Abstract
Aim
We studied the use of methyl-CpG binding domain (MBD) protein-enriched genome sequencing (MBD-seq) as a cost-effective screening tool for methylome-wide association studies (MWAS).
Materials & methods
Because MBD-seq has not yet been applied on a large scale, we first developed and tested a pipeline for data processing using 1500 schizophrenia cases and controls plus 75 technical replicates with an average of 68 million reads per sample. This involved the use of technical replicates to optimize quality control for multi- and duplicate-reads, an in silico experiment to identify CpGs in loci with alignment problems, CpG coverage calculations based on multiparametric estimates of the fragment size distribution, a two-stage adaptive algorithm to combine data from correlated adjacent CpG sites, principal component analyses to control for confounders and new software tailored to handle the large data set.
Results
We replicated MWAS findings in independent samples using a different technology that provided single base resolution. In an MWAS of age-related methylation changes, one of our top findings was a previously reported robust association involving GRIA2. Our results also suggested that owing to the many confounding effects, a considerable challenge in MWAS is to identify those effects that are informative about disease processes.
Conclusion
This study showed the potential of MBD-seq as a cost-effective tool in large-scale disease studies.
Keywords: MBD, methylome-wide association studies, next-generation sequencing, principal component analysis, pyrosequencing
Epigenetic modifications to chromatin provide stability and diversity to the cellular phenotype. These modifications are largely preserved or regenerated during cell division [1,2]. Among the different forms of epigenetic modifications, the most intensively studied is the methylation of DNA cytosine residues at the carbon 5 position. This common epigenetic mark is most often, although not exclusively, found in the sequence context CpG and is typically associated with transcriptional repression.
Methylation studies are a promising complement to genetic studies of variation in DNA sequence and structure. First, because methylation directly affects gene expression, it may capture additional individual variation in disease susceptibility [3]. Indeed, dysregulation of DNA methylation has been associated with a wide variety of human diseases, including neurological disorders such as Alzheimer’s and Parkinson’s diseases [4], autoimmune disorders such as systemic lupus erythematosus [5], cardiovascular disease [6] and several types of cancer [7–9]. Second, methylation can account for a wide variety of phenomena that characterize complex diseases [8,10] such as sex differences [11,12], genotype– environment interactions [13,14], and age-related patterns associated with the disease course [15]. Third, methylation sites are potential new drug targets as they are modifiable by pharmaceutical interventions [16] and have good properties from a translational perspective such as being stable and enabling cost-effective assays in biosamples that can be relatively easy to collect [17].
The potential importance of DNA methylation in the etiology of complex diseases has led to interest in the development of methylome-wide association studies (MWAS; as our study only considers methylation and not histone modification and other epigenetic marks, it is not an epigenome-wide association study) [1,18] aimed at interrogating all possible methylation sites in the human genome [19]. The most comprehensive laboratory method for ascertaining methylation (DNA cytosine residues at the carbon 5 position) status at each nucleotide position is bisulfite sequencing [20], where unmethylated cytosines in genomic DNA are converted to uracil and then converted to thymine in post-bisulfite PCR [21]. This single base resolution is attractive because it allows precise mapping of disease relevant sites [22]. However, owing to the combination of high costs of sequencing entire genomes and the large numbers of samples needed to provide adequate statistical power, whole-genome bisulfite sequencing is not currently economically feasible as a screening tool for disease association studies [18]. An alternative is reduced representation bisulfite sequencing that applies bisulfite sequencing to a reduced portion of the genome. It captures approximately 12% of all CpGs [23] that tend to be located in CpG dense regions [24].
Other methods have been developed that differentiate methylation status using enzymatic digestion or affinity-based capture methods, followed by detection on arrays or using next-generation sequencing (NGS). Examples of array-based methods include the Infinium system from Illumina (CA, USA) [19], but even their largest array interrogates only 450,000 loci. Genome-wide tiling arrays such as the 2.1 million probe array set from NimbleGen (WI, USA) or the 45 million probe array set from Affymetrix (CA, USA) [25] offer more comprehensive coverage. However, NGS seems more promising than tiling arrays [18,26]. Not only can NGS, in principle, detect all methylation sites in the human genome, the relatively lower amount of starting material also reduces the need for extensive DNA amplification and hence errors and bias. Furthermore, the increasing data volume generated per run, the decrease in reagent costs and fast semi-automated sample preparation have already made NGS a very competitive option in terms of cost per measured site.
Methods for detection of methylation that are adaptable to NGS include HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) that uses the methylation sensitive enzyme HpaII [27], methylated DNA immunoprecipitation (MeDIP) that uses an antibody for 5-methylcytosine [28], and methyl-CpG binding domain (MBD)-based capture that uses the methyl binding domain of methyl binding proteins MBD2 or MBD3L1 to capture methylated DNA fragments [26]. Ultimately, enzyme-based methods are restricted by the need for a specific recognition sequence, meaning they can never be unbiased and representative of the whole genome. The affinity-based capture methods, while lacking the single base resolution of bisulfite sequencing, arguably provide a good compromise. Among affinity-based capture methods, there are differences in assay properties depending on the DNA binding protein/antibody used. Both MeDIP sequencing and MBD-isolated DNA sequencing (MBD-seq) are capable of detecting differentially methylated regions (DMRs) [29] and capture approximately the same proportions of the methylome [23]. However, whereas the proteins used for MBD-based capture strictly bind to methylated CpGs, the antibody used in MeDIP captures DNA fragments containing any methylated C. Non-CpG methylation may be of great importance for complex diseases. However, taking association studies of sequence variants as an example, the different polymorphisms (e.g., common SNPs, rare SNPs and CNVs) are typically not examined in a single study as different study designs and techniques are more suitable for some polymorphisms than for others. Similar arguments may apply here. For example rather than simply calculating the number of reads covering a CpG to quantify methylation, the use of peak finding algorithms may be considered for MeDIP and it could also be the case that there may be more rare methylation sites requiring different kinds of association tests. Furthermore, because sporadically methylated sequences can comprise a significant portion of the data generated by MeDIP [26], (many) more reads may be needed to accurately measure all methylation signals. To some extent, we view MBD-seq as being similar to genome-wide association study (GWAS) with common SNPs as a first pass at examining the methylome by only considering sites where methylation is likely to occur. Perhaps this more focused technology may be one of the factors contributing to the observation that MBD-seq may detect substantially more DMRs compared with MeDIP-seq [30].
MBD-seq has already been demonstrated to be highly specific, sensitive and applicable to identifying DMRs [23,26,29,31,32]. Laboratory automation can facilitate MBD-seq on a large scale. However, because MBD-seq has not yet been applied on a large scale, an efficient and accurate pipeline for data processing and analysis that capitalizes on the inherent strengths of the approach is currently not available. The purpose of this article is to study the potential of MBD-seq for large-scale MWAS and develop a robust pipeline for analysis. For this purpose we will make use of a schizophrenia case–control data set where an average of 68 million reads were sequenced in 1500 individuals, amounting to over 100 billion reads.
Materials & methods
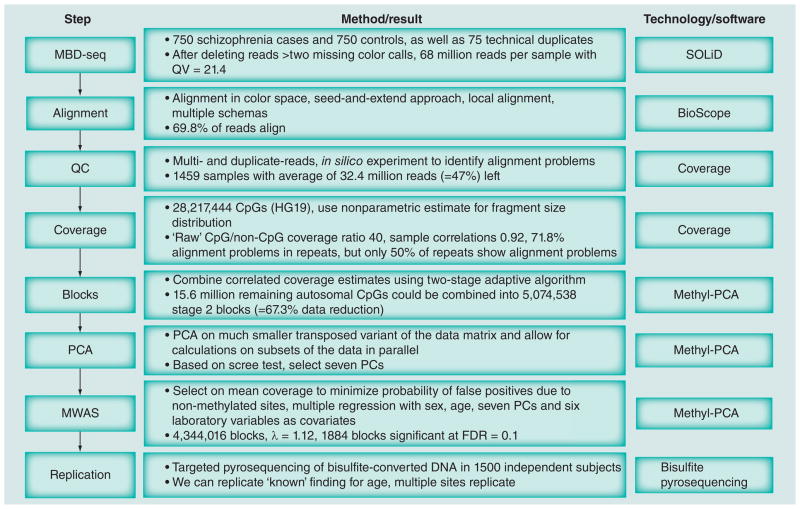
Figure 1 gives an overview of the ‘Materials & methods’ section of this paper by summarizing the data analysis pipeline plus key results from each step. Full details can be found in the Supplementary Material (see www.futuremedicine.com/doi/suppl/10.2217/epi.12.59).
Figure 1. Overview of the data processing pipeline and key results at each step.
FDR: False-discovery rate; MBD-seq: Methyl-CpG binding domain sequencing; MWAS: Methylome-wide association studies; PC: Principal component; PCA: Principal component analysis; QC: Quality control; QV: Quality value.
Sample description
Our study includes 750 schizophrenia cases and 750 controls, as well as 75 technical duplicates. This study is part of a large ongoing project entitled ‘A Large-Scale Schizophrenia Association Study in Sweden’. The project is supported by grants from National Institute of Mental Health (NIMH) and the Stanley Foundation and aims at improving our understanding of the etiology of schizophrenia and bipolar disorder plus their clinical and epidemiological correlates using high dimensional biological investigations and proper analysis. For details on the project see [33–35]. Cases with schizophrenia were identified via the Swedish Hospital Discharge Register. Population controls, who had never received a discharge diagnosis of schizophrenia, were selected at random from the national population registers and then group matched to the cases on age, gender and county of residence. All procedures were approved by ethical committees in Sweden and in the USA, and all subjects provided written informed consent (or legal guardian consent and subject assent). DNA was extracted from peripheral blood donated at the local medical facilities of the participants. Samples were then shipped to the Broad Institute where they were fingerprinted using a 24-plex fingerprint iPLEX assay (Sequenom, CA, USA) prior to shipping to the service provider that did the sequencing for this project.
Laboratory procedures
We used the MethylMiner™ kit from Invitrogen (CA, USA) that employs MBD protein-based enrichment of the methylated DNA fraction, followed by single end sequencing (50 bp read length) on the Applied Biosystems SOLiD next-generation sequencing platform (Life Technologies, CA, USA). Methods were standard and based upon manufacturer’s recommendations (see Supplementary Material). Genomic regions that are CpG poor can be difficult to capture by MBD-seq [30]. To improve methylome- wide coverage, we used an existing protocol variant that increases the relative number of fragments from CpG poor regions by eluting the captured methylated fraction with 0.5 M NaCl.
For replication purposes we used targeted pyrosequencing (Qiagen, CA, USA). This quantitative method allows for targeted sequencing of bisulfite converted DNA with high accuracy. We included several controls to ensure the assays worked properly (e.g., a standard curve of five DNA samples in duplicates with known methylation levels). Furthermore, each additional set of study samples (e.g., each additional 96-well plate) contains two plate controls, in duplicates, of DNA from the standard curve. Assay designs and further details are available in the Supplementary Material.
Data analysis
Alignment
The sequenced reads were aligned to the human genome (build hg19/GRCh37) using BioScope 1.2 (Life Technologies) that aligns in color– space and takes full advantage of the increased ability of SOLiD two-base encoding to identify sequencing errors [36]. We used a seed-and-extend approach combined with local alignment and multiple schemas. Specifically, our seed was 25 bases. Rather than considering the entire extension, local alignment may improve sensitivity by finding the maximum similarity score between the reference sequence and a substring of the extension. A maximum of two color space mismatches were allowed in the seed (e.g., as two color call matches are required to change the base call, a SNP will have two color call mismatches). If the seed could not be mapped, a second schema was attempted by moving the seed from base 1 to base 15.
Coverage estimation
In the case of MBD-seq, only fragments with methylated CpGs can be pulled down. Given that we know exactly where the CpGs are located, there is no need to search for peaks and to obtain a methylation measurement. We therefore calculated coverage for the 28,217,444 CpG sites in the reference genome (hg19/GRCh37). A standard procedure is to count the number of sequence reads covering the CpG. Because the methylation of any CpG in the entire fragment could lead to its capture by the MBD protein binding, the read length is sometimes extended to the expected fragment length. However, because not all fragments will have exactly the same specified size and the fragment pool obtained after shearing may not be identical to the pool that gets successfully sequenced (e.g., smaller fragments may be more likely to get pulled down by the enrichment protocol), this procedure can be imprecise. Furthermore, particularly in large-scale studies there may be (stochastic) variation in the fragment size distribution across samples. Thus, rather than assuming an identical predetermined fragment size for all fragments and samples, we estimated the fragment size distribution for each sample from the empirical sequencing data.
A limitation of commonly used methods for estimating fragment size distributions, for example those used in ChIP-seq peak finding algorithms [37,38], is that they make strong parametric assumptions about these distributions and do not take advantage of the specific features of MBD-seq data where we know exactly where methylation can occur. We therefore developed a nonparametric method that uses isolated CpGs (no other CpG site within 400 bp) to estimate the fragment size distribution empirically from the sequencing data. To validate our method we performed extensive simulations and sequenced paired-end libraries in eight mice [Van den Oord E, Bukszar J, Rudolf G et al. Estimation of CpG coverage in whole methylome next-generation sequencing studies (2012), Submitted]. Using the successfully aligned read pairs, we obtained the ‘observed’ fragment size distribution by subtracting the start positions of the two reads. We then performed coverage calculations using the fragment size distributions as observed in paired-end read data. Next we performed a ‘traditional’ coverage calculation where we counted the number of sequence reads covering the CpGs. Results showed that the mean coverage was merely 29.2% of that obtained after analyzing the paired-end data. Furthermore, when we correlated these coverage estimates with those obtained from the paired-end data, we only obtained a very modest Pearson correlation of 0.606. The DNA samples were fragmented by ultrasonication (Covaris, MA, USA) to a target median size of 150 bp. We also performed coverage calculations by extending the read length from 50 bp to this 150 bp target. Results improved but coverage was still underestimated by 13% and the correlation with paired-end coverage estimates was 0.934. Finally, we excluded one read from each pair and used our estimator. Results were now very similar to those obtained with the paired-end data with only a slight coverage overestimation of 0.6%. In addition, our coverage estimates correlated 0.999 with those from the paired-end data suggesting almost identical results.
The sample-specific estimated fragment size distributions were used to calculate a probability test for each read. This probability indicates how likely it is that the fragment, tagged by the read, covers the CpG under consideration. Coverage for each CpG can then be calculated by taking the sum of the probabilities that all fragments in its neighborhood cover the CpG. For example, this probability equals 1.0 for fragments with reads starting within 50 bp of the CpG, but will be ≤1.0 for fragments with reads starting further away. Coverage is affected by the total number of used reads per sample that is a function of the laboratory protocol (e.g., sequencer output and degree of multiplexing) rather than methylation. Our estimates were therefore standardized using the total number of reads that remained after quality control (QC).
When using methylation enrichment based approaches such as M BD-seq, the number of fragments covering a particular CpG depends not only on the methylation status of that site but also the number of methylated CpGs in the region [23]. To make coverage estimates more comparable across sites and improve the correlation with actual methylation levels, coverage estimates can be further normalized using the local CpG density as a proxy for the number of methylated CpGs in the region [39,40]. However, in association analyses we essentially compare the means of cases and controls. As this mean difference remains unchanged by corrections based on CpG density (i.e., the same constant will be added for cases and controls), for sake of simplicity we did not use such a normalization step.
Duplicate- & multi-reads
Many reads map to multiple locations of the genome. Often a single alignment can be selected because it is clearly better than the others. In the case of multireads, multiple alignments are about equally good. Selecting only the single best alignment for each multiread carries along the danger of alignment errors (e.g., alignments to regions with SNPs are less likely to be best alignments because SNPs cause mismatches). On the other hand, excluding all multireads may affect accuracy in a negative way [41]. Duplicate-reads are reads that start at the same nucleotide positions. When sequencing a whole genome duplicate-reads often arise from template preparation or amplification artifacts. In our context of sequencing an enriched genomic fraction, duplicate-reads are increasingly likely to occur by chance because reads are expected to align to a much smaller fraction of the genome.
We examined empirically whether it would be better to allow for high-quality multi- and duplicate-reads or to exclude all such reads. To compare these strategies we calculated Pearson correlations between the technical replicates using the data from all CpG sites. As this correlation decreases when measurement error increases, it reflects the precision of coverage estimates. To select high-quality multireads, any read that mapped to more than ten loci was excluded from further consideration. From the remaining multireads, we selected only those that aligned almost equally well to a few number of loci. Specifically, we selected the multireads that had fewer than five alignments with alignment scores (read length – 3 × the number of mismatches) within five points of the best score. Multireads were weighted proportional to the number of alignments. This essentially accounts for the uncertainty of not knowing which part of the genome the multiread comes from by using a simple probability estimate that the read aligns to that specific location. Thus, multireads were treated in a different way than reads with a single best alignment in the coverage calculations.
In all instances where >3 (duplicate) reads started at the same position, we reset the read count to one for the coverage calculations assuming that these reads all tagged a single fragment. If two or three reads started at the same position, we looked for other reads in the region of ±25 bp. If other reads mapped to this area, we retained the read count of two or three in the coverage calculations assuming that the duplicate-reads occurred by chance owing to enrichment of fragments caused by methylated CpG in the region. If no other reads were found, we assumed again that the duplicate reads were artifacts and reset the read count to one for the coverage calculations.
Eliminating sites affected by alignment problems
CpG sites in loci that are problematic in terms of alignment need to be eliminated prior to analysis as coverage estimates will be confounded with alignment errors. For example, repetitive elements constitute approximately 45% of the human genome. Reads may be difficult to align to these loci because of their high sequence similarity. To identify problematic sites, we conducted an in silico experiment. We first generated all 2.86 billion possible 50 bp single-end reads for the human reference genome (build hg19/GRCh37), with each 50 bp sequence starting 1 bp downstream of the previous sequence. For example, the sequence of in silico read one was identical to the sequence of positions 1–50 on chromosome 1 of the reference, read two was identical to positions 2–51 and so on. In the perfect scenario, aligning these reads to the reference genome should show that each CpG is covered by 50 reads. CpG sites with coverage <40 or >60 were eliminated from further analyses.
Data reduction
We combined highly intercorrelated coverage estimates at adjacent CpG sites into a single mean coverage estimate. The sum of substantially intercorrelated measurements is a more reliable indicator of the underlying signal than the individual measurements separately [42]. Furthermore, reducing the total number of sites has computational and statistical advantages (e.g., decreased risk of false discoveries). Rather than using a sliding window of a predetermined fixed length, sites were combined adaptively based on the observed Pearson correlations between them [25]. In MBD-seq intercorrelations occur because neighboring CpGs are largely covered by the same 100–200 bp fragments, and because of biological phenomena [43]. To account for these different causes, we used a two-stage algorithm. We first combined CpGs that were largely covered by the same fragments by specifying a very high threshold for the intercorrelations of r > 0.9. We then used the combined ‘block’ data from the first stage using a somewhat smaller intercorrelation of r > 0.6 to capture ‘biological’ correlations. We evaluated the robustness of the algorithm we used to create blocks. In these studies we randomly deleted 5, 10, 25 and 50% of the CpGs and compared these results with those obtained using the complete data. Block structures were very similar suggesting this algorithm was robust.
Case–control association testing
We used the block data obtained after the data reduction stage as input for multiple regression analyses to test for association with case–control status. Blocks with low coverage were excluded to minimize the probability of analyzing regions that were not methylated and therefore would produce false-positive MWAS findings. To identify such sites, we calculated baseline noise level in our data by selecting sites in the genome that were at least 400 bp away from the nearest CpG. The 99th percentile of the coverage estimated at these non-CpGs was used as a threshold below which we declare a site as having low coverage. In addition to sex and age, to eliminate possible assay related artifacts, we regressed out variables such as amount of starting material for MethylMiner, the quantity of methylation-enriched DNA captured and sample batch. We also considered ancestry as a possible covariate. These ancestral dimensions were calculated using the multidimensional scaling (MDS) implemented in PLINK [44] with genome-wide SNP data as input (Affymetrix 5.0 and Affymetrix 6.0 arrays, see [33] for details). Two dimensions (MDS1 and MDS2) were sufficient to characterize the ancestral structure in this sample. As SNP data was missing for 20% of the sample and their use as covariates would reduce sample size and statistical power, we examined whether their inclusion was critical by studying their correlations with methylation patterns.
Controlling for confounders to avoid false-positive findings presents a major challenge in MWAS. In addition to technical factors associated with processing samples and the risk of false positives due to population structure, in MWAS there are many possible differences between cases and controls that may affect the methylome and consequently produce significant association results. Examples include differences in lifestyle, diet and medication use. Although these effects represent real differences between cases and controls, they are not informative about the underlying disease processes. If assessed, such variables can be regressed out. However, the list of potential confounders is long, only a subset of these variables will have been measured, and many confounders may simply be unknown. Statistical methods that first capture the major sources of variation in the methylome, and then regress out these components when performing the MWAS may provide an effective solution for handling confounders. We choose principle component analysis (PCA) for this purpose because it is well developed and commonly used in high-dimensional biological investigations [45,46]. To validate the PCA [Chen W, Gao G, Aberg K et al. Methyl-PCA: a toolkit for principal component analysis in methylome-wide association studies (2012), Submitted] we simulated MWAS data for 500 cases and 500 controls with five confounding factors. After regressing out principal components (PCs), the inflation factor λ (i.e., the observed median test statistic value divided by the expected median assuming no effect for any site), dropped from 8.93 to 1.01. This indicated that the PCA accurately controlled for confounders. The same assay-related variables that were included in the MWAS were also regressed out here to ensure that PCA captured a distinct set of confounders.
Software
We have experienced several problems when attempting to analyze the MBD-seq data using existing software packages. First, existing packages tend to store all data in working memory. Because of the size of the data sets (i.e., 30 million CpGs × 1500 subjects) this becomes problematic. Second, because multiple steps of the analyses involve processing data from all CpG sites, central processing unit time may be prohibitive. Finally, in our data there are many more variables (i.e., sites) than samples. Consequently, standard algorithms for performing PCA are no longer suitable. To address the above challenges we developed two packages called COVERAGE and Methyl-PCA. Whereas COVERAGE is specific for MBD-seq, Methyl-PCA can be used with any MWAS technology. The source code, Windows plus Linux executables and documentation are freely available online [101].
All computational and input/output intensive parts in these two packages were implemented in C++ where the R package was used for other purposes such as plotting and to provide a user interface. Where relevant, the code avoids reading all data points into working memory by processing the raw data one CpG at the time. For clusters there is the option to speed up calculations by processing data from different chromosomes simultaneously or partition data in subsets so that computations can be done in parallel. Statistics that are used repeatedly (e.g., the mean across the entire sample) are calculated only once and stored to further increase efficiency. Finally, following Gower the PCA is performed through eigen-decomposition of a much smaller transposed variant of the data matrix [47]. All programs (parts) were double coded either by a different programmer or in another language. For example, to check our PCA implementation, we simulated a small methylation data set that was analyzed with Methyl-PCA as well as the R function prcomp() that performs PCA on a regular observed covariance/correlation matrix. Results were identical.
Bioinformatics
To annotate results we downloaded or calculated a variety of CpG features using data from the UCSC genome browser [102]. Features included: CpG islands, shores (2000 bp flanking a CpG island [24]), transcription factor binding sites, within gene boundaries, exon, intron or untranslated regions, within potential gene promoters (8000 bp upstream from known transcription start), within evolutionarily conserved regions or within regions overlapping with known repetitive elements (RepeatMasker).
Results
Based on observations that 30–60 million reads per sample may be sufficient to reveal valuable information for whole-genome methylation analysis [30,40], we aimed for more than 60 million reads per sample with a minimum of 40 million reads for each of the 1575 methylomes sequenced in this investigation. The 205 samples for which we got fewer than 40 million reads were rerun to supplement reads. After these reruns only 25 samples still had fewer than 40 million reads. The SOLiD system essentially reads each base twice thereby producing two ‘color calls’ for each base. We deleted all reads with >2 missing color calls after which we observed an average of 68.0 million (standard deviation [SD]: 26.8 million) reads per sample (see Supplementary Figure 1 for box and whisker plot). The mean quality value (quality value: −10log10(p), with p being the probability of an error) per color call was 21.4 (SD: 1.1). Assuming these estimates are accurate, the probability of an incorrect color call would be 0.72%. Because it takes two wrong adjacent color calls to make a base call error, the risk of a wrong base call is predicted to be approximately 1 in 20,000 (= 0.72 × 0.72%).
Starting with an average of 68 million reads and 1500 samples, we first dropped all reads from runs with <40% alignment or that produced a very small number of reads. These involved mainly reruns. The percentage of mapped reads in the remaining samples was 69.9% (SD: 6.3). Of these mapped reads, we eliminated 32.5% because they were low quality multi- or duplicate- reads (see above for definition). Thirty eight subjects were excluded because <15 million reads remained after all QC and another three subjects were excluded because they withdrew their consent during the study. This left a sample of 1459 subjects with on average 32.4 million quality controlled reads (SD: 13.7 million). The percentage of uniquely mapped reads was 85.0%, with remaining reads being high-quality multireads.
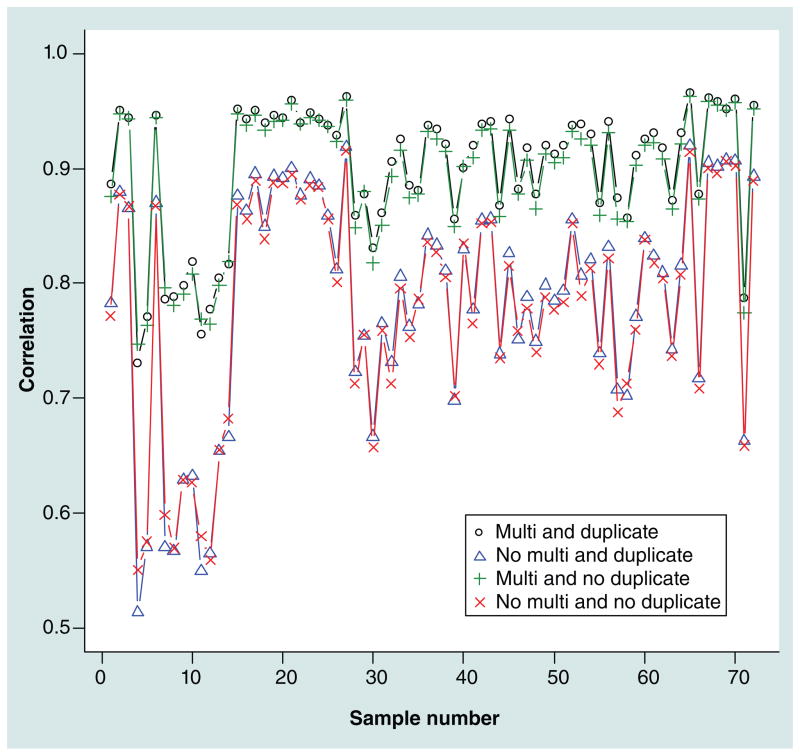
Figure 2 shows the correlations between 73 technical replicates (two technical replicates with reruns were dropped for the sake of simplicity) used to study duplicate- and multi-reads. The highest correlations between technical replicates (median: 0.92) were obtained when high-quality multireads and duplicate-reads were included. These correlations decreased markedly after excluding high-quality multireads (0.80), and decreased modestly after excluding high-quality duplicate-reads (0.91). Thus, these results supported the inclusion of both high-quality multi-and duplicate-reads, where the multireads are most critical. We also dichotomized the methylation measures using ‘non-CpG’ coverage (see the ‘Case–control association testing’ section) and calculated concordance rates. This would, for example, reduce possible effects of multireads aligning to the same location in the replicates or outliers. Results in Supplementary Figure 2 show the same pattern as for the correlations with the inclusion of high-quality multireads improving concordance rates between technical replicates.
Figure 2.
Correlations calculated for 73 duplicates after different quality control procedures for duplicate- and multi-reads.
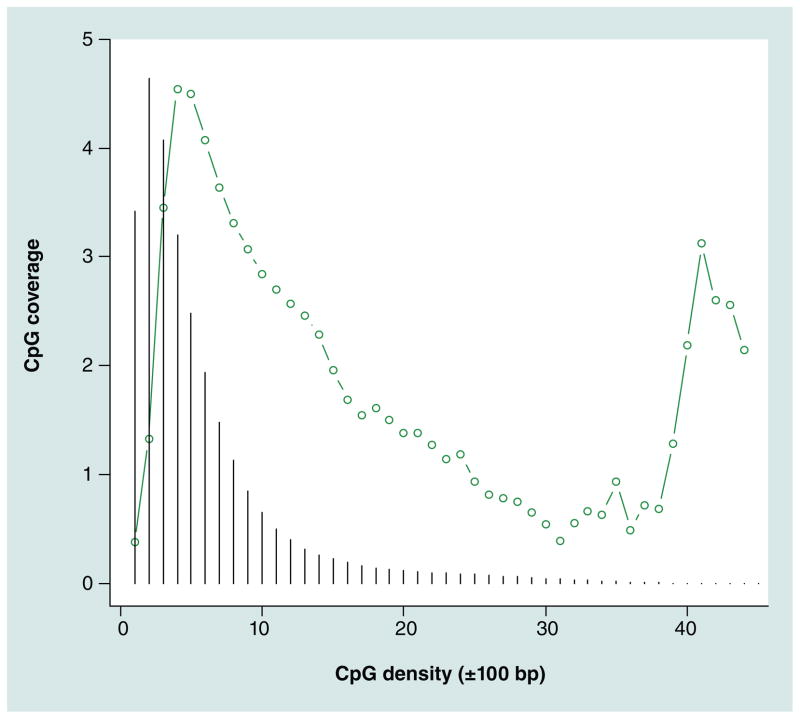
To obtain an indication of the efficiency of the enrichment protocol, we compared the coverage estimates of the autosomal CpGs with ‘noise’ coverage levels estimated for loci that were at least 400 bp away from the nearest CpG. The ratio of the median coverage of CpG versus non-CpG coverage was 40.6. Because for samples that are not enriched the expected ratio is one, this suggested the enrichment worked properly. Next, for each CpG we calculated the local CpG density (also called coupling factors [39]) by counting all other CpGs within ± 100 bp. Figure 3 confirms the well known relation between coverage and local CpG density [39,48]. The relationship is the result of multiple phenomena such as different MBD2 binding efficiencies and regions of higher CpG densities being less likely to be methylated. Isolated CpGs seem relatively difficult to capture using MBD-seq, while very dense regions (e.g., 40 CpGs within 200 bp) are relatively easy. For optimal methylome-wide coverage it is important that fragments cover the vast majority of CpGs. Figure 3 therefore also depicts the relative frequencies (black vertical lines) of CpGs with a certain density. These frequencies show that the vast majority of CpGs have local densities in the 1–10 range. As the majority of sequenced fragments also cover CpGs in this range, this suggests that our MBD protocol provided relatively good coverage of the regions where most of the CpGs are located.
Figure 3. CpG density versus ‘raw’ coverage.
For CpG density a simple ‘coupling’ factor was calculated by counting the number of CpGs within ±100 bp. The vertical lines indicate the relative frequencies of CpGs with that specific density.
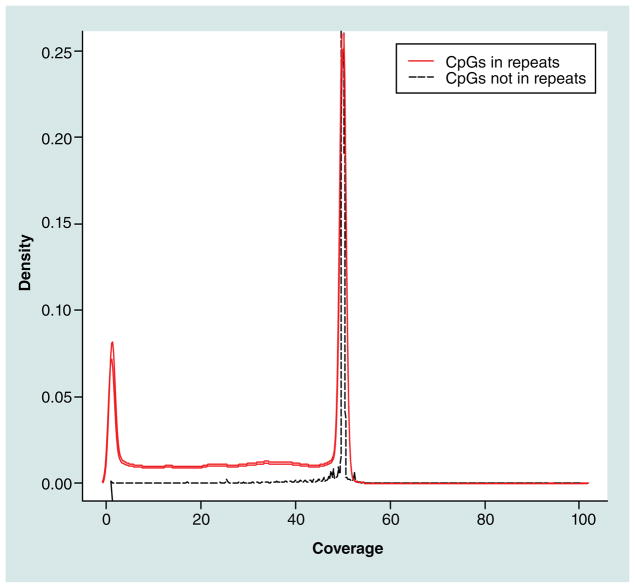
Results from our in silico alignment experiment indicated that, except for 23 reads, all 2.86 billion reads aligned back to the reference genome. The distribution of the in silico CpG coverage in Figure 4 shows that the alignment worked well for the majority of CpGs. A total of 36% (10.5 million) of the CpGs showed alignment problems (defined as ±10 the expected coverage of 50). The majority of these CpGs (71.8%) were located in regions flagged as repetitive elements by RepeatMasker. Alignment problems were highly dependent on the repeat class with, for example, DNA transposons aligning well and long interspersed nuclear elements aligning poorly. However, of the 15.0 million CpGs located in repeats, only 50.2% showed alignment problems. Thus, although repeats are a major source of alignment problems and CpGs in problematic repeats need to be eliminated, a considerable number of CpGs in repeats align well and can be retained for subsequent association analyses.
Figure 4.
Distribution of in silico coverage for CpGs in loci overlapping and not overlapping with RepeatMasker.
Prior to combining intercorrelated coverage estimates at adjacent sites, we eliminated all sites with alignment problems. In addition, sites with low coverage were eliminated to avoid numerical problems when calculating correlations between adjacent CpGs. The remaining 15.6 million CpGs could be combined into 8,822,240 stage 1 blocks, and these stage 1 blocks could in turn be combined into 5,074,538 stage 2 blocks. These results imply overall data reduction of 67.3% where only 10.9% of the stage 2 blocks consisted of a single CpG. Table 1 shows that the stage 1 blocks were small (mean = 15.6 bp) with high intercorrelations (r = 0.95) indicating they indeed involved CpGs in close proximity that were most likely covered by the same fragments. The stage 2 blocks comprised, on average, 3.1 CpGs with the largest blocks consisting of >18 CpGs and spanning over 500 bp. This indicated longer range biological correlations between the methylation levels of CpGs.
Table 1.
Descriptive statistics of stage 1 and 2 blocks.
| Descriptive information | Stage 1
|
Stage 2
|
||||
|---|---|---|---|---|---|---|
| CpGs (n) | Length (bp) | Cor. | CpGs (n) | Length (bp) | Cor. | |
| Mean | 1.8 | 15.6 | 0.95 | 3.1 | 73.7 | 0.73 |
|
| ||||||
| SD | 1.8 | 306.9 | 0.03 | 5.1 | 246.5 | 0.09 |
|
| ||||||
| Median | 1 | 0 | 0.95 | 2 | 42 | 0.71 |
|
| ||||||
| 95th percentile | 4 | 68 | 0.99 | 8 | 221 | 0.88 |
|
| ||||||
| 99th percentile | 6 | 102 | 1 | 18 | 500 | 0.9 |
Cor.: Correlation; SD: Standard deviation.
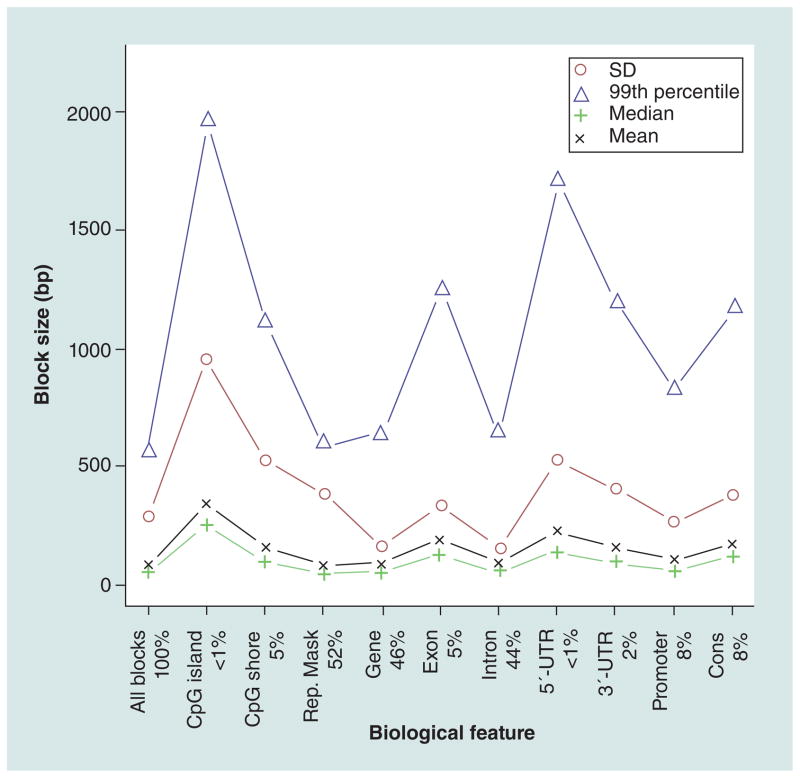
Figure 5 shows how the sizes of the stage 2 blocks vary across biological features. There is a clear trend where blocks are larger compared with the methylome-wide average in the majority of the annotated features. In particular, blocks are larger in CpG islands and shores, exons, and in UTRs with 5′-UTRs on average being larger than 3′-UTRs. The smallest average blocks were observed in introns and in regions overlapping with well aligning repeats.
Figure 5. Size of stage 2 blocks across biological features.
The mean, median, SD and 99th percentile for the stage 2 block size, in base pairs, is given for all blocks included and for blocks overlapping with CpG islands, CpG shores, regions marker by Rep. Mask, genes, exons, introns, 5′-UTRs, 3′-UTRs, regions within 8 kb upstream of transcriptional start sites corresponding to potential promoter regions (promoter) and conserved regions (cons). The percentage of blocks overlapping with the biological features are given in the legend on the x-axis. Note that a single block can overlap with multiple biological features.
Rep. Mask: RepeatMasker; SD: Standard deviation.
Association testing & replication
To minimize the probability of false-positive findings due to sites that may not be methylated, a total of 730,522 stage 2 blocks were excluded because the mean coverage level was less than the coverage of the 99th percentile of non-CpGs (see the ‘Case–control association testing’ section). This left 4,344,016 blocks for the MWAS where the mean number of fragments covering these blocks was 12.9 (SD: 454.3).
In Table 2 we present correlations between the first ten PCs and clinical, demographic and ancestral dimensions (the intercorrelations among these variables are given in Supplementary Table 1). In general, correlations were very modest suggesting that most of the variation in the methylome cannot be attributed to these variables. Four PCs showed small but significant correlations with case–control status. A number of PCs were correlated with potential confounders such as smoking and alcohol use. Among the 20 correlations, only two were significant for the MDS dimensions, suggesting that ancestry did not substantially contribute to variation in the methylome. As SNP data was missing for 20% of the sample and their use as covariates would reduce sample size and statistical power, they were not included in the MWAS.
Table 2.
Correlates of principal component scores.
| PC | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Clinical | ||||||||||
| Schizophrenia | 0.13† | −0.03 | −0.03 | 0.07† | 0.03 | −0.05 | −0.07† | 0.01 | 0.05† | 0.02 |
| Alcohol use | −0.03 | 0 | 0.03 | −0.01 | −0.01 | 0.01 | 0.08† | 0.05 | 0.03 | −0.03 |
| Smoking | −0.01 | 0.06 | −0.1 | −0.06 | 0.04 | −0.16† | 0.01 | 0.03 | −0.04 | −0.1 |
| Narcotics | 0 | −0.03 | −0.03 | 0.01 | 0 | −0.04 | 0.02 | −0.03 | −0.04 | −0.02 |
| Epilepsy | 0.04 | −0.02 | −0.06† | 0.03 | 0.03 | −0.02 | −0.02 | −0.03 | 0.07† | 0.01 |
| Diabetes | 0.03 | −0.01 | −0.03 | −0.01 | 0.02 | −0.03 | −0.04 | −0.04 | 0.05† | 0.06† |
| Hyperthyroid | 0.01 | −0.06† | 0.02 | 0.03 | 0.04 | −0.01 | 0.03 | 0 | 0.02 | 0.07† |
| Hypothyroid | −0.02 | 0.01 | −0.01 | −0.02 | −0.01 | −0.02 | −0.03 | −0.01 | 0.05 | −0.02 |
| Autoimmune | −0.05 | −0.03 | −0.1† | 0.03 | 0 | 0.02 | 0.03 | −0.04 | 0.04 | 0.03 |
| Demographic | ||||||||||
| Male | −0.04 | 0.06† | 0.03 | 0.02 | −0.04 | 0.04 | −0.1† | −0.02 | −0.1† | −0.01 |
| Age | 0.03 | −0.06† | 0.03 | −0.05 | −0.01 | 0.03 | 0 | 0.01 | 0.02 | 0.03 |
| Parent origin | −0.03 | −0.01 | 0.01 | −0.05 | −0.08† | 0.01 | −0.04 | 0.04 | 0 | 0.07 |
| Ancestry | ||||||||||
| MDS1 | −0.02 | −0.06 | −0.06† | 0.05 | 0.05 | −0.02 | 0.01 | −0.07† | 0.01 | −0.03 |
| MDS2 | −0.02 | 0.04 | 0.02 | −0.02 | 0.01 | 0.04 | 0.04 | 0.06 | 0.01 | −0.02 |
Significant correlation with p < 0.05.
PC: Principal component.
Based on a scree test (Supplementary Figure 3), we selected the first seven PCs for inclusion in the association analyses. Prior to the inclusion of the PCs, the quantile–quantile plot showed considerable inflation (Supplementary Figure 4A: λ = 7.32). This inflation was effectively dealt with by regressing out the PCs (Supplementary Figure 4B: λ = 1.12).
To study the ability of MBD-seq to identify sites that can be replicated in independent samples with a technology that provides single base resolution, we designed targeted pyrosequencing assays (see Supplementary Table 2 for assay designs) to replicate two of the top findings from the MWAS where we did not regress out the PCs and also a negative control. The substantive results and replication findings from the MWAS after regressing out the PCs will be reported in a separate paper. The two genes were FNDC3B (p < 1.7 × 10−12) and DCTN (p < 7.8 × 10−13). The replication was carried out in independent samples from the same study population as the MWAS samples. In these analyses we regressed out sex and age as well as possible plate effects. Supplementary Figure 5 shows the raw data. Table 3 summarizes results and shows that the two replication sites comprised a total of five CpGs that replicated with p-values between 8.5 × 10−4 and 6.2 × 10−3. In addition, the direction of effects in the MBD-seq and replication study was identical. Cohen’s d [49], indicated that the case–control difference in the replication study was about half the standard deviation, which may be considered a medium effect size according to Cohen’s criteria. The negative control was not significant. Overall, these findings suggested that MBD-seq identified effects that could be replicated using a technology that provides single base resolution.
Table 3.
Replication results.
| Gene | Chr. | Position | n | β | T-value | p-value | Cohen’s d |
|---|---|---|---|---|---|---|---|
| Replication sites | |||||||
|
| |||||||
| FNDC3B | 3 | 171788002 | 189 | −0.06 | 0.02 | 9.58E-04 | −0.48 |
| 17188032 | 188 | −0.06 | 0.02 | 3.65E-03 | −0.40 | ||
|
| |||||||
| DCTN2 | 12 | 57939307 | 187 | −0.06 | 0.02 | 1.09E-03 | −0.47 |
| 57939275 | 185 | −0.06 | 0.02 | 6.73E-04 | −0.47 | ||
| 57939245 | 183 | −0.07 | 0.02 | 2.06E-03 | −0.42 | ||
|
| |||||||
| Negative control | |||||||
|
| |||||||
| PARK2 | 6 | 162054935 | 333 | 0.04 | 0.07 | 0.52 | 0.02 |
| 162054989 | 330 | 0.02 | 0.02 | 0.32 | −0.06 | ||
n is sample size and β the standardized regression coefficient.
Chr.: Chromosome.
The replication also suggested that association findings obtained if PCs are not regressed out may reflect real differences between cases and controls and are not type I errors or technology driven artifacts. To further study possible confounders captured by the PCs, we ran an MWAS where we regressed out all PCs in the top 30 that were associated with schizophrenia except the three PCs (1, 4 and 7) that were among the seven main PCs selected based on the scree test. The top finding in this analysis with a p-value of 1.5 × 10−12 was HTRA3. HTRA3 is known to be hyper-methylated in smokers and this effect can be demonstrated empirically in cell lines following cigarette smoke carcinogen treatment [50]. It is well established that patients with schizophrenia have an extremely high prevalence of smoking [51] so this finding is likely the result of the confounding effects of smoking. Similarly, another top finding is that CAMK2D (p < 1.5 × 10−12) has been suggested in a large GWAS meta-analysis as a susceptibility gene for metabolic syndrome [52], a well-known side effect of antipsychotics used to treat schizophrenia.
Epigenetic changes, including DNA methylation, have emerged as key contributors to the genomic alterations that accompany aging [53]. As several studies of age-related changes in methylation exist, this outcome provides another opportunity to examine the ability of MBD-seq to detect associations. To make the most appropriate comparisons, we limit ourselves to comparisons between our control sample (n = 750) and published exploratory studies in healthy adult samples. Three such studies used the Illumina Infinium arrays enabling association testing of 27K sites [54–56]. Among the most robust associations across these studies involved GRIA2. This gene was one of five loci consistently found to be hypermethylated with age in several different tissue types in a study by Koch et al. [54], with a mean correlation of GRIA2 methylation with age across tissues of 0.62. GRIA2 was also associated with age in the study by Bell et al. [56] using whole blood DNA (p = 4.66 × 10−6), and was among the top findings (p = 1.37 × 10−6) in a study by Bocklandt et al. [55] using buccal DNA. In our MWAS for age, GRIA2 was among the top 20 most significant blocks that reached methylome-wide significance (p-value = 2.78 × 10−8). This validated the use of MBD-seq to detect association through MWAS.
Discussion
We have tested and validated the use of MBD-seq as a screening tool for MWAS. This method has the advantages of being cost effective compared with whole-genome bisulfite shotgun sequencing and, compared with methods based on enzymatic digestion, capable of assessing methylation levels across the vast majority of sites in the genome. Using a schizophrenia case–control sample of 1500 subjects, we show that we can replicate top findings from the MWAS in independent samples using a different technology that provides single base resolution. Through an MWAS of age-related methylation changes we were also able to detect a previously reported robust association involving GRIA2. This supported the potential of MBD-seq as a cost-effective tool in large-scale methylome-wide disease association studies.
Repetitive elements constitute approximately 45% of the human genome. Reads may be difficult to align to these loci because of their high sequence similarity. The methylation of repeats may be biologically meaningful. For example, transposable elements may be methylated to prevent them from copying and reinserting, thereby causing possible disruptions to functional genes. Indeed, methylation markers in repeats have been found to be associated with diseases [57–59]. Rather than simply eliminating all repeats, we used an in silico alignment study to discard only those reads with alignment problems. Results showed that reads mapped well to approximately 50% of the CpGs overlapping with repeats in RepeatMasker. We also observed alignment problems for 30% of the CpGs that were not in repeats. These findings suggest that simply excluding all CpGs in repeats may not be an optimal solution because of the loss of information and the fact that it does not solve all alignment problems.
Approximately 57.5% of the stage one blocks could be combined into stage two blocks. This suggested that larger regions of the genome can be similarly methylated, possibly indicating coordinated epigenetic regulation of gene expression. Consistent with prior observations, we found that block sizes were larger in CpG dense regions such as island and shores. A possible explanation [43] may be that above a critical CpG density, neighboring CpGs may influence each other’s DNA methylation states so that individual CpGs cannot stably maintain different states. From a biological perspective, this may serve as a bistable epigenetic switch, in which multiple CpGs collectively maintain a CpG-island-wide ‘on’ or ‘off ’ state. The possible technological implication of this observation is that the single base resolution offered by bisulfite sequencing may be less critical in CpG dense regions. That is because if CpG sites have very similar methylation status, it may not be possible to more precisely locate association signals.
To study how to best handle multireads (reads that align to multiple loci with equal or similar numbers of mismatches) and duplicate-reads (reads starting at the same base), we compared different read QC strategies. The use of particularly high-quality multireads improved data quality. This finding was robust. For example, after dichotomizing (e.g., this would eliminate possible outlier effects) the methylation measures, we observed the same pattern where the inclusion of high-quality multireads improved concordance rates between technical replicates. In addition, two other studies arrived at the same conclusion but examined the value of multireads in a different context for other outcome measures [41] [Van den Oord E, Bukszar J, Rudolf G et al. Estimation of CpG coverage in whole methylome next-generation sequencing studies (2012), Submitted]. Two additional points are worth noting. First, the majority of multi- and duplicate-reads were deleted and only a limited set of high-quality reads was retained. For example, there were several areas where high numbers of duplicate-reads mapped to heterochromatin and pericentromeric regions that are rich in satellite DNA and other repeats. An explanation is that centromeric DNA from the physical DNA template is pulled down and sequenced. However, because the reference genome is only representative of the euchromatic portion, these sequences that are in very high copy numbers in the sample align to the few sites in the reference genome where they are represented. Clearly, such reads should be eliminated prior to association testing. Second, to account for the uncertainty that you do not know from which part of the genome the multiread comes, we essentially use a simple probability estimate that the read aligns to that specific locus. Thus, multireads are treated differently compared with reads with a single best alignment.
Previous studies have suggested that 30–60 million sequenced reads per sample may be sufficient to reveal valuable information for whole-genome methylation analysis [30,40]. We obtained on average 68.0 million reads per sample, which is at the high end of these estimates. After stringent QC, 32.4 million high-quality reads (47.6%) per sample remained. The association analyses we performed on the block data where, after QC, an average of 12.9 reads covered each block. This appeared sufficient to detect methylation markers that replicated in independent samples or previously reported associations from age-related methylation studies. In studies aimed at calling DNA sequence variants the number of sequenced bases covering the target is the main determinant of accuracy. By contrast, in MBD-seq studies the number of fragments sequenced is critical because the number of fragments covering the site is used as a measure of methylation. Factors such as read length or the use of single- versus paired-end libraries are important in the sense that they improve the accuracy of the alignment. However, because the number of fragments sequenced remains unchanged, they are less critical. This has implications for the design of MBD-seq studies. Rather than increasing read length or using paired-end libraries that are more expensive and almost double the sequencing run time, it may be more efficient to maximize the number of fragments sequenced by using single-end libraries and relatively short reads.
MBD-seq has the critical advantage of being cost-effective compared with whole-genome bisulfite sequencing and even arrays. The method, however, does not provide single base resolution. Since replication is standard in high dimensional biological investigations, an obvious approach is to first use MBD-seq to screen for regions of interest and then use a technology that does provide single base resolution in the replication stage. Our results provided a proof-of-concept by showing that effects identified with MBD-seq could be replicated using a technology that provided single base resolution. In this study we used pyrosequencing for this replication. However, in principle custom array based approaches as well as approaches using targeted capture of regions of interest followed by bisulfite sequencing are becoming possible as well.
MBD-seq is semiquantitative in the sense that it does not yield direct estimates of methylation levels. For the purpose of assessing methylation levels of sites, methods have been developed to remedy this problem by normalizing the data based on CpG density [39]. These methods are, however, less relevant for MWAS where we focus on a mean difference between cases and controls. As this mean difference remains unchanged after such normalization (i.e., for each site a similar constant is added for cases and controls), these methods will not alter results. Because each site is (one average) covered by multiple reads MBD-seq is quantitative in the sense that some subjects will have zero reads covering a CpG and other subjects may have many (e.g., ‘Scores’ could range from, for example, 0–20). As there is extensive literature showing that dichotomizing quantitative variables may result in a substantial loss of statistical power [60], it seems better to analyze the data as a quantitative variable rather than as a binary variable, where data is transformed to methylation versus no methylation.
Prior to regressing out PCs, λ (the observed median test statistic value divided by the expected median assuming no effect for any site) was 7.32. We were able to replicate these pre-PCA findings in independent samples using a different technology. This suggests that these significant results were not sampling fluctuations or technical artifacts but more likely caused by the many possible differences between cases and controls (e.g., lifestyle, diet and medication use) that affect the methylome. We have seen similar dramatic effects previously when studying the methylation pattern in blood from mice treated with the antipsychotic haloperidol [Aberg K, Xie L, McClay J et al. A next-generation sequencing study to compare genome-wide methylation patterns in whole blood and brain (2012), Submitted]. In methylation studies where a limited number of sites are assayed (e.g., studying specific candidate genes), controlling for confounders using PCA or related tools is not possible. An implication is that such (candidate gene) methylation studies are at high risk for producing false discoveries. Some caution seems to be required when using tools such as PCA. If confounders can have such pervasive effects on the methylome, the pathogenic pathways that cause the disease may also affect many methylation sites. A careful inspection of the loadings generated by the PCA may be important to identify possible disease pathways and prevent such components from being regressed out in the MWAS.
The fact that we observed many effects and could replicate the two top findings with smaller samples sizes seems to suggest, that in MWAS, the challenge may not be to identify replicable effects but rather to identify those effects, among the many, that are biologically meaningful for the disease that is being studied. Data integration may be among the tools that could prove valuable to parse biologically meaningful effects. For example, confounders cannot affect sequence variation and, with proper control for population stratification, the direction of effects in GWAS is always from the SNP to the outcome. Integrating methylation and GWAS results could be useful because MWAS signals in genes also implicated by GWAS may be more likely to represent causal disease processes.
Methylation can be tissue specific and analyses are therefore ideally performed in the relevant disease tissue [8]. However, this may not always be possible. Examples are studies of psychiatric conditions where most of the pathogenic processes are likely to involve brain or clinical settings where it is important that biosamples can be collected in a multiobtrusive fashion. In these situations peripheral tissues such as (whole) blood is typically used as a surrogate. The use of surrogate tissues seems supported by studies suggesting that tissue-specific DMRs) [61,62] constitute only a limited proportion of all methylated sites. This can be explained by three possible reasons. First, peripheral tissues may reveal methylation marks predating or resulting from the epigenetic reprogramming events affecting germline and embryogenesis [63–66]. As the epigenetic profile of somatic cells is mitotically inherited, these epigenetic mutations could potentially be found in multiple tissues. Second, blood contains cells that may be modified as they circulate through diseased tissues, and can also include cell-free DNA from those tissues [67]. As such, traces of the aberrant methylation in disease-targeted regions may be present in blood. Finally, and perhaps most importantly, environmental influences such as diet, drugs and lifestyle factors, as well as genetic polymorphisms can change methylation levels [68– 71]. Although these changes may only have a functional effect in specific tissues, it is very well possible that the changes themselves are more global and cause similarities in methylation profiles across tissues. For example, after administering an antipsychotic drug to mice we observed high correlations between changes in methylation in blood, cortex and hippocampus [Aberg K, Xie L, McClay J et al. A next-generation sequencing study to compare genome-wide methylation patterns in whole blood and brain (2012), Submitted]. Interestingly, correlations between blood and brain versus the two brain tissues were very similar, suggesting that blood can predict the methylation patterns in a specific brain tissue to a similar degree as another brain tissue.
As is true for most tissues, blood is a complicated biological system consisting of a variety of cell types. This heterogeneity creates challenges for whole-methylome studies. For example, if the methylation status of a CpG differ across the various blood cell types in the same individual, interindividual differences will be reduced in whole blood as the cell type differences may average out across subjects. Another example is that diseases may alter the relative abundance of specific cell types. In the presence of heterogeneity in methylation patterns, this will produce differences between cases and controls at many CpG sites. However, as these case–control differences are related to shifts in the general cell type population, results for individual CpGs would not provide any specific biological insight into the disease process. Studying cell types adds costs and labor as whole methylome assays may need to be performed for each cell type and the cells need to be isolated. To some extent PCA or related techniques provide a statistical solution to this problem. That is because cases will have different profiles across many methylation sites, these differences will show up as a PC. If the PCs are regressed out as covariates in multiple regression analyses, association results will no longer be confounded by these general cell type differences.
A wide variety of other existing methods can, in principle, be used to analyze MWAS data (e.g., to remove batch effects [72]). However, a major practical problem is the size of the data set. We found, for example, that even calculating simple means and standard deviations using the R package was problematic. To be able to analyze the data, we needed to create new software that employs parallel computing, uses a low level programming language for central processing unit intensive calculations, stores intermediate results to avoid computing the same statistics multiple times or storing results in working memory, and uses algorithms specifically designed for high dimensional data (e.g., for the PCA). Thus, efficient analysis of MWAS data is likely to require tailored computational tools.
Conclusion
Owing to the combination of high costs of sequencing entire genomes and the large numbers of samples needed to provide adequate statistical power, whole-genome bisulfite sequencing is not currently economically feasible for disease-association studies. MBD-seq may have limitations but we demonstrate in this paper that not all limitations are relevant for MWAS (e.g., difficult to estimate actual methylation levels) and that others (e.g., batch effects) can be remedied. As only a small proportion of the genome (e.g., CpGs constitute only approximately 1% of the genome) can be methylated, MBD-seq is very cost effective as it essentially sequences only this potentially methylated proportion while still providing comprehensive coverage of the methylome. Furthermore, because short range correlations in the methylation status of CpGs seem pervasive, in many cases it may not be possible to improve the resolution obtained with MBD-seq owing to these biological constraints. These properties make MBD-seq a very efficient screening tool for large-scale methylome-wide disease studies to identify differentially methylated regions that can then be followed up with more targeted studies.
Future perspective
Because of the promise of methylation studies and the fact that such studies are possible using biomaterial that is easy to collect, MWAS are likely to become a standard tool to study complex diseases. Owing to their cost–effectiveness, affinity-based capture followed by massively parallel sequencing is likely to become a commonly used method.
Supplementary Material
Executive summary.
Methylation studies are a promising complement to genetic studies of variation in DNA sequence because methylation may capture additional individual variation in disease susceptibility, can account for a wide variety of phenomena that characterize complex diseases, and methylation sites are potential new drug targets plus have good properties from a translational perspective.
We studied the use of methyl-CpG binding domain protein-enriched genome sequencing as a cost-effective screening tool for methylome-wide association studies (MWAS).
Because methyl-CpG binding domain protein-enriched genome sequencing has not yet been applied on a large scale, we first developed and tested a pipeline for data processing using 1500 schizophrenia cases and controls plus 75 technical replicates with an average of 68 million reads/sample.
Optimization of the data analysis pipeline included the use of technical replicates to optimize quality control for multi- and duplicate-reads, an in silico experiment to identify CpGs in loci with alignment problems, CpG coverage calculations based on multiparametric estimates of the fragment size distribution, a two stage adaptive algorithm to combine data from correlated adjacent CpG sites and principal component analyses to control for confounders.
We replicated MWAS findings in independent samples using a different technology that provided single base resolution.
In an MWAS of age-related methylation changes one of our top findings was a previously reported robust association involving GRIA2.
Our results also suggested that due to the many confounding effects, a considerable challenge in MWAS is to identify those effects that are informative about disease processes.
Acknowledgments
Institutions involved in this project are: Karolinska Institutet, University of North Carolina at Chapel Hill, Virginia Commonwealth University, Broad Institute and the US National Institute of Mental Health. The authors are indebted G Rudolf for suggesting the ‘in silico’ experiment to identify alignment problems. MBD-enrichment, library construction and next-generation sequencing was carried out by EdgeBio, MD, USA.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
This work was supported by the National Institutes of Mental Health (RC2MH089996), and it is part of a larger project entitled ‘A Large-Scale Schizophrenia Association Study in Sweden’ that is supported by grants from NIMH (MH077139) and the Stanley Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
- 1.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 2.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 4.Kwok JB. Role of epigenetics in Alzheimer’s and Parkinson’s disease. Epigenomics. 2010;2:671–682. doi: 10.2217/epi.10.43. [DOI] [PubMed] [Google Scholar]
- 5.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol. 2010;7:510–519. doi: 10.1038/nrcardio.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkies P, Sale JE. Cellular epigenetic stability and cancer. Trends Genet. 2012;28:118–127. doi: 10.1016/j.tig.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 9.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology – analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 11.Jost JP, Saluz HP, Pawlak A. Estradiol down regulates the binding activity of an avian vitellogenin gene repressor (MDBP-2) and triggers a gradual demethylation of the mCpG pair of its DNA binding site. Nucleic Acids Res. 1991;19:5771–5775. doi: 10.1093/nar/19.20.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokomori N, Moore R, Negishi M. Sexually dimorphic DNA demethylation in the promoter of the Slp (sex-limited protein) gene in mouse liver. Proc Natl Acad Sci USA. 1995;92:1302–1306. doi: 10.1073/pnas.92.5.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland JE, Costa M. Epigenetics and the environment. Ann NY Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 14.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Cooney CA. Are somatic cells inherently deficient in methylation metabolism? A proposed mechanism for DNA methylation loss, senescence and aging. Growth Dev Aging. 1993;57:261–273. [PubMed] [Google Scholar]
- 16.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 17.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 18▪.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. Paper discussing methylome-wide association studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Zhu J, Tian G, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aberg K, Khachane AN, Rudolf G, et al. Methylome-wide comparison of human genomic DNA extracted from whole blood and from EBV-transformed lymphocyte cell lines. Eur J Hum Genet. 2012;20(9):953–955. doi: 10.1038/ejhg.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪.Serre D, Lee BH, Ting AH. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. Paper discussing methyl-CpG binding domain protein-enriched genome sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M, Greally JM. DNA methylation profiling using HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) Methods. 2010;52:218–222. doi: 10.1016/j.ymeth.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 29▪.Nair SS, Coolen MW, Stirzaker C, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. Paper comparing affinity-based capture methods. [DOI] [PubMed] [Google Scholar]
- 30.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogart A, Lichtenberg J, Ajay SS, Anderson SM, Margulies EH, Bodine DM. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal over-representation of ETS transcription factor binding sites. Genome Res. 2012;22(8):1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan X, Adams C, Landers M, et al. High resolution detection and analysis of CpG dinucleotides methylation using MBD-Seq technology. PLoS One. 2011;6:e22226. doi: 10.1371/journal.pone.0022226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergen SE, O’Dushlaine CT, Ripke S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared to bipolar disorder. Mol Psychiatry. 2012;17(9):880–886. doi: 10.1038/mp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study of schizophrenia identifies five novel loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKernan KJ, Peckham HE, Costa GL, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19(9):1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6(11 Suppl):S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavez L, Jozefczuk J, Grimm C, et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20(10):1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji Y, Xu Y, Zhang Q, et al. BM-Map: Bayesian mapping of multireads for next-generation sequencing data. Biometrics. 2011;67(4):1215–1224. doi: 10.1111/j.1541-0420.2011.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bollen KA. Structural Equations With Latent Variables. Wiley; NY, USA: 1989. [Google Scholar]
- 43.Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36(10):e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97(18):10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 47▪.Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–338. Paper proposing an algorithm that makes principle component analysis on methylome-wide association studies possible. [Google Scholar]
- 48.Pelizzola M, Koga Y, Urban AE, et al. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18(10):1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen J. Statistical Power Analysis for the Behavioral Sciences. Erlbaum; NJ, USA: 1988. [Google Scholar]
- 50.Beleford D, Liu Z, Rattan R, et al. Methylation induced gene silencing of HtrA3 in smoking-related lung cancer. Clin Cancer Res. 2010;16(2):398–409. doi: 10.1158/1078-0432.CCR-09-1677. [DOI] [PubMed] [Google Scholar]
- 51.Lohr JB, Flynn K. Smoking and schizophrenia. Schizophr Res. 1992;8(2):93–102. doi: 10.1016/0920-9964(92)90024-y. [DOI] [PubMed] [Google Scholar]
- 52.Kristiansson K, Perola M, Tikkanen E, et al. Genome-wide screen for metabolic syndrome susceptibility loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5(2):242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalo S. Epigenetic alterations in aging. J Appl Physiol. 2010;109(2):586–597. doi: 10.1152/japplphysiol.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koch CM, Wagner W. Epigenetic-aging-signature to determine age in different tissues. Aging (Albany NY) 2011;3(10):1018–1027. doi: 10.18632/aging.100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bocklandt S, Lin W, Sehl ME, et al. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell JT, Tsai PC, Yang TP, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8(4):e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ushida H, Kawakami T, Minami K, et al. Methylation profile of DNA repetitive elements in human testicular germ cell tumor. Mol Carcinog. 2011;51(9):711–722. doi: 10.1002/mc.20831. [DOI] [PubMed] [Google Scholar]
- 58.Bollati V, Galimberti D, Pergoli L, et al. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav Immun. 2011;25(6):1078–1083. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bollati V, Fabris S, Pegoraro V, et al. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis. 2009;30(8):1330–1335. doi: 10.1093/carcin/bgp149. [DOI] [PubMed] [Google Scholar]
- 60.Cohen J. The cost of dichotomization. Applied Psychological Measurement. 1983;7:249–253. [Google Scholar]
- 61.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 64.Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994;4(2):265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 65.Yeivin A, Razin A. Gene methylation patterns and expression. EXS. 1993;64:523–568. doi: 10.1007/978-3-0348-9118-9_24. [DOI] [PubMed] [Google Scholar]
- 66.Rakyan VK, Preis J, Morgan HD, Whitelaw E. The marks, mechanisms and memory of epigenetic states in mammals. Biochem J. 2001;356(Pt 1):1–10. doi: 10.1042/0264-6021:3560001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lavon I, Refael M, Zelikovitch B, Shalom E, Siegal T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro Oncol. 2010;12(2):173–180. doi: 10.1093/neuonc/nop041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGowan PO, Meaney MJ, Szyf M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Res. 2008;1237:12–24. doi: 10.1016/j.brainres.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pilsner JR, Liu X, Ahsan H, et al. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86(4):1179–1186. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 70.Sutherland JE, Costa M. Epigenetics and the environment. Ann NY Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 71.Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40(7):904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 72.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.Center for Biomarker Research and Personalized Medicine. www.biomarker.vcu.edu.
- 102.UCSC Genome Bioinformatics. http://genome.ucsc.edu.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.