Abstract
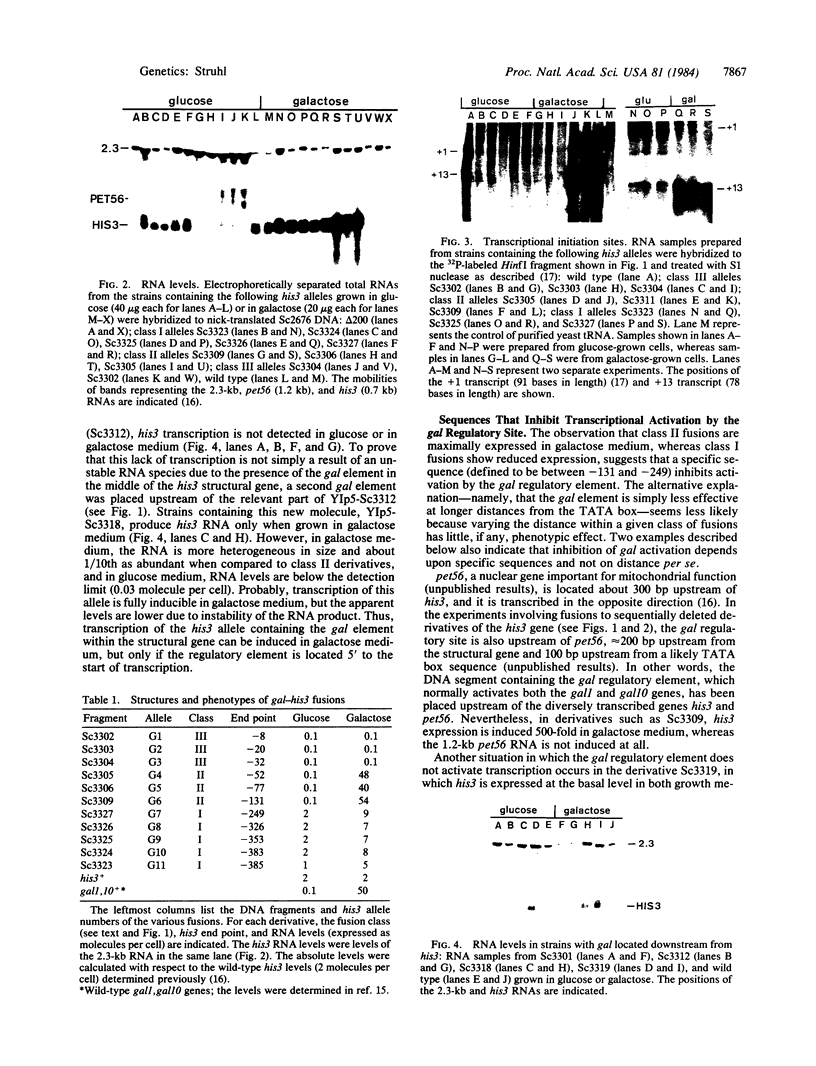
DNA molecules created by fusing a 365-base-pair segment of yeast DNA encoding the galactose-regulated upstream promoter element (gal) to a set of derivatives that systematically delete sequences upstream from the his3 gene are introduced in single copy back into the yeast genome precisely at the his3 locus and then assayed for transcription. Fusions of the gal regulatory element to his3 derivatives containing all normal mRNA coding sequences but lacking essentially the entire promoter region fail to express his3 under any growth conditions. Fusions to derivatives lacking the his3 upstream promoter element but containing the "TATA box" place his3 expression under gal control--i.e., extremely high RNA levels in galactose-containing medium and essentially no his3 RNA in glucose-containing medium. However, of the two normal his3 initiation sites, only the downstream one is activated by the gal element. In fusions of this type, neither the orientation of the gal element nor the distance between the element and the his3 TATA box affects the level or the initiation points of transcription. However, the gal element does not influence transcription when placed 100 or 300 base pairs downstream from the normal mRNA start sites. Fusions to derivatives containing the entire his3 promoter region restore the basal level of his3 transcription in glucose-grown cells, and both transcriptional initiation sites are used. Furthermore, RNA levels in galactose-grown cells, although somewhat higher than in glucose-grown cells, are significantly below the fully induced level. The distance from his3 coding sequences does not affect RNA levels, suggesting that specific sequences, possibly corresponding to the his3 upstream promoter element, reduce the ability of the gal element to activate transcription. Analysis of chromatin from some of these strains indicates a DNase I-hypersensitive site(s) in the middle of the gal element. However, this structural feature is not correlated with transcriptional initiation because it is found when cells are grown in glucose medium and also in derivatives lacking a TATA box. Thus, the gal upstream element possesses most, but not all, of the properties of viral and cellular enhancer sequences of higher eukaryotes. In addition, it appears that the his3 and gal upstream sequences represent two distinct classes of promoter elements, which activate transcription from different initiation sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Beier D. R., Young E. T. Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature. 1982 Dec 23;300(5894):724–728. doi: 10.1038/300724a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Maler B. A., Yamamoto K. R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983 Jun;33(2):489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Promotor mutants of the yeast his3 gene. J Mol Biol. 1981 Nov 5;152(3):553–568. doi: 10.1016/0022-2836(81)90268-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Transcription of the his3 gene region in Saccharomyces cerevisiae. J Mol Biol. 1981 Nov 5;152(3):535–552. doi: 10.1016/0022-2836(81)90267-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoter elements, regulatory elements, and chromatin structure of the yeast his3 gene. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):901–910. doi: 10.1101/sqb.1983.047.01.104. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]