ml. Yields from DNTD MEFs were not changed significantly with increasing IFN treatment until a concentration of 10 U/ml was used, resulting in a >1,000-fold reduction in VSV titers (p<0.0001) relative to untreated MEFs. In contrast, yields from DDBD MEFs were not changed with increasing IFN treatment up to 1U/ml, with a non-statistically significant 8-fold reduction in titer at 10 U/ml of IFNβ. Notably the control of VSV replication did not alter further in DDBD MEFs even at a higher concentration of 100U/ml (data not shown). Taken together, these data demonstrate that DDBD and DNTD MEFs differ significantly from each other (p<0.0001) in their abilities to control VSV replication in response to IFN treatment, with DNTD MEFs being more responsive to IFN than DDBD. As expected, both Stat1−/− strains were significantly less responsive to IFN than control MEFs. The rank order of these MEFs to respond to IFN and control VSV replication, (control ≫ DNTD > DDBD) was recapitulated when infected with HSV-1 strain KOS (Fig. 1B). Together, these data largely dispel the hypothesis that it is the differential capability of HSV-1 to counter any residual activity of the DNTD and DDBD Stat1 alleles that results in disparate viral growth in these two mouse strains. Rather, these data support the notion that it is simply the intrinsic differential ability of these two mouse lines to respond to IFN that results in unequal control of virus replication.
FIGURE 1.
A. Titers of VSV 12 hours postinfection from MEFs infected at MOI 0.01. MEFs (derived from control 129 mice, or DDBD and DNTD Stat1-deficient mice) were pre-treated for 18 hours with varying concentrations of IFNβ. B. Titers of HSV-1 24 hours postinfection from MEFs infected at MOI 0.01 pretreated for 18 hours with varying concentrations of IFNβ. C. Real-time PCR analysis using the 2−DDCT method for IFIT1 transcript 6 hours post-treatment with indicated amounts of IFNβ. Data shown for each panel are derived from two or more independent experiments performed in duplicate. Dashed lines indicate the limit of detection, * indicates statistically significantly different from untreated controls (p<0.05, ≥0.0001), § indicates statistically significantly different from wild-type cells (p<0.05, ≥0.004). Data shown in panels A and B are averaged from a minimum of 2 experiments performed in duplicate, or in panel C in triplicate.
IFN-treated primary MEFs derived from DNTD relative to DDBD mice differentially up-regulate IFIT1
To further address the hypothesis that DNTD and DDBD mice differ in their ability to respond to IFN, we measured ISG gene expression in response to IFN treatment in the absence of viral infection. We used IFIT1 as a representative ISG since it is strongly up-regulated by IFN in a Stat1-dependent fashion (Bluyssen et al., 1994). At all concentrations of IFN used, MEFs derived from 129SVEV mice responded strongly (Fig. 1C). Indeed, addition of 1U/ml of IFNβ appeared sufficient to saturate the induction of IFIT1. In contrast, the addition of IFN induced a dose-dependent increase in IFIT1 expression from MEFs derived from both DNTD and DDBD mice. Interestingly, and in concordance with the VSV and HSV growth data (Figs 1A and 1B), induction of IFIT1 was higher in the DNTD MEFs relative to DDBD. Furthermore, despite an overall lower trend, the induction of IFIT1 RNA in DNTD MEFs was statistically indistinguishable from 129SVEV following treatment with 10 and 100 U/ml IFNβ. Together, these data show that primary cells derived from DNTD and DDBD mice differ significantly at the level of control of virus replication and ISG induction.
DNTD and DDBD mice exhibit equivalent susceptibility to VSV
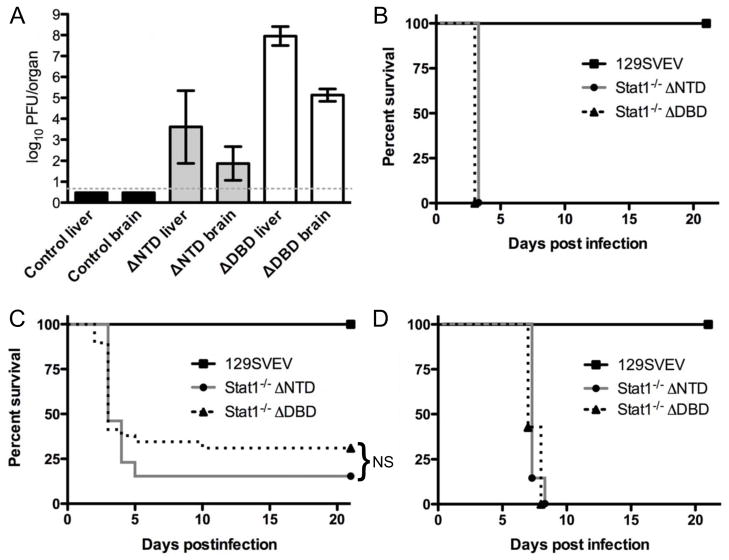
The results above showing differential replication of VSV and HSV-1 in DNTD and DDBD MEFs led us to speculate whether the rank order of resistance (control ≫ DNTD > DDBD) would recapitulate in vivo. The original publications describing DNTD and DDBD mice showed high mortality following VSV infection for both strains compared to controls (Durbin et al., 1996; Meraz et al., 1996), but no direct or quantitative comparisons of the Stat1−/− mouse strains were performed. Several studies have shown that DNTD mice are also highly susceptible to HSV-1, but direct comparison to DDBD mice has been limited to a single study of hepatotropism (Pasieka et al., 2011). We therefore wished to compare these mice in more detail following infection with VSV and HSV-1. We infected mice ip with 100pfu VSV and measured time for mice to reach to endpoint criteria, as well as assessing titers in liver and brain on day 2, one day before the onset of mortality (Fig. 2). There were significantly higher VSV titers in both the brains and livers of DDBD relative to DNTD and control mice (p<0.001), consistent with the in vitro data shown above (Fig 2A). Based on these replication data, we anticipated that the DDBD mice would be more susceptible to VSV-induced mortality than DNTD mice. Surprisingly, following ip infection with VSV there was rapid and synchronous mortality observed in both DNTD and DDBD mice at 3 days post-infection, while the wild-type mice all survived out to the 21 day cutoff time point (Fig. 2B). Given this early simultaneous VSV-induced mortality we postulated that the 100 pfu inoculum might be too high to parse differences between the mouse strains. We therefore infected mice ip with a lowered dose of 20 pfu VSV and again examined mortality (Fig. 2C). At this lower dose we observed a more step-wise pattern of mortality, with survival of a total of 11/34 (32%) DDBD mice, and 3/19 (16%) DNTD mice, with 7/7 (100%) of control mice surviving. While Kaplan-Meier plots for both DNTD and DDBD mice were significantly different (p>0.0001) from control mice, they were not significantly different from each other. That stated, there was a non-significant trend for DNTD to be more susceptible to VSV infection relative to DDBD mice. This slight change in susceptibility notwithstanding, these results were surprising given the significantly greater replication of VSV in DDBD MEFs relative to DNTD, and the higher titers in the livers and brains of DDBD relative to DNTD mice. To examine this further we infected mice with 2 × 106 pfu/eye of KOS and monitored time to reach endpoint criteria (Fig. 2D). Similar to the experiments using VSV, there was no discernable difference between DDBD and DNTD mice in their mortality following infection with HSV-1. This further extends the idea that these mice are equivalent in terms of general susceptibility to infection, regardless of the IFN sensitivity of the pathogen, and despite demonstrable significant differences in the responses of primary cells to IFN.
FIGURE 2.
Replication and lethality of VSV and HSV in DDBD and DNTD mice. A. VSV titers in brain and liver of control (129), DDBD and DNTD mice 2 days post ip infection with 100pfu of VSV. Data were collected from a total of 42 mice over 2 experiments with a minimum of 5 mice per group. B. Time taken to reach endpoint mortality criteria for control (129), DDBD and DNTD mice following ip infection with 100pfu VSV. Data were collected from a total of 16 mice (n=7 for control, n=5 for DDBD and n=6 for DNTD). C. Time taken to reach endpoint mortality criteria for control (129), DDBD and DNTD mice following ip infection with 20pfu VSV. Data were collected from a total of 16 mice (n=7 for control, n=34 for DDBD, n=19 for DNTD). NS = not statistically significant. D. Time taken to reach endpoint mortality criteria for control (129), DDBD and DNTD mice following corneal infection with 2 × 106 pfu/eye. Data were collected from a total of 23 mice (n=9 for control, n=7 for DDBD, n=7 for DNTD).
DISCUSSION
The data of this study underscore the complexity of interpretation of data from knockout mouse models in general, and also further give caution to making direct comparisons between these two Stat1−/− mouse strains. The in vitro data described herein demonstrate that MEFs derived from DDBD and DNTD mice respond differentially to IFN in terms of their ability to control VSV, and HSV-1 -- two very different viruses, especially with regard to their IFN sensitivities. HSV-1 encodes for several specific functions that interfere directly with Stat1 and the antiviral activities of IFN, and is generally resistant to IFN (Mossman and Ashkar, 2005). While VSV does antagonize the type I IFN response through blocking RNA export via the matrix protein (Waibler et al., 2007), VSV remains highly susceptible to the effects of IFN. Despite their differences in IFN sensitivity, both viruses replicate with a similar pattern in DDBD and DNTD MEFs in the presence of IFN. Importantly, this demonstrates that it is an inherent difference in the ability of these Stat-deficient cells to mount an antiviral response, rather than a difference in the ability of these viruses to counter any residual functions of DDBD and DNTD alleles. This conclusion is further supported by the observation that the induction of IFIT1 RNA synthesis by IFNβ treatment in the absence of virus infection, clearly differs between primary cells derived from the 2 mouse strains. Previous work eliminated the possibility that mouse background account for the phenotypic disparity (Pasieka et al., 2011), so these data thereby show formally that primary cells derived from these mouse lines differ in their molecular and functional antiviral responses. One additional caveat is that only fibroblasts were examined, and it is possible that IFN-driven responses in DDBD and DNTD mice may also differ by cell type. The responses of each cell type must therefore be determined empirically.
The relative equivalence in susceptibility to VSV of DDBD and DNTD mice was surprising given the differences in VSV production between the two mouse lines. This pattern of equivalent lethality but non-equivalent viral replication, however, is not exclusive to VSV, since HSV-1 also induced similar mortality in these two mouse strains despite the disparate replication patterns in vitro shown in this study, and in vivo (Pasieka et al., 2011). These data suggest that the susceptibility of both strains to viral infection is sufficiently high such that even relatively large differences in viral titers in critical organs (such as liver and brain) do not significantly alter the timing with which these mice reach endpoint clinical criteria. It is likely that once the innate immune response is compromised below a certain functional threshold (as in DNTD), further compromise (as in DDBD) does not result in further detectable susceptibility as judged by endpoint analysis following a pathogenic challenge. This is consistent with data showing that VSV kills 100% of Ifit2−/− mice within 6 days following intra-nasal infection of VSV regardless of input dose (Fensterl et al., 2012). That study also showed that mortality and viral titers were often not correlated for VSV. This also emphasizes the relative crudeness of endpoint-type experiments, which although informative, do not reveal underlying important differences in biology, tropism and pathogenesis. Another confounding issue is route of infection, which likely also plays a critical role in the relative susceptibilities of these mouse strains.
These Stat1-deficient mouse lines created by Meraz et al., and Durbin et al. (Durbin et al., 1996; Meraz et al., 1996) have been used in a wide variety of in vitro and in vivo studies, with almost 600 citations of these original papers in the primary literature. The data of this study show that for in vitro experiments at least, data acquired from these two lines cannot be directly compared. In addition, Stat1-independent effects should be equivalent in both lines, so the data further support the idea that the Stat1 mutation in DDBD mice represents a more complete ablation of Stat1 function than mutation in the DNTD strain. The DDBD strain, therefore seems preferable for studies in which it is necessary for the mice to lack any residual Stat1 activity which may confound interpretation of the data. The DNTD strain, however, is a closer model of human Stat1 insufficiency, since, humans deficient in Stat1 and DNTD mice both succumb to HSV-1 encephalitis (Boisson-Dupuis et al., 2012; Pasieka et al., 2009), whereas DDBD mice acquire fulminant hepatitis (Pasieka et al., 2011). Clearly both mouse strains have strengths and weaknesses for investigation of IFN-driven innate immunity to viruses, but it is apparent that these mouse strains differ significantly in terms of their innate responses, regardless of virus type and IFN sensitivity.
METHODS AND MATERIALS
Cells, viruses, and animal infection procedures
Mouse embryo fibroblast (MEF) cultures were generated from 129 Sv/Ev, and Stat1−/− mice at embryonic day 15 and passaged once before being plated for infection. MEFs were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 0.1 mM sodium pyruvate, 250 U/ml penicillin, 250 μg/ml streptomycin, and 250 ng/ml amphotericin B. Isogenic MEFs were also utilized. For multiple-step growth curves, cells were pretreated overnight with the appropriate concentration of IFN (Sigma, St. Louis, MO), and cells were infected at a multiplicity of infection (MOI) of 0.01. All titering and viral stock preparation was performed on Vero cells as previously described. The VSV used was strain Indiana, and the HSV-1 strain was KOS. Mouse strains used included the control 129S6 as wild type mice (Taconic Farms, Germantown, NY), 129S6 Stat1-deficient mice lacking the N-terminal domain (referred to here as DNTD or Stat1−/−NTD), and 129 Stat1-deficient mice lacking the DNA binding domain (referred to here as DDBD or Stat1−/−DBD). Mice were genotyped by PCR and housed in the barrier facility in the Center for Comparative Medicine and Research at The Geisel School of Medicine at Dartmouth and were infected intraperitoneally or corneally in the biohazard facility between the ages of 6–8 weeks (Rader et al., 1993; Strelow and Leib, 1995). Mice were harvested at appropriate times and organs dissected and titered as previously described, and times to reaching of endpoint criteria recorded. Sentinel mice were screened every 3 months and determined to be negative for adventitious mouse pathogens, in particular mouse norovirus. Mice were housed, infected, and euthanized when necessary in accordance with all Federal and University policies.
Quantitative real-time PCR
MEFs from the three mouse strains were cultured as above and were treated with IFNβ (0, 1, 10 or 100 U/ml) for 6 hours, then RNA was collected using the RNeasy kit (Quiagen). The RNA was treated with DNase (New England Biolabs, MA), and cDNA was synthesized using the SuperScript III kit (Life Technologies) with random hexamers (Promega, WI). For QPCR, SYBR Green (Life Technologies, NY) was used with primers for IFIT1 (Fw: TGC TTT GCG AAG GCT CTG AAA GTG, Rv: TGG ATT TAA CCG GAC AGC CTT CCT, 200nM) and GAPDH (GAPDH Fw: CAT CTT CCA GGA GCG AGA TCC C Rv: CAA ATG AGC CCC AGC CTT CTC C 400nM). IFIT1 values were calculated by the 2−DDCT method (Livak and Schmittgen, 2001) normalized to GAPDH, and values for IFNβ-treated MEFs were normalized to untreated MEFs of the same strain.
We compared 2 strains of Stat1---deficient mice by infection with VSV and HSV---1.
Significant differences in antiviral activity and ISG induction were seen in vitro.
Significant differences in virus replication were observed in vivo.
The susceptibilities of these mice to lethal infection are comparable.
Acknowledgments
This study was supported by National Institutes of Health grant to D.L. (RO1 EY10707). The project was also supported by P20RR016437 from the National Center for Research Resources to Dartmouth.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bente DA, Alimonti JB, Shieh WJ, Camus G, Stroher U, Zaki S, Jones SM. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. Journal of virology. 2010;84:11089–11100. doi: 10.1128/JVI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluyssen HA, Vlietstra RJ, Faber PW, Smit EM, Hagemeijer A, Trapman J. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics. 1994;24:137–148. doi: 10.1006/geno.1994.1591. [DOI] [PubMed] [Google Scholar]
- Boisson-Dupuis S, Kong XF, Okada S, Cypowyj S, Puel A, Abel L, Casanova JL. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Current opinion in immunology. 2012;24:364–378. doi: 10.1016/j.coi.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowick GC, Airo AM, Bente DA. Expression of interferon-induced antiviral genes is delayed in a STAT1 knockout mouse model of Crimean-Congo hemorrhagic fever. Virology journal. 2012;9:122. doi: 10.1186/1743-422X-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM, Lonardi S, Arthur C, Young LJ, Levy DE, Welch MJ, Cardiff RD, Schreiber RD. STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast cancer research : BCR. 2012;14:R16. doi: 10.1186/bcr3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford WP, Weisend C, Grace J, Soboleski M, Carr DJ, Balliet JW, Imai Y, Margolis TP, Gebhardt BM. ICP0 antagonizes Stat 1-dependent repression of herpes simplex virus: implications for the regulation of viral latency. Virology journal. 2006;3:44. doi: 10.1186/1743-422X-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hix LM, Karavitis J, Khan MW, Shi YH, Khazaie K, Zhang M. Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells. The Journal of biological chemistry. 2013;288:11676–11688. doi: 10.1074/jbc.M112.441402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer MJ, Li W, Manders P, Terry R, Lim SL, King NJ, Campbell IL. Mice deficient in STAT1 but not STAT2 or IRF9 develop a lethal CD4+ T-cell-mediated disease following infection with lymphocytic choriomeningitis virus. Journal of virology. 2012;86:6932–6946. doi: 10.1128/JVI.07147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia. 2010;12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Decker T, Darnell JE., Jr Alpha interferon and gamma interferon stimulate transcription of a single gene through different signal transduction pathways. Molecular and cellular biology. 1989;9:5404–5411. doi: 10.1128/mcb.9.12.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Mossman KL, Ashkar AA. Herpesviruses and the innate immune response. Viral immunology. 2005;18:267–281. doi: 10.1089/vim.2005.18.267. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Gadina M, Kanno Y. Cytokine signaling: birth of a pathway. Journal of immunology. 2011;187:5475–5478. doi: 10.4049/jimmunol.1102913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasieka TJ, Cilloniz C, Lu B, Teal TH, Proll SC, Katze MG, Leib DA. Host responses to wild-type and attenuated herpes simplex virus infection in the absence of Stat1. Journal of virology. 2009;83:2075–2087. doi: 10.1128/JVI.02007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasieka TJ, Collins L, O’Connor MA, Chen Y, Parker ZM, Berwin BL, Piwnica-Worms DR, Leib DA. Bioluminescent imaging reveals divergent viral pathogenesis in two strains of Stat1-deficient mice, and in alphassgamma interferon receptor-deficient mice. PloS one. 2011;6:e24018. doi: 10.1371/journal.pone.0024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasieka TJ, Lu B, Leib DA. Enhanced pathogenesis of an attenuated herpes simplex virus for mice lacking Stat1. Journal of virology. 2008;82:6052–6055. doi: 10.1128/JVI.00297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader KA, Ackland-Berglund CE, Miller JK, Pepose JS, Leib DA. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. The Journal of general virology. 1993;74 ( Pt 9):1859–1869. doi: 10.1099/0022-1317-74-9-1859. [DOI] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. The Journal of biological chemistry. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Schindler U, Wu P, Rothe M, Brasseur M, McKnight SL. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- Shresta S, Sharar KL, Prigozhin DM, Snider HM, Beatty PR, Harris E. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. Journal of immunology. 2005;175:3946–3954. doi: 10.4049/jimmunol.175.6.3946. [DOI] [PubMed] [Google Scholar]
- Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelow LI, Leib DA. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. Journal of virology. 1995;69:6779–6786. doi: 10.1128/jvi.69.11.6779-6786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. The EMBO journal. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- Waibler Z, Detje CN, Bell JC, Kalinke U. Matrix protein mediated shutdown of host cell metabolism limits vesicular stomatitis virus-induced interferon-alpha responses to plasmacytoid dendritic cells. Immunobiology. 2007;212:887–894. doi: 10.1016/j.imbio.2007.09.003. [DOI] [PubMed] [Google Scholar]