The authors show by thermal denaturation and SHAPE studies that tRNA folding cooperativity increases in physiologically low concentrations of Mg2+ (0.5–2 mM) and K+ (140 mM) if the solution is supplemented with physiological amounts (∼20%) of a water-soluble neutral macromolecular crowding agent such as PEG or dextran. Low-molecular-weight cosolutes show varying effects on tRNA folding cooperativity, increasing or decreasing it based on the identity of the cosolute.
Keywords: folding cooperativity, tRNA, macromolecular crowding, biological cosolutes, temperature-dependent structure mapping
Abstract
Folding mechanisms of functional RNAs under idealized in vitro conditions of dilute solution and high ionic strength have been well studied. Comparatively little is known, however, about mechanisms for folding of RNA in vivo where Mg2+ ion concentrations are low, K+ concentrations are modest, and concentrations of macromolecular crowders and low-molecular-weight cosolutes are high. Herein, we apply a combination of biophysical and structure mapping techniques to tRNA to elucidate thermodynamic and functional principles that govern RNA folding under in vivo–like conditions. We show by thermal denaturation and SHAPE studies that tRNA folding cooperativity increases in physiologically low concentrations of Mg2+ (0.5–2 mM) and K+ (140 mM) if the solution is supplemented with physiological amounts (∼20%) of a water-soluble neutral macromolecular crowding agent such as PEG or dextran. Low-molecular-weight cosolutes show varying effects on tRNA folding cooperativity, increasing or decreasing it based on the identity of the cosolute. For those additives that increase folding cooperativity, the gain is manifested in sharpened two-state-like folding transitions for full-length tRNA over its secondary structural elements. Temperature-dependent SHAPE experiments in the absence and presence of crowders and cosolutes reveal extent of cooperative folding of tRNA on a nucleotide basis and are consistent with the melting studies. Mechanistically, crowding agents appear to promote cooperativity by stabilizing tertiary structure, while those low molecular cosolutes that promote cooperativity stabilize tertiary structure and/or destabilize secondary structure. Cooperative folding of functional RNA under physiological-like conditions parallels the behavior of many proteins and has implications for cellular RNA folding kinetics and evolution.
INTRODUCTION
The last decade has witnessed remarkable advances in understanding RNA folding from a physical–chemical perspective. Tertiary folding of complex RNAs begins with rapid electrostatic collapse, followed by search for the native state (Thirumalai et al. 2001; Russell et al. 2002; Moghaddam et al. 2009). This occurs on a rough landscape (Woodson 2010; Takamoto et al. 2012) wherein misfolds are broken prior to native folding (Thirumalai et al. 2001). These features can lead to very slow (minutes to hours) and multiphasic RNA folding (Zarrinkar and Williamson 1994; Sclavi et al. 1998; Swisher et al. 2002). Advances have been made in predicting RNA secondary and tertiary structures: Thermodynamic parameters have been measured for many motifs (Mathews and Turner 2006), and structure predictions have been interfaced with experimental techniques such as NMR (Hart et al. 2008) and SHAPE (Wilkinson et al. 2008; Deigan et al. 2009) to improve prediction of secondary structure. While the above studies contribute greatly to establishing physical principles of RNA folding, almost all have been conducted under “standard literature conditions” of dilute solution and folding-favorable nonphysiological ionic conditions of ∼10 mM Mg2+ or 1 M Na+/K+.
Physiological conditions are quite different. Typical K+ concentrations in prokaryotic and eukaryotic cells are only ∼140 mM, while free Mg2+ concentrations are just 1.5–3.0 mM in prokaryotic cells (Lusk et al. 1968; Truong et al. 2013) and 0.5–1.0 mM in eukaryotic cells (London 1991; Alberts et al. 1994; Feig and Uhlenbeck 1999; Grubbs 2002; Romani 2007). Eukaryotic and prokaryotic cells also contain 20%–40% (w/v) macromolecules, as well as lower-molecular-weight cosolutes, which can exclude volume, interact with the RNA, and lead to altered solvent properties (Ellis 2001; Minton 2001). Differences between standard literature folding conditions and in vivo folding conditions have the potential to profoundly affect RNA folding mechanisms, which is the focus of this study.
Steric crowding and variation of solvent properties via a variety of water-soluble neutral cosolutes, including polyethyleneglycol (PEG) and dextran, have been shown to affect, and in many cases promote, the stability and function of proteins (Zhou et al. 2008). These species also affect RNA folding. They promote ribozyme cleavage (Nakano et al. 2009; Strulson et al. 2012, 2013), drive compaction of large RNAs (Kilburn et al. 2010), and affect folding free energy (Spink and Chaires 1998; Lambert and Draper 2007; Spink et al. 2007; Feng et al. 2010; Denesyuk and Thirumalai 2011; Kilburn et al. 2013). While these studies provide insight into ways in which biological conditions affect RNA folding and function, there has been little focus on how they affect RNA folding cooperativity.
Folding of RNA is a largely hierarchical process (i.e., secondary then tertiary structure formation) (Brion and Westhof 1997; Leontis et al. 2006; Greenleaf et al. 2008) because most tertiary structure assembles from preformed secondary structures. For example, tRNA folds through five intermediates involving combinations of four different helices prior to formation of tertiary structure (Riesner et al. 1973; Crothers et al. 1974), while the HDV ribozyme folds through numerous base-pairing states and misfolds (Isambert and Siggia 2000; Chadalavada et al. 2002; Brown et al. 2004). Some variations in overall RNA folding mechanisms have been reported, e.g., tertiary structure or proteins driving native secondary structure (Thirumalai 1998; Wu and Tinoco 1998; Duncan and Weeks 2010; Woodson 2010); however, even in these cases, native secondary structure typically precedes tertiary structure. Although helical intermediates do exist in the folding pathway for functional RNAs, it remains unclear if they populate appreciably. Dill defined a cooperatively folding system as one in which obligate folding intermediates are poorly populated, or “hidden” (Ozkan et al. 2002; Weikl et al. 2004). If secondary structural helical intermediates are intrinsically unstable and only the initial and final states populate, then folding is defined as apparent two-state (all-or-none) and fully cooperative. On the other hand, a high population of intermediates leads to noncooperative folding (Kwok et al. 2013). In the present study, we probe the folding cooperativity of a functional RNA under biological solution conditions.
To assess the effects of molecular crowders and cosolutes on RNA folding cooperativity, we chose to study the folding of tRNAPhe (Fig. 1A). This choice was made for several reasons. First, the structure, folding, and function of tRNA has been characterized in the literature, with many studies detailing the folding of tRNA under standard literature folding conditions (Yang et al. 1972; Stein and Crothers 1976a,b; Fang et al. 2000; Misra and Draper 2000; Pulukkunat and Gopalan 2008). Second, tRNA has a well-folded native tertiary structure that is relevant to many functional RNAs in eukaryotic and prokaryotic cells. Third, the T7 transcript of tRNA has similar properties as the naturally occurring modified RNA (Sampson and Uhlenbeck 1988; Nobles et al. 2002; Wilkinson et al. 2005; Whitman 2011) but is easily mutated to provide weakened tertiary structure so that effects of molecular crowders and cosolutes on secondary and tertiary structure individually can be assessed (Fig. 1B). The tRNA chosen is unmodified tRNAPhe from yeast, which was selected because the base-pairing in the various helices has a relatively even distribution of GC and AU/GU base pairs. The latter was important so as not to bias the RNA toward or away from cooperative folding of secondary and tertiary structure.
FIGURE 1.
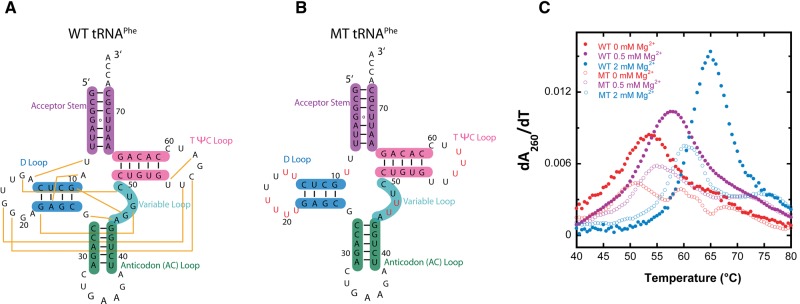
Secondary structure and first derivative melt curves of WT and MT tRNAPhe. (A) Secondary structure of wild-type (WT) tRNAPhe. WT tRNAPhe was prepared by in vitro T7 transcription. (B) Secondary structure of mutationally weakened tertiary structure tRNAPhe, referred to as MT tRNAPhe. (Red) Nucleotides mutated to remove tertiary contacts. (C) First derivative melt curves parameteric in Mg2+concentration for WT tRNAPhe (closed symbols) and MT tRNAPhe (open symbols).
Herein, we reveal that macromolecular crowding agents promote RNA folding cooperativity at biological concentrations of magnesium. Conversely, low-molecular-weight cosolutes either promote or diminish folding cooperativity in a manner dependent on the identity of the cosolute. We show that crowding agents promote cooperativity by stabilizing tertiary structure, while those low molecular cosolutes that promote cooperativity do so by stabilizing tertiary structure and/or destabilizing secondary structure. Temperature-dependent SHAPE analysis reveals RNA unfolding properties on a nucleotide basis and confirms increase in cooperativity in the presence of crowder and certain cosolutes.
RESULTS
Folding cooperativity of wild-type tRNAPhe in the absence of additives
We first evaluated the folding cooperativity of tRNAPhe in 0, 0.5, and 2 mM Mg2+ in the absence of crowding and cosolute additives, all in the background of 140 mM KCl. We chose to evaluate RNA folding in 0.5 and 2 mM Mg2+, as these are within the range of free Mg2+ concentrations reported in eukaryotic and prokaryotic cells, respectively, and in 150 mM K+, as this is near physiological (Lusk et al. 1968; London 1991; Alberts et al. 1994; Feig and Uhlenbeck 1999; Grubbs 2002; Romani 2007; Truong et al. 2013). Additionally, we examined folding in 140 mM KCl without Mg2+ added in order to later determine if molecular crowders and cosolutes could promote folding cooperativity in the absence of divalent ions.
We began by performing melts of the wild-type (WT) tRNAPhe in 0, 0.5, and 2 mM Mg2+ in the absence of additives. Melting data are provided in Figure 1C (closed symbols) and summarized in Table 1. In general, a cooperative melt is expected to display a sharpened two-state-like unfolding transition with the absence of shoulders, while a less cooperative or noncooperative melt is expected to display a broad unfolding transition where multiple shoulders may be observed. With no Mg2+ present, WT tRNAPhe has a broad melting profile that contains a noticeable shoulder and thus consists of at least two transitions (Fig. 1C). These properties are indicative of multiple folding intermediates and little to no folding cooperativity. This outcome is consistent with significant heterogeneity in the structure, which is expected with no divalent ions and only relatively low monovalent ions. In 0.5 mM Mg2+, the major unfolding transition shifts to higher temperature, from 53.5°C to 57.5°C, and is sharpened, as evidenced by narrowing and heightening of the transition. Indeed, the max dA/dT of the derivative plot increases from 0.0084 to 0.0106 (Table 1); nonetheless, the transition is not highly cooperative, as evidenced in part by the long tail remaining in the transition (Fig. 1C). In the presence of 2 mM Mg2+, there is just a single significant unfolding transition, which is noticeably cooperative when compared with the unfolding transitions in 0 and 0.5 mM Mg2+. The unfolding transition is shifted to much higher temperatures, from 57.5°C in 0.5 mM Mg2+ to 65.0°C in 2 mM Mg2+, and is much steeper, with a max dA/dT value of 0.0154 as compared with 0.0106 in 0.5 mM Mg2+. These observations are consistent with earlier studies, which demonstrated that the unfolding of tRNAfMet is multistate at 0.5 mM Mg2+ but approximately two-state in 3 mM Mg2+ (Stein and Crothers 1976a).
TABLE 1.
Tm and max dA/dT values for WT and MT tRNAPhe unfolding with increasing Mg2+ concentration
Impact of macromolecular crowders on the folding cooperativity of wild-type tRNAPhe
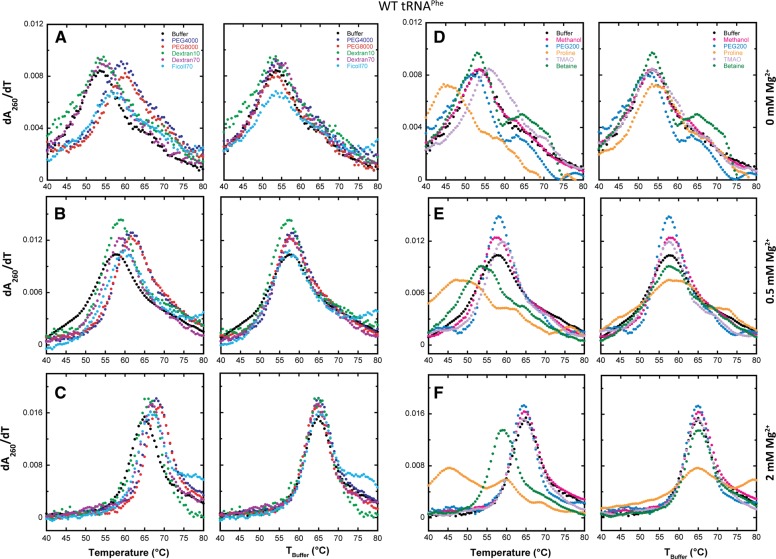
Having evaluated the impact of magnesium ion concentration on the folding of tRNAPhe in the absence of additives, we describe the effects of macromolecular crowders on RNA folding. We expected that the crowders would promote RNA folding cooperativity since they favor a more compact form of the RNA. The molecular crowders—PEG4000, PEG8000, Dextan10, Dextran70, and Ficoll70 with molecular weights of 4 kDa, 8 kDa, 8 kDa, 10 kDa, 70 kDa, and 70 kDa, respectively—were chosen because they allow exploration of different chemical compositions and sizes. Concentrations of these were 20% (w/v) to approximate the lower estimates of crowding in vivo (Minton 2001). Melting experiments on the WT tRNAPhe in the presence of the macromolecular crowders were performed at 0, 0.5, and 2 mM Mg2+. These data are provided in Figure 2 both as a function of actual temperature and as a function of an offset temperature referred to as “TBuffer,” in which a constant was added to or subtracted from the actual temperature to give a Tm matched to the tRNA's Tm in buffer alone. This analysis was implemented to align the various melting maxima, facilitate comparison of melts, and aid generation of difference plots. Additionally, this treatment decouples thermostability and cooperativity, as they do not always parallel each other.
FIGURE 2.
Melts of WT tRNAPhe in additives with increasing Mg2+ concentration. All melts were performed in the background of 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl. (A–C, both columns) The effects of various crowding agents (20% [w/v]) in 0, 0.5, and 2 mM Mg2+, respectively. No additive (black), PEG4000 (blue), PEG8000 (red), Dextran10 (green), Dextran70 (purple), and Ficoll70 (cyan). (D–F, both columns) The effects of low-molecular-weight cosolutes (2 m, except PEG200, which was 20% [w/v]) in 0, 0.5, and 2 mM Mg2+, respectively. No additive (black), methanol (pink), PEG200 (cobalt blue), proline (orange), TMAO (light purple), and betaine (dark green). In certain panels, data were adjusted to have the same Tm as the buffer, designated as “TBuffer,” so that the effects on cooperativity are clear.
In the absence of magnesium, the crowding agents did not have a significant effect on folding cooperativity. For example, the max dA/dT in PEG8000 and Ficoll70 are somewhat lower than in buffer alone, while for PEG4000, Dextran10, and Dextran70, the max dA/dT values are slightly elevated above those in buffer alone (Fig. 2A; Table 1). The lack of any appreciable gains by the crowders on cooperativity in the absence of Mg2+ is visualized in the bar graphs in Figure 3A (left set of bars), where the max dA/dT for the first (i.e., lowest temperature) unfolding transition has been plotted as a function of additive.
FIGURE 3.
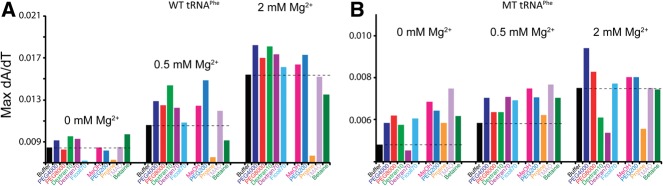
Max dA/dT plot of WT and MT tRNAPhe with crowding and low-molecular-weight cosolute agents in increasing Mg2+ concentration. Max dA/dT values were calculated for the first (i.e., lowest temperature) unfolding transition from the first derivative melt curves for (A) WT and (B) MT. Experimental conditions and coloring of plots are identical to those in Figure 2. The black dashed line across each subset of conditions shows the max dA/dT in buffer alone for that condition. Bars that extend greater than the dashed line indicate an increase in folding cooperativity. Note the difference in the y-axis scales for panels A and B.
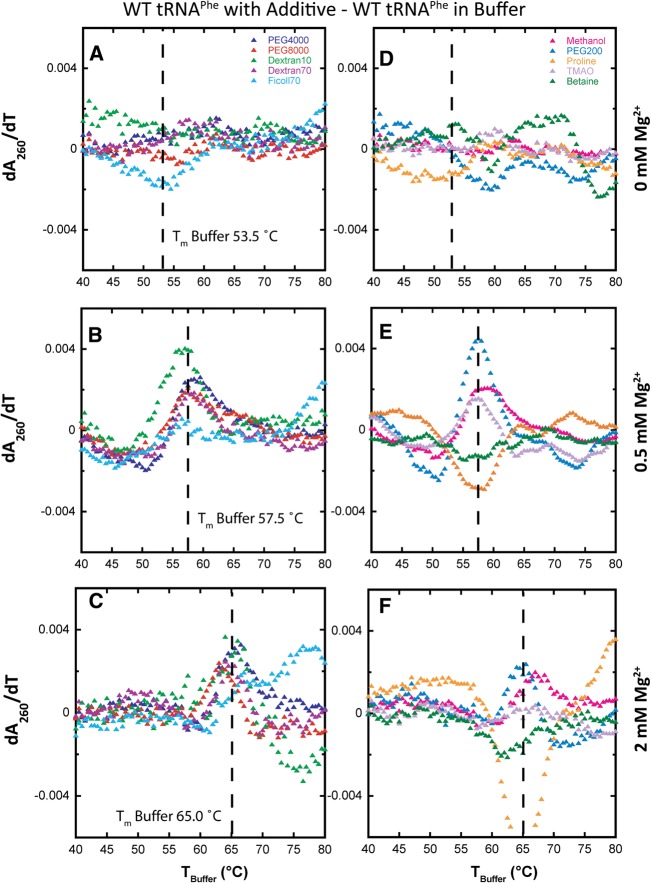
Difference plots show melt data with additive present minus melt data in buffer alone and help reveal additive-specific effects (Fig. 4). For generation of these plots, the melt data as a function of TBuffer were used. Difference melts in the absence of Mg2+ are relatively flat (Fig. 4A), consistent with a minimal effect on cooperativity in the absence of divalent ions. These plots are flat because the parent melts with and without additive (Fig. 2A, right panel) overlay very closely for 0 mM Mg2+, with a small difference observed for Ficoll70.
FIGURE 4.
Difference plots of melts of WT tRNAPhe in additives minus in buffer alone with increasing Mg2+ concentration. All melts were performed in the background of 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl. All data were adjusted to have the same Tm, designated as “TBuffer.” (A–C) The effects of various crowding agents (20% [w/v]) in 0, 0.5, and 2 mM Mg2+, respectively. PEG4000 (blue), PEG8000 (red), Dextran10 (green), Dextran70 (purple), and Ficoll70 (cyan). (D–F) The effects of low-molecular-weight cosolutes (2 m, except PEG200, which was 20% [w/v]) also in 0, 0.5, and 2 mM Mg2+, respectively. Methanol (pink), PEG200 (cobalt blue), proline (orange), TMAO (light purple), and betaine (dark green). Difference plots show the effects of the additives on WT folding cooperativity.
In the presence of 0.5 mM Mg2+, the crowding agents lead to a substantial increase in RNA folding cooperativity (Fig. 2B). Remarkably, every crowding agent tested led to an increase in the max dA/dT value from that of buffer alone (Fig. 3A; Table 1) and to sharpening of the folding transition. When melt data in the presence of additive were again offset to TBuffer, it is evident that the crowder melts have higher dA/dT max than buffer alone (Fig. 2B, right panel). This effect is accentuated in the difference melts presented in Figure 4B, where there are distinct bell-shaped derivate plots in the presence of all crowders, with a positive node centered at the Tm in the presence of buffer denoted “Tm,Buffer.” Additionally, there is a negative feature that precedes the positive node for several crowders leading to a so-called differential feature (Fig. 4B; Gong et al. 2008). The negative feature arises because the melting of the tRNA in the presence of the crowder is over a narrower temperature range, reflective of the sharpening of a transition as associated with RNA folding cooperativity.
In the presence of 2 mM Mg2+, all crowders promote folding cooperativity (Fig. 2C). Every crowding agent again leads to an increase in the value of max dA/dT over that in buffer alone (Fig. 3A; Table 1) and to sharpening of the folding transition. Examination of the difference melts in Figure 4C reveals bell-shaped derivative plots, similar to observations in 0.5 mM Mg2+. Taken together, the 0.5 and 2 mM Mg2+ tRNA unfolding data indicate that biological magnesium concentrations allow folding cooperativity to be enhanced by crowding agents.
Next, we examined the apparent enthalpy change for the first unfolding transition in the melt because steeper (i.e., more cooperative) melting transitions are typically associated with greater enthalpy changes (Puglisi and Tinoco 1989). We fit the melting data to a two-state model to extract an apparent van't Hoff enthalpy, ΔHVH. In several cases, the melting is clearly non-two-state, as discussed above; in these instances, we fit the first (i.e., lowest melting) apparent unfolding transition. The rationale for treating the data in this manner is that the apparent ΔHVH from such a fit serves as a marker of the number of RNA interactions that unfold in the first transition and so describes in a semi-quantitative manner the degree of folding cooperativity. Values for ΔHVH and ΔHVH/ΔHVH,buffer, which provide a comparison to buffer alone, are provided in Supplemental Table S1. In 0 mM Mg2+, nearly all crowders led to a ΔHVH for unfolding that is smaller in magnitude than in the absence of additive. This is revealed in relative ΔHVH/ΔHVH,buffer factors that are less than unity, ranging from 0.7 to 0.8. These data provide no evidence for enhancement of folding cooperativity by the additive in the absence of Mg2+, consistent with the analysis above.
We then extended this analysis to melts in the background of 0.5 mM Mg2+. The value of ΔHVH in the absence of additive increased from 39.7 to 47.6 kcal/mol upon increasing the Mg2+ from 0 to 0.5 mM, supporting a gain in folding cooperativity in the presence of 0.5 mM Mg2+ over no Mg2+, consistent with the analysis above (Supplemental Table S1). Remarkably, in 0.5 mM Mg2+, all of the crowders led to a ΔHVH for unfolding that is greater in magnitude than in buffer alone, which is a trend opposite to that in the 0 mM Mg2+ background. This effect is clearly revealed in ΔHVH/ΔHVH,buffer ratios that are substantially greater than unity, ranging from 1.1 to 1.4. This observation supports the above analysis that additives increase cooperativity in the background of 0.5 mM Mg2+.
Evaluation of the 2 mM Mg2+ melt data reveals ΔHVH/ΔHVH,buffer ratios that are greater than unity, again supporting the conclusion that most crowders promote cooperativity. The effect, however, is somewhat diminished relative to 0.5 mM Mg2+ effects, with maximal ΔHVH/ΔHVH,buffer ratios near 1.1 rather than 1.4. This smaller effect may arise because the cooperativity has already been enhanced by the higher Mg2+.
Impact of low-molecular-weight cosolutes on the folding cooperativity of wild-type tRNAPhe
We next consider the effect of various low-molecular-weight cosolutes on RNA folding cooperativity. These measurements were conducted on tRNAPhe in the same Mg2+ concentrations of 0, 0.5, and 2 mM Mg2+ in a background K+ of 140 mM. Methanol, PEG200, proline, TMAO, and betaine were chosen for the low-molecular-weight cosolutes since they have been shown to stabilize or destabilize RNA, and in cases of proline, TMAO, and betaine are biologically relevant (Di Domenico and Lavecchia 2002; Lambert and Draper 2007; Nakano et al. 2009; Knowles et al. 2011). Concentrations of these were chosen to be 2 m, which is similar to concentrations used in other RNA folding studies (Lambert and Draper 2007).
We analyzed tRNA melting in the presence of cosolutes in 0, 0.5, and 2 mM Mg2+ (Fig. 2D). In the absence of Mg2+, the small-molecule cosolutes do not have a significant effect on cooperativity, similar to that observed with the macromolecular crowders. For example, in the absence of added Mg2+, the max dA/dT is quite similar in buffer alone and in each of the cosolutes with at least two transitions present in the presence of each cosolute (Table 1). This is also apparent in the plot of dA/dT maximum versus additive (Fig. 3A). In addition, the difference plots offset to the buffer-alone temperature show no distinct features (Fig. 4D), supporting no clear effect of cosolutes on folding cooperativity in the absence of divalent ions. We note that cosolutes lead both to increases and decreases in thermostability in the presence of the additive, which is distinct from the effect of macromolecular crowders, none of which decrease thermostability (Table 1).
In the presence of 0.5 mM Mg2+, substantial effects of cosolute on RNA folding cooperativity are observed (Fig. 2E). As discerned from effects on the value of max dA/dT, PEG200, methanol, and TMAO all enhance folding cooperativity, while proline and betaine decrease folding cooperativity (Table 1, cf. max dA/dT values to buffer). These effects are especially apparent in the difference melts in Figure 4E. In the case of PEG200, methanol, and TMAO, their difference plots are bell-shaped with a positive feature centered at Tm,Buffer and the max dA/dT greatest for PEG200. Moreover, the low-temperature negative differential feature is especially distinct for PEG200, which also shows a negative feature at higher temperature. The two negative features arise because melting of tRNA in the presence of PEG200 is especially sharp, reflecting enhanced folding cooperativity. In contrast, in proline and betaine, reverse effects are seen. The bell-shaped difference plots for betaine and especially proline are approximately mirror images of those for PEG200, methanol, and TMAO, in which the largest magnitude node is the negative node (Fig. 4E). This observation is consistent with a decrease in folding cooperativity arising from these additives. Last, in the presence of 2 mM Mg2+, a gain in folding cooperativity still occurs upon addition of PEG200, methanol, and TMAO, but the stimulation is diminished from what it was in 0.5 mM Mg2+, likely due to the same reasons discussed above for macromolecular crowders.
Consideration of enthalpy changes in the presence of cosolute strengthens the conclusions reached above. In particular, ΔHVH/ΔHVH,buffer factors are ≤1 in the absence of Mg2+, providing no evidence for cooperativity enhancement under these conditions (Supplemental Table S1). In the presence of 0.5 mM Mg2+, ΔHVH/ΔHVH,buffer factors range from 1.2 to 1.5 for PEG200, methanol, and TMAO, consistent with the abovementioned gains in folding cooperativity, while ΔHVH/ΔHVH,buffer factors are 0.7 and 0.9 for proline and betaine, supporting the decrease in folding cooperativity in these cosolutes. In 2 mM Mg2+, the ΔHVH/ΔHVH,buffer factors are only 1.1–1.2 for PEG200, methanol, and TMAO, again consistent with smaller gains in folding cooperativity in higher Mg2+. Proline and betaine ratios of ΔHVH/ΔHVH,buffer remain similarly destabilizing at 0.6 and 0.9, respectively.
It is of interest to note that for the macromolecular crowders, increases in cooperativity were generally associated with increases in thermostability. For instance, thermostability increased in the presence of all crowders in 0, 0.5, and 2.0 mM Mg2+ by an average of 2.7°C, and by as much as 6.5°C. However, this trend does not hold for low-molecular-weight cosolutes, which had little or no effect on thermostability, except for proline and betaine, which decreased it. The differential effects of crowders and cosolutes on thermostability appear to be associated with their impacts on secondary and tertiary structure (see Discussion).
Finally, we conducted melts of tRNA in 10 mM Mg2+ in representative macromolecular crowders and low-molecular-weight cosolutes. These experiments were carried out in order to provide a comparison between the effects of physiological Mg2+ concentrations and folding-favorable, nonphysiological Mg2+ concentrations on RNA folding, as well as to test the effects of crowder and cosolute additives in these two Mg2+ backgrounds. These data can be found in Supplemental Figure S1. While crowders enhanced thermostability at biological Mg2+ concentrations, as discussed above, they had no appreciable effect on thermostability in the background of 10 mM Mg2+. This is consistent with the notion that effects of additives are lessened in nonphysiologically high Mg2+ concentrations. On the other hand, the crowders did enhance folding cooperativity slightly in both Mg2+ concentrations.
Low molecular crowders had differential effects on thermostability at biological Mg2+, as discussed above; however, they all decreased thermostability in the background of 10 mM Mg2+ (Supplemental Fig. S1). Likewise, while some cosolutes enhanced folding cooperativity and some decreased cooperativity at biological Mg2+ concentrations, in 10 mM Mg2+ slight enhancements or no effects on folding cooperativity were found. Overall, the effects of additives in 10 mM Mg2+ provide some biophysical insight for understanding how additives affect RNA folding; however, we focus herein on the effects of additives under biological Mg2+ concentrations.
Impact of crowders and cosolutes on the folding cooperativity of mutated tertiary tRNAPhe
In an effort to dissect effects of additives on RNA secondary and tertiary structure formation, we prepared a mutated tertiary structure tRNAPhe, referred to as MT, in which key tertiary interactions were abolished by changing D loop, variable loop, and TΨC loop residues to Us (Fig. 1B). The tertiary interactions targeted are represented by yellow lines in the WT secondary structure (Fig. 1A). The folding behavior of MT tRNAPhe was examined through perpendicular temperature-gradient gel electrophoresis (TGGE) (Bevilacqua and Bevilacqua 1998; Chadalavada and Bevilacqua 2009) displayed in Figure 5. The MT tRNAPhe migrates slower than WT at low temperatures, has a single lower-melting unfolding transition associated with loss of secondary structure, and then migrates identically to WT at higher temperatures. Slower migration than WT at low temperature is consistent with less compaction in MT and thus removal of much of the tertiary structure, while comigration with WT at high temperature supports identical unfolded structures for MT and WT.
FIGURE 5.
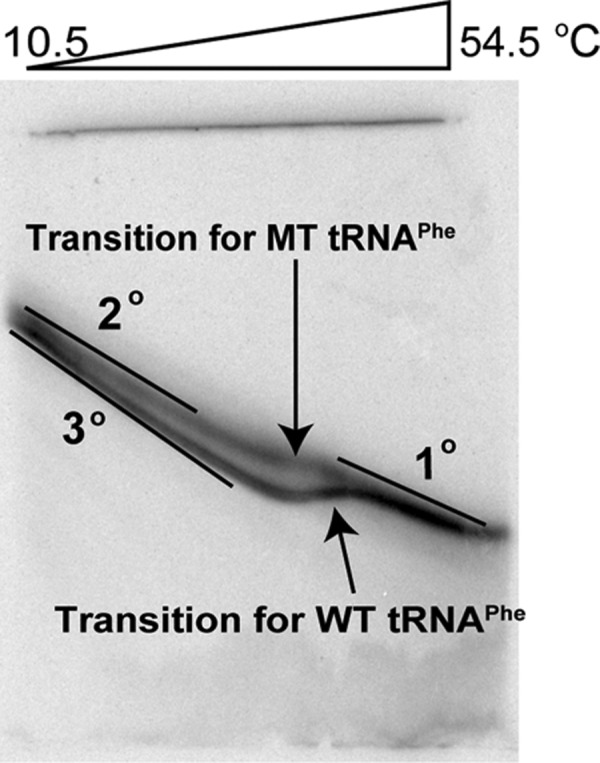
Perpendicular TGGE plot for WT and MT tRNAPhe melting transitions. Electrophoresis is 13% PAGE and 1× THEN10M0.9 (pH 7.5) (= 33 mM Tris, 66 mM HEPES, 0.1 mM EDTA, 10 mM NaCl, 0.9 mM MgCl2) and 4 M urea (to facilitate melting in the TGGE range). Baselines slant to the bottom right-hand corner owing to the temperature gradient, which was also observed in the blue tracking dyes (data not shown).
We first tested MT tRNAPhe for any folding cooperativity by conducting melts in 0, 0.5, and 2 mM Mg2+ in the absence of additives in the background of 140 mM KCl (Fig. 1C, open symbols). In the absence of Mg2+, MT tRNAPhe has a very broad melting profile that consists of at least three very distinct transitions with their own local maximum. In comparison to WT, MT tRNAPhe melts over a much wider temperature range and has a substantially smaller dA/dT maximum (0.0048 vs. 0.0084 for MT and WT, respectively) (Table 1). These observations clearly support multiple folding intermediates and absence of cooperativity.
When Mg2+ is added to a final concentration of 0.5 or 2 mM, the transitions shift to higher temperature, consistent with known effects of divalent ions on secondary structure (Fig. 1C; Serra et al. 2002). However, the melting profiles remain much broader than those in WT, show less thermostability, and contain multiple unfolding transitions. These results suggest that MT tRNAPhe does not fold with any significant cooperativity even in the presence of Mg2+.
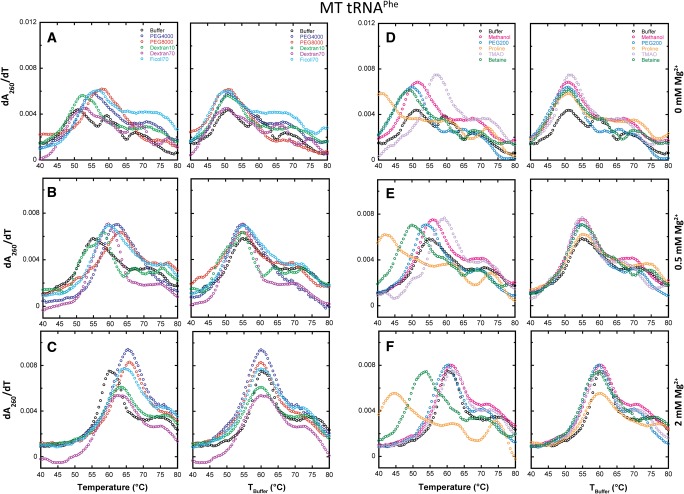
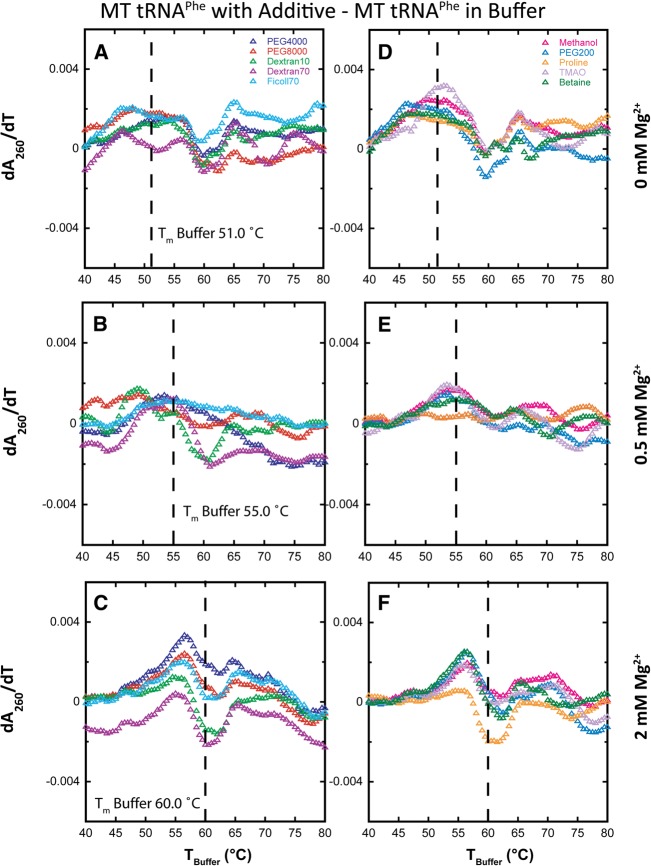
Next, we consider the effects of macromolecular crowders and low-molecular-weight cosolute additives on the folding of MT tRNAPhe in the presence of 0, 0.5, and 2 mM Mg2+. The same additives at the same concentrations were studied as in WT tRNAPhe. In the absence of Mg2+, the crowding agents increase the max dA/dT relative to buffer (Fig. 6A). This change is not consistent with an increase in cooperativity, however, because the transition does not narrow and because two or three transitions are clearly evident (Table 1). Absence of narrowing is evident both in the similarity of the melts in the TBuffer plots (Fig. 6A, right panel), as well as in the difference plots, which do not show a sharp feature (Fig. 7A); compare to Figure 4B, for example. Given that crowders are known to stabilize extended secondary structures but destabilize short secondary structures (Nakano et al. 2004), the increase in max dA/dT relative to buffer may be due to the cumulative effect of concomitantly melting secondary structural elements.
FIGURE 6.
Melts of MT tRNAPhe in additives with increasing Mg2+ concentration. All melts were performed in the background of 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl. (A–C) The effects of various crowding agents (20% [w/v]) in 0, 0.5, and 2 mM Mg2+, respectively. No additive (black), PEG4000 (blue), PEG8000 (red), Dextran10 (green), Dextran70 (purple), and Ficoll70 (cyan). (D–F) The effects of low-molecular-weight cosolutes (2 m, except PEG200, which was 20% [w/v]) in 0, 0.5, and 2 mM Mg2+, respectively. No additive (black), methanol (pink), PEG200 (cobalt blue), proline (orange), TMAO (light purple), and betaine (dark green). In certain panels, data were adjusted to have the same Tm as the buffer, designated as “TBuffer,” so that the effects on cooperativity are clear.
FIGURE 7.
Difference plots of melts of MT tRNAPhe in additives minus in buffer alone with increasing Mg2+ concentration. All melts were performed in the background of 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl. All data were adjusted to have the same Tm, designated as “TBuffer.” (A–C) The effects of various crowding agents (20% [w/v]) in 0, 0.5, and 2 mM Mg2+, respectively. PEG4000 (blue), PEG8000 (red), Dextran10 (green), Dextran70 (purple), and Ficoll70 (cyan). (D–F) The effects of low-molecular-weight cosolutes (2 m, except PEG200, which was 20% [w/v]) also in 0, 0.5, and 2 mM Mg2+, respectively. Methanol (pink), PEG200 (cobalt blue), proline (orange), TMAO (light purple), and betaine (dark green). Difference plots show the effects of the additives on MT folding cooperativity.
In the absence of Mg2+, the cosolute agents show an even greater increase in the max dA/dT for MT tRNAPhe relative to buffer than do crowding agents (Fig. 6D). However, this change again is not consistent with an increase in cooperativity because the transition does not narrow and because two or three transitions are clearly evident (Table 1). Absence of narrowing is evidenced both in the similarity of the melts with and without cosolute in the TBuffer plots (Fig. 6D, right panel) and the difference plots (Fig. 7D). Given that cosolutes are known to strongly destabilize secondary structure (Lambert and Draper 2007), the increase in max dA/dT for cosolutes relative to buffer for MT tRNAPhe may again be due to overlapping of the melting of many secondary structural elements.
In the presence of 0.5 mM Mg2+, crowding additives have relatively little effect on the maximal value of dA/dT or the number of transitions, which remains at two or three (Fig. 6B; Table 1). Moreover, the difference melt plots show little or no regular features (Fig. 7B), consistent with little effect on folding cooperativity. Cosolutes again lead to an increase in max dA/dT relative to buffer, albeit by a smaller amount (Figs. 6E, 7E).
In the presence of 2 mM Mg2+, there are more noticeable effects of crowders and cosolutes including a differential feature in the difference plots (Fig. 7C,F). The differential feature, however, has a negative node at Tm,Buffer, rather than a positive node as seen with WT (e.g., cf. Fig. 4B,E). Moreover, the positive node appears at a temperature lower than the Tm,Buffer. These features arise from broadening of the transition and are compatible with loss rather than gain of cooperativity. Observation that the number of unfolding transitions is two or three with most crowders and cosolutes (Table 1) further strengthens the conclusion that folding cooperativity of MT tRNA is not promoted by additives.
Last, we note that the thermostability of MT was affected by additives in a similar fashion as WT. Macromolecular crowders increased thermostability by an average of 4.7°C, while low molecular cosolutes had little or no effect on thermostability, with the same exceptions of proline and betaine.
Mapping of tRNAPhe structure by SHAPE as a function of temperature in the absence and presence of additives
The above thermodynamic results show that cooperativity of tRNA folding is induced by all macromolecular crowders tested as well as by certain low-molecular-weight crowders. To gain structural insight into these results, we assessed the structure of the tRNA by SHAPE in the absence and presence of PEG8000, methanol, and betaine, in the presence of 0.5 mM Mg2+. The SHAPE method reagent acylates accessible and dynamic 2′-hydroxyls, which can be detected by reverse transcription (RT) (Merino et al. 2005; Wilkinson et al. 2006). We chose SHAPE because this method has been validated as providing detailed information on the secondary structure of folded RNAs under a wide range of conditions. To facilitate SHAPE experiments, a 30-nucleotide (nt) overhang was appended to the 3′ end of the tRNA to provide a primer binding site, with a 6-nt linker between the 3′ end of the tRNA and the extension (Supplemental Fig. S2B). This extension was designed not to interact with the tRNA, which was validated by free-energy minimization calculations as well as by the results presented below, which reveal a native fold to the tRNAPhe. Structure mapping by SHAPE was done as a function of temperature every ∼2.7°C between 41°C and 70°C, which spans the melting temperature. In terms of additives, we chose a macromolecular crowder and a cosolute that promoted cooperativity, as well as a cosolute that diminished cooperativity. PEG8000 was chosen as the crowder because it has an intermediate molecular weight and it should have minimal reactivity with the SHAPE reagent. Methanol was chosen as a cosolute to promote cooperativity, while betaine was chosen as a cosolute that diminished cooperativity. We tested 0.5 mM Mg2+ because the additives have the biggest effect of cooperativity under these conditions as described above.
We first mapped the structure of the tRNAPhe in buffer alone and PEG8000 at 42.3°C, which is well below the Tm values in 0.5 mM Mg2+ in these conditions. SHAPE reactivity as a function of nucleotide position is provided in Supplemental Figure S2. SHAPE reactions in buffer alone and PEG8000 have nearly identical patterns under these conditions, with moderate to high reactivity in the D-loop, AC-loop, and TψC-loop regions, as well as in the linker region between the primer binding site and the 3′ end of the tRNA. These data were mapped onto the secondary structure of tRNA, as shown in Supplemental Figure S2, and agree with the established secondary and tertiary structure of tRNA. This result provides confirmation that this T7 transcript of tRNAPhe folds natively in the presence of buffer and PEG8000 in 0.5 mM Mg2+.
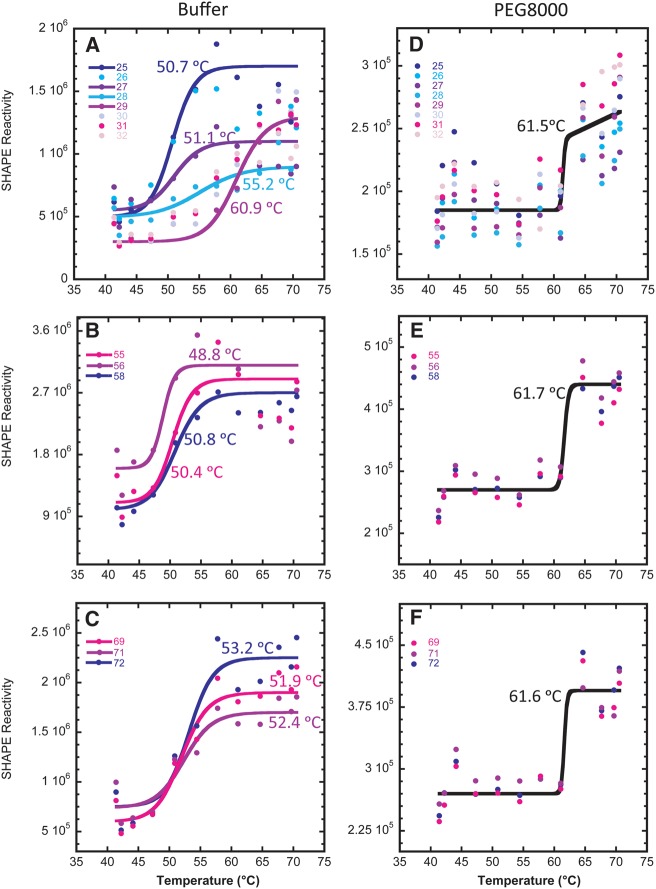
Next, SHAPE experiments were conducted as a function of temperature. We analyzed nucleotides in the D stem and loop, TψC loop, and acceptor stem. Nucleotides 25–32 (D stem and AC stem) were chosen because this stretch contains both secondary and tertiary structure; nucleotides 55–56 and 58 (TψC loop) were chosen because they participate only in tertiary interactions; and nucleotides 69 and 71–72 (acceptor stem) were chosen because they participate only in secondary interactions (Fig. 1A). Each of the nucleotide SHAPE temperature profiles was fit to a standard two-state melting equation, with sloping baselines as needed (see Materials and Methods).
As shown in Figure 8 (melts of individual nucleotides) and Supplemental Figure S3 (polyacrylamide gels), in the presence of buffer alone in 0.5 mM Mg2+, the tRNA melts out in multiple transitions. For example, Figure 8A displays nucleotides that melt in the D stem and AC stem with Tms ranging from 50.7°C to 60.9°C, while Figure 8B,C show nucleotides that melt in the TψC loop and acceptor stem with Tms ranging from 48.8°C to 53.2°C. The Tm values observed span the Tm observed in UV melting in 0.5 mM Mg2+ and no additive of 57.5°C (Table 1). It is also noteworthy that some nucleotides transition from unmodified to modified to unmodified with increasing temperature, such as residue 55 in the TψC loop, supporting a population of a folding intermediate. Together, these data support low folding cooperativity in the presence of buffer alone, consistent with the UV-detected melting data.
FIGURE 8.
Temperature-dependent SHAPE analysis of WT tRNAPhe in the presence and absence of PEG8000 in 0.5 mM Mg2+. (A–C) SHAPE analysis of WT tRNAPhe in buffer alone. (D–F) SHAPE analysis of WT tRNAPhe in 20% (w/v) PEG8000. (A,D) Nucleotides 25–32 in the D stem and AC stem (secondary and tertiary structure interactions). (B,E) Nucleotides 55, 56, and 58 in the TψC loop (tertiary structure interactions). (C,F) Nucleotides 69, 71, and 72 in the acceptor stem (secondary structure interactions). For plots A–C buffer alone, each nucleotide was fit separately to a two-state folding model, since there was poor correlation in Tms for global analysis. For plots D–F in PEG8000, all nucleotides were fit globally since there was excellent agreement of Tms within each data set and panel.
In contrast to buffer alone, SHAPE data in the presence of 20% PEG8000 reveal that the tRNA melts out largely in a single transition. In this case, the same two-state transition was able to fit all the nucleotides in each of the above regions, with a Tm of ∼61.6°C (Fig. 8D–F). Notably, the single Tm from each of these SHAPE-mapped regions agrees well with the Tm from the UV melts in 20% PEG8000 and 0.5 mM Mg2+ of 61.5°C (Table 1). Overall, the agreement among the Tm values derived from temperature-dependent SHAPE data for the different nucleotides in secondary and tertiary regions as well as the Tm from the UV melting experiments support the conclusion that cooperativity is induced by addition of crowder in biological Mg2+.
Next, we conducted SHAPE experiments on tRNAPhe at 42.3°C in the presence of either methanol or betaine, which is below their Tm values in 0.5 mM Mg (Supplemental Figs. S4, S5). The SHAPE reactivities were nearly identical in both additives, with reactivity primarily localized to the D-loop, AC-loop, and TψC-loop regions, as well as the linker region. These data mapped well onto the established secondary and tertiary structure of tRNA, indicating that this tRNA folds natively in the presence of both cosolutes in 0.5 mM Mg2+. Mapping was then conducted as a function of temperature and plotted in the same regions of the tRNA described for buffer alone and PEG8000. In the presence of betaine, which is a destabilizing cosolute, the melting appears noncooperative (Supplemental Fig. S6). There is a pattern showing increased reactivity with temperature, followed by a decrease in reactivity, and followed by another increase in reactivity (Supplemental Fig. S6A–C). This pattern of reactivity is consistent with multiple unfolding transitions in certain regions of the RNA and suggests that the unfolding transition is noncooperative, which agrees with results from the melts. The temperature-dependent SHAPE reaction in the presence of betaine was therefore not fit to a two-state model.
In contrast, in the presence of methanol, which is a stabilizing cosolute, the melting appears cooperative (Supplemental Fig. S6). There is a pattern showing decreased reactivity with temperature, followed by an increase in reactivity, and followed by another decrease in reactivity. The decrease in reactivity with temperature is likely due to the SHAPE reagent 1M7 being consumed through reacting with methanol in an increasing fashion with temperature. In this case, the same two-state transition with negatively sloping baselines fits all the nucleotides in each of these three regions, and the thermodynamics parameters agree between the different regions with a Tm of ∼60.2°C. This Tm is in reasonable agreement with the Tm from the melts of 57.0°C, especially given the difficulty in assigning baselines in the temperature-dependent SHAPE. In sum, the temperature-dependent SHAPE data in the presence of cosolutes agree well with the conclusions made from the UV melting experiments, which is that certain cosolutes, such as methanol, induced cooperative folding of the RNA, while others, such as betaine, diminish cooperative folding. Overall, the degree of cooperativity observed in the SHAPE analysis in the presence and absence of crowder and cosolutes parallels that inferred from the max dA/dT values of betaine < buffer < methanol < PEG8000 (Table 1), providing structural support to the above conclusions.
DISCUSSION
In this study, we examined the folding cooperativity of tRNA in the absence and presence of Mg2+. The concentrations of Mg2+ tested, 0.5 and 2 mM, are similar to those typically found in eukaryotic and prokaryotic cells, respectively. Furthermore, we tested the effects of crowders and cosolutes on RNA folding cooperativity at concentrations typical of both cells. Melts revealed that increasing Mg2+ concentration increased RNA folding cooperativity, as expected, and that folding cooperativity is further enhanced in backgrounds of biological Mg2+ concentration by the presence of all macromolecular crowders tested. This was revealed in increased sharpness of single melting transitions. Melts in the presence of small-molecule cosolutes revealed that certain cosolutes, such as methanol and PEG200, increase folding cooperativity, whereas other cosolutes, such as betaine and proline, reduce folding cooperativity. Moreover, for those crowders and cosolutes that enhance cooperativity, the enhancements were similar in magnitude to the enhancements in going from no Mg2+ to 0.5 mM Mg2+, as inferred from max dA/dT values. Probing the base-pairing status of individual nucleotides with SHAPE provided structural support for these conclusions, revealing enhanced cooperativity of tRNA folding in crowders and certain cosolutes in biological Mg2+ concentrations.
Origins of folding cooperativity and its relationship to thermostability
Folding cooperativity can be considered to arise from two limiting scenarios: (1) strengthening of tertiary structure interactions more than secondary structure interactions, or (2) weakening secondary structure interactions more than tertiary structure interactions. In this section, we consider the 0.5 mM Mg2+ data followed by 2 mM Mg2+. In the absence of Mg2+, essentially no cooperativity was observed, so this condition is not considered in detail.
The effects of crowders and cosolutes on WT and MT tRNAPhe thermostability and folding cooperativity in the presence of 0.5 mM Mg2+ are summarized in Scheme 1. In the presence of 0.5 mM Mg2+, thermostability of both WT and MT tRNAPhe increased in various crowders relative to buffer alone (Table 1). However, the cooperativity of only WT tRNAPhe increased in the presence of crowder. Moreover, MT unfolded with at least two transitions but WT had one transition. Because thermostability increased for both WT and MT tRNAPhe, with a somewhat greater increase for MT, it appears that the first scenario, in which tertiary structure interactions are strengthened more than secondary structure interactions, is responsible for the gain in folding cooperativity in the presence of crowders. Strengthening of tertiary structure is expected from the presence of macromolecular crowders (Nakano et al. 2009).
SCHEME 1.
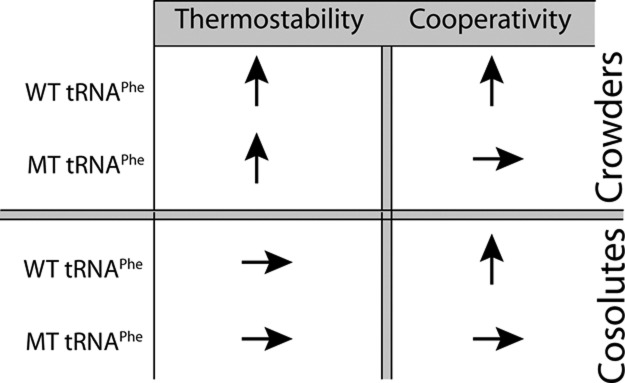
Summary of effects of cosolutes and crowders on thermostability and RNA folding cooperativity in the presence of eukaryotic free magnesium concentration of 0.5 mM Mg2+. A vertical arrow signifies an increase, and a horizontal arrow signifies no change.
In general, the effects of cosolutes on cooperativity are specific to the given additive chosen. These effects of cosolutes on secondary structure stability are consistent with other results (Lambert and Draper 2007; Lambert et al. 2010), where TMAO was slightly stabilizing or destabilizing depending on the RNA, methanol was only slightly destabilizing, while proline and betaine were strongly destabilizing.
We first consider TMAO in the presence of 0.5 mM Mg2+. Because TMAO both enhances folding cooperativity, as evidenced by a max dA/dT increase over buffer alone of 0.0106 to 0.0119, and thermostability, as evidenced by Tm increases from buffer alone for both WT and MT tRNAPhe, it appears to enhance folding cooperativity by the first scenario in which tertiary structure interactions are strengthened more than secondary structure interactions.
Methanol and PEG200 both increase cooperativity, as evidenced by max dA/dT increases over buffer of 0.0106 to 0.0124 and 0.0149, respectively. In the presence of PEG200, which leads to the most cooperative unfolding, thermostability is enhanced in WT and diminished in MT, albeit by small effects. It thus appears that cooperativity arises in PEG200 due to a combination of both scenarios: strengthening of tertiary structure and weakening of secondary structure. It is likely the combination of these two mechanisms that leads to PEG200 being the additive that has the most cooperative unfolding. Studies on melting behavior of model oligonucleotides representing various tRNAPhe helices support this conclusion (data not shown), as do data on shorter oligonucleotides (Nakano et al. 2004). The molecular origin of this effect appears to be related to water activity being reduced in the presence of PEG200, which disfavors secondary structure formation but favors higher-order structure formation (Nakano et al. 2004; Knowles et al. 2011). With methanol, thermostability is changed only slightly in both the WT and MT, diminished in WT and enhanced in MT relative to buffer alone. The origin of the folding cooperativity increase in the presence of methanol thus appears to be related to increase in tertiary structure stability, which is consistent with other observations (Shiman and Draper 2000).
Proline and betaine both strongly destabilize thermostability, to the point where cooperativity is lost as revealed by max dA/dT decreases relative to buffer of 0.0106 to 0.0075 and 0.0091, respectively, and by the presence of multiple transitions (Table 1). The origin of the loss of cooperativity could be due to interactions between the additive and the RNA and/or Mg2+ ions (Lambert and Draper 2007).
Next, we consider folding cooperativity in the presence of 2 mM Mg2+. In the presence of crowders, thermostability of both WT and MT tRNAPhe increases relative to buffer alone, as in the case of 0.5 mM Mg2+. Regarding cosolutes, folding cooperativity in 2 mM Mg2+ is still enhanced by PEG200 to a significant extent and by methanol to a smaller extent. Proline and betaine continue to diminish cooperativity, while TMAO no longer enhances cooperativity. Overall, cosolutes have similar, but not identical, effects on cooperativity in 2 and 0.5 mM Mg2+. We do note, however, that the cooperativity gains in 2 mM Mg2+, both in the presence of crowders and cosolutes, are somewhat smaller than those in 0.5 mM Mg2+, as revealed by smaller gains in max dA/dT, in difference plots, and ΔHVH/ΔHVH,buffer. Diminished enhancement of cooperativity by additive is likely caused in large part because the background cooperativity in buffer alone is considerably greater in 2 mM Mg2+ than 0.5 mM Mg2+. These results are qualitatively similar to those from Lambert and Draper, who observed that Mg2+ diminishes the stabilizing contributions of osmolytes for RNAs that bind Mg2+ diffusely (Lambert and Draper 2007).
Evidence that the unfolding of RNA is only partially cooperative
It appears that folding cooperativity gains in tRNAPhe are only partial. The largest ΔH for unfolding that we observe is ∼100 kcal/mol, under conditions of 2 mM Mg2+ and all crowders as well as the cosolutes methanol, TMAO, and PEG200 (Supplemental Table S1). Turner rules suggest that melting of all the secondary structure should lead to a ΔHVH for unfolding of ∼208 kcal/mol (mFold with version 2.3 energies). Comparison of the ΔH values measured herein to those from Turner and coworkers is somewhat indirect as their melts are typically in 1 M NaCl, while ours are in 0.5 or 2.0 mM Mg2+ and 140 mM KCl, as well as the presence of various additives. Nonetheless, it appears that even in the more cooperative unfolding transitions observed herein, the extent of structure lost in the transition is not due to an all-or-none transition. Thus, while we clearly see that additives lead to increases in the magnitude of ΔH, it is likely the case that not all of the RNA is melting at once and that the gains in cooperativity are partial, at least under the conditions tested herein.
Weeks and colleagues conducted SHAPE studies on the folding of a different tRNA as a function of temperature (Wilkinson et al. 2005). Their experiments had several differences from ours, including studies based on a (unmodified) tRNAAsp from yeast, which is much more GC-rich than the yeast tRNAPhe studied herein, and a background of 10 mM Mg2+. Nonetheless, they made a number of observations that are similar to ours. In particular, they found that certain tertiary structure and secondary structure elements melt simultaneously, while others melt separately. The similarities may come about because of the enhanced secondary structure stability from the GC-rich helices being offset by the enhanced tertiary structure stability from 10 mM Mg2+.
CONCLUSIONS
Under standard literature conditions, intermolecular interactions in RNA are strong. For example, a helix of just 8 GC bp has a t1/2 of ∼16 h at 37°C and 1 M NaCl (Turner and Bevilacqua 1993). As such, RNA has the potential to be extraordinarily stable, with long-lived intermediates and misfolded intermediates. This has led to the notion that secondary structure is stronger than tertiary structure and that RNA folds through highly populated helical intermediates. This does not have to be the case, however. Base-pairing is typically much weaker in mesophilic than thermophilic functional RNAs (Galtier and Lobry 1997; Lu et al. 2006), and strong tertiary interactions can hold together intrinsically unstable helices (Stein and Crothers 1976a; Blose et al. 2007). Thus, it is possible that secondary and tertiary structure could melt simultaneously leading to partial or full unfolding cooperativity.
Herein, we established that physiological concentrations of crowding and cosolutes influence the folding cooperativity of RNA in biological Mg2+ concentrations. Future studies will be needed to dissect the molecular identity of the cooperativity through such methods as double and triple mutant cycles (Siegfried and Bevilacqua 2009), which we and others have applied under dilute solution conditions (Moody and Bevilacqua 2003; Moody et al. 2004; Sattin et al. 2008; Behrouzi et al. 2012). Moreover, studies will be needed on large, multidomain RNAs to see if these respond differently to crowders and cosolutes. In addition, studies will be needed to probe the extent of folding cooperativity in living cells to determine if RNA folds cooperatively, paralleling protein folding under these conditions.
MATERIALS AND METHODS
Chemicals
PEG4000 was from Alfa Aesar; PEG8000 was from Research Organics; PEG200, proline, TMAO, betaine, Dextran10, Dextran70, and Ficoll70 were from Sigma-Aldrich; methanol was from Mallinckrodt. MgCl2 was obtained from J.T. Baker; KCl was from EMD Chemicals; and sodium cacodylate was from Sigma-Aldrich. 1M7 was a gift from the Showalter laboratory. Polynucleotide kinase was from New England Biolabs. SuperScript III reverse transcriptase was from Invitrogen.
RNA preparation
All tRNAPhe substrates were prepared by in vitro T7 transcription using 10% T7 RNA polymerase in 40 mM Tris (pH 8.0), 25 mM MgCl2, 2 mM DTT, 1 mM spermidine, and 4 mM NTPs for 4 h at 37°C. The RNA was purified by 10% PAGE and recovered by a crush-and-soak, ethanol precipitation procedure. DNA oligonucleotide templates to make T7 tRNAPhe were purchased from Integrated DNA Technologies (IDT) and used without further purification.
The sequences of tRNA are as follows:
Wild-type tRNAPhe, 5′-GCGGAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCA;
Mutated tertiary (MT) tRNAPhe, 5′-GCGGAUUUUGCUCUUUUUUUUGAGCGCCAGACUGAAGAUCUGGAUUUCCUGUGUUUUUUCCACAGAAUUCGCACCA;
SHAPE tRNAPhe, 5′-GCGGAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCAAGAAAACCAAAUCGGGCUUCGGUCCGGUUC.
WT and MT tRNAPhe were exchanged into 10 mM sodium cacodylate (pH 7.0) buffer using an Amicon Ultracentrifugal filter (MWCO, 3 kDa).
RNA thermal denaturation monitored by UV absorbance
WT and MT tRNAPhe were first renatured by heating for 3 min to 90°C in buffer and KCl and cooled to room temperature over 10 min. The final concentration of RNA in all melts was 0.5 µM. The MgCl2 was then added to RNA, and the samples were heated for 3 min to 55°C and cooled to room temperature over 10 min. The MgCl2 was not included in the high-temperature renaturation to avoid cleavage to the RNA backbone. Crowder or low-molecular-weight cosolute was added during the second renaturation step with the Mg2+. Following addition of the Mg2+ and/or crowding or cosolute agent, the sample was centrifuged at 16,000g for 10 min to degas the sample.
All crowding and cosolute agents were prepared as 2× stock solutions and centrifuged at 16,000g for 20 min after preparation. Macromolecular crowding agents (PEG4000, PEG8000, Dextran10, Dextran70, and Ficoll70) were present at a final concentration of 20% (w/v), and low-molecular-weight cosolutes (methanol, PEG200, proline, TMAO, and betaine) were present at a final concentration of 2 m, with the exception of PEG200, which was present at 20% (w/v). Melting experiments were performed in the background of 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl with either 0, 0.5, or 2 mM MgCl2. A Gilford Response II spectrophotometer was used for melting experiments, with a data point acquired every 0.5°C and a heating rate of ∼0.6°/min at 260 nm. All melts were performed in duplicate, and excellent agreement between these melts was observed.
Melts of all WT and MT tRNAPhe transcripts were normalized by dividing all absorbance readings by the maximum absorbance value. Monophasic melt data were fit to a two-state model using sloping baselines and analyzed using a Marquadt algorithm for nonlinear curve fitting in KaleidaGraph v. 3.5 (Synergy software) (Siegfried and Bevilacqua 2009). Since excellent agreement in melts was observed in duplicate measurements, only one melt from each trial was used for data analysis. Errors from these fits are reported. Derivative plots were of normalized data and were smoothed using an 11-point window prior to taking the derivative. Tm values were calculated by identifying the max dA/dT in the derivative plot; when multiple maxima were present, the reported Tm and max dA/dT are for the first transition, which involves tertiary structure and also had the largest dA/dT. Tm values for subsequent transitions are not reported as these are likely due to contributions of multiple transitions. The number of transitions reported in Table 1 is equal to the number of local maxima. All difference plots were generated from normalized melt curves.
Temperature gradient gel electrophoresis
Electrophoresis was performed in 13% polyacrylamide, 1× THEN10M0.9 (pH 7.5) (33 mM Tris, 66 M HEPES, 0.1 mM EDTA, 10 mM NaCl, 0.9 mM MgCl2), and a semi-denaturing background of 4 M urea in order to facilitate melting in the TGGE temperature range (Bevilacqua and Bevilacqua 1998; Chadalavada and Bevilacqua 2009). Briefly, the experiment was conducted in perpendicular format in which samples were layered across a single wide well to which a temperature gradient of ∼10°C–60°C was applied. The temperature was measured directly in the gel using a digital thermometer and a long narrow probe. The gel was then dried and imaged on the PhosphorImager. Melting temperatures were determined graphically by interpolation, and comparisons between Tms were carried out for samples fractionated on the same gel. WT and MT were loaded at different concentrations to allow traces to be assigned.
Temperature-dependent SHAPE
The SHAPE tRNAPhe transcript was prepared as described above. The sequence of the RNA was identical to the WT tRNAPhe but contained a 3′-extension AGAAAACCAAAUCGGGCUUCGGUCCGGUUC-3′, which provided a 6-nt spacer (AGAAAA) and a binding site for the RT primer. Free-energy minimization calculations with mFold (Zuker 2003) supported absence of interaction of this extension with the tRNA itself, which was further supported by the SHAPE results reported herein. The RT primer (IDT, HPLC purified) was labeled on its 5′ end with [γ-32P]ATP and polynucleotide kinase. The labeled primer was purified using a GE Healthcare G-50 microspin column to remove unincorporated ATP.
The WT tRNAPhe sample for SHAPE analysis was diluted to a final concentration of 150 nM in buffer conditions of 100 mM HEPES (pH 7.6)/0.5 mM Mg2+/100 mM KCl. Temperature-dependent SHAPE was performed in 0.5 mM Mg2+ both with and without additives, and each had a total reaction volume of 20 μL. RNA was first renatured by heating for 1 min to 90°C in buffer and KCl only, and then cooled to room temperature over 2 min. The MgCl2 was then added to the RNA, and the samples were heated for 2 min to 55°C. Samples were cooled to room temperature for 2 min during which time the additive was added, and then incubated for 1 min at 4°C. PEG8000 was present at a final concentration of 20% (w/v), and betaine and methanol were present at a final concentration of 2 m to mirror melting experiments. All heating and cooling were performed on a Biometra Tcycler thermocycler. Samples were then brought to the temperature of the experiment and incubated at that temperature for 2 min prior to addition of the SHAPE reagent to ensure that the sample had reached that temperature. A temperature window of 41°C–70°C was used with an ∼2.7°C temperature step between wells. Temperature in the gradient cycler was calibrated using a thermocouple.
The chemical probing step of SHAPE was performed similar to that previously described (Wilkinson et al. 2006; Gherghe et al. 2008). Briefly, the electrophile 1M7 (dissolved in DMSO) was diluted to a final concentration of ∼10 mM to allow single-hit kinetics conditions (Gherghe et al. 2008). Reactions were carried out for 2 min for the lower temperature samples and 1 min for the higher temperature samples, which allowed for ∼5 half-lives of the 1M7 reagent. An experiment containing no SHAPE reagent was also performed, and it was found that the background reaction is not highly temperature dependent (Supplemental Fig. S7). Ethanol was added after reactions were complete, and the tubes were immediately placed on dry ice for precipitation. Reverse transcription was then performed for 10 min at 55°C, using SuperScript III (Invitrogen) along with dideoxy sequencing. Next, RNA was hydrolyzed by addition of 0.5 μL of 2 M NaOH and heating for 1 min at 95°C. Equal volume of formamide loading buffer was added, and samples were fractionated on a denaturing 10% polyacrylamide gel. Gels were visualized using a PhosphorImager and analyzed using ImageQuant software (Molecular Dynamics).
SHAPE data analysis
SHAPE data were first analyzed to assess the overall fold of the tRNAPhe. This analysis was performed for SHAPE reactions in the presence and absence of additives to test whether the additives altered the native fold of the RNA. For these plots, the overall fold of the RNA was determined at 42.3°C. Background reaction (RT stops without SHAPE reagent present) was subtracted from the buffer alone and additive SHAPE reactions. Data were normalized using SAFA before subtraction for these plots (Das et al. 2005). These data were normalized to the most intense nucleotide on the gel, excluding the full-length band, following subtraction of the no SHAPE reagent reaction.
For analysis of temperature-dependent SHAPE data, the plotted regions were chosen to represent a diverse set of RNA motifs. All data are under single-hit conditions, as judged by the large amount of full-length band at the top of the SHAPE gels. To account for loading differences on the gel, differences in yield of RNA following precipitation and temperature dependence of the SHAPE reagent, the data were normalized to nucleotides 34–36 in the anticodon loop, as these nucleotides are single-stranded throughout the temperature range. The data were fit to a two-state melting model using either a sloping or nonsloping baseline depending on the data (Siegfried and Bevilacqua 2009). A Marquadt algorithm for nonlinear curve fitting in KaleidaGraph v. 3.5 (Synergy software) was used. Baselines were assigned by minimizing the error in the fit. When possible the folding of multiple nucleotides was fit to a single set of thermodynamic parameters corresponding to a two-state transition; otherwise, data were fit separately to two-state transitions.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Kaycee Quarles, Chris Wostenberg, and Scott Showalter for providing the SHAPE reagent as a gift. We also thank Chun Kit Kwok and Rao Nallagatla for their assistance in SHAPE and TGGE experiments, and Christine Keating for helpful discussions. This work was supported by NASA grant NNX13AI01G.
REFERENCES
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD 1994. Molecular biology of the cell, 3rd ed Garland Science, New York [Google Scholar]
- Behrouzi R, Roh JH, Kilburn D, Briber RM, Woodson SA 2012. Cooperative tertiary interaction network guides RNA folding. Cell 149: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua JM, Bevilacqua PC 1998. Thermodynamic analysis of an RNA combinatorial library contained in a short hairpin. Biochemistry 37: 15877–15884 [DOI] [PubMed] [Google Scholar]
- Blose JM, Silverman SK, Bevilacqua PC 2007. A simple molecular model for thermophilic adaptation of functional nucleic acids. Biochemistry 46: 4232–4240 [DOI] [PubMed] [Google Scholar]
- Brion P, Westhof E 1997. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct 26: 113–137 [DOI] [PubMed] [Google Scholar]
- Brown TS, Chadalavada DM, Bevilacqua PC 2004. Design of a highly reactive HDV ribozyme sequence uncovers facilitation of RNA folding by alternative pairings and physiological ionic strength. J Mol Biol 341: 695–712 [DOI] [PubMed] [Google Scholar]
- Chadalavada DM, Bevilacqua PC 2009. Analyzing RNA and DNA folding using temperature gradient gel electrophoresis (TGGE) with application to in vitro selections. Methods Enzymol 468: 389–408 [DOI] [PubMed] [Google Scholar]
- Chadalavada DM, Senchak SE, Bevilacqua PC 2002. The folding pathway of the genomic hepatitis δ virus ribozyme is dominated by slow folding of the pseudoknots. J Mol Biol 317: 559–575 [DOI] [PubMed] [Google Scholar]
- Crothers DM, Cole PE, Hilbers CW, Shulman RG 1974. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol 87: 63–88 [DOI] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB 2005. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denesyuk NA, Thirumalai D 2011. Crowding promotes the switch from hairpin to pseudoknot conformation in human telomerase RNA. J Am Chem Soc 133: 11858–11861 [DOI] [PubMed] [Google Scholar]
- Di Domenico R, Lavecchia R 2002. Stabilization of human haemoglobin by naturally occurring osmolytes. Biochem Eng J 10: 27–30 [Google Scholar]
- Duncan CD, Weeks KM 2010. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry 49: 5418–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ 2001. Macromolecular crowding: Obvious but underappreciated. Trends Biochem Sci 26: 597–604 [DOI] [PubMed] [Google Scholar]
- Fang X, Littrell K, Yang X, Henderson SJ, Siefert S, Thiyagarajan P, Pan T, Sosnick TR 2000. Mg2+-dependent compaction and folding of yeast tRNAPhe and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle X-ray scattering. Biochemistry 39: 11107–11113 [DOI] [PubMed] [Google Scholar]
- Feig AL, Uhlenbeck OC 1999. The role of metal ions in RNA biochemistry. In The RNA world (ed. Gesteland RF, Cech TR, Atkins JF), pp. 287–319 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Feng B, Frykholm K, Norden B, Westerlund F 2010. DNA strand exchange catalyzed by molecular crowding in PEG solutions. Chem Commun 46: 8231–8233 [DOI] [PubMed] [Google Scholar]
- Galtier N, Lobry JR 1997. Relationships between genomic G + C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J Mol Evol 44: 632–636 [DOI] [PubMed] [Google Scholar]
- Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM 2008. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc 130: 8884–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Chen Y, Christian EL, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR 2008. Detection of innersphere interactions between magnesium hydrate and the phosphate backbone of the HDV ribozyme using Raman crystallography. J Am Chem Soc 130: 9670–9672 [DOI] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs RD 2002. Intracellular magnesium and magnesium buffering. Biometals 15: 251–259 [DOI] [PubMed] [Google Scholar]
- Hart JM, Kennedy SD, Mathews DH, Turner DH 2008. NMR-assisted prediction of RNA secondary structure: Identification of a probable pseudoknot in the coding region of an R2 retrotransposon. J Am Chem Soc 130: 10233–10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isambert H, Siggia ED 2000. Modeling RNA folding paths with pseudoknots: Application to hepatitis δ virus ribozyme. Proc Natl Acad Sci 97: 6515–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA 2010. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc 132: 8690–8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D, Roh JH, Behrouzi R, Briber RM, Woodson SA 2013. Crowders perturb the entropy of RNA energy landscapes to favor folding. J Am Chem Soc 135: 10055–10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT 2011. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc Natl Acad Sci 108: 12699–12704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Sherlock ME, Bevilacqua PC 2013. Decrease in RNA folding cooperativity by deliberate population of intermediates in RNA G-quadruplexes. Angew Chem Int Ed 52: 683–686 [DOI] [PubMed] [Google Scholar]
- Lambert D, Draper DE 2007. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA–Mg2+ interactions. J Mol Biol 370: 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D, Leipply D, Draper DE 2010. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: Evidence for a large barrier to folding from phosphate dehydration. J Mol Biol 404: 138–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E 2006. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol 16: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London RE 1991. Methods for measurement of intracellular magnesium: NMR and fluorescence. Annu Rev Physiol 53: 241–258 [DOI] [PubMed] [Google Scholar]
- Lu ZJ, Turner DH, Mathews DH 2006. A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation. Nucleic Acids Res 34: 4912–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk JE, Williams RJ, Kennedy EP 1968. Magnesium and the growth of Escherichia coli. J Biol Chem 243: 2618–2624 [PubMed] [Google Scholar]
- Mathews DH, Turner DH 2006. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol 16: 270–278 [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231 [DOI] [PubMed] [Google Scholar]
- Minton AP 2001. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem 276: 10577–10580 [DOI] [PubMed] [Google Scholar]
- Misra VK, Draper DE 2000. Mg2+ binding to tRNA revisited: The nonlinear Poisson–Boltzmann model. J Mol Biol 299: 813–825 [DOI] [PubMed] [Google Scholar]
- Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, Thirumalai D, Woodson SA 2009. Metal ion dependence of cooperative collapse transitions in RNA. J Mol Biol 393: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody EM, Bevilacqua PC 2003. Folding of a stable DNA motif involves a highly cooperative network of interactions. J Am Chem Soc 125: 16285–16293 [DOI] [PubMed] [Google Scholar]
- Moody EM, Feerrar JC, Bevilacqua PC 2004. Evidence that folding of an RNA tetraloop hairpin is less cooperative than its DNA counterpart. Biochemistry 43: 7992–7998 [DOI] [PubMed] [Google Scholar]
- Nakano S, Karimata H, Ohmichi T, Kawakami J, Sugimoto N 2004. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J Am Chem Soc 126: 14330–14331 [DOI] [PubMed] [Google Scholar]
- Nakano S, Karimata HT, Kitagawa Y, Sugimoto N 2009. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J Am Chem Soc 131: 16881–16888 [DOI] [PubMed] [Google Scholar]
- Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF 2002. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res 30: 4751–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan SB, Dill KA, Bahar I 2002. Fast-folding protein kinetics, hidden intermediates, and the sequential stabilization model. Protein Sci 11: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JD, Tinoco I 1989. Absorbance melting curves of RNA. Methods Enzymol 180: 304–325 [DOI] [PubMed] [Google Scholar]
- Pulukkunat DK, Gopalan V 2008. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res 36: 4172–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner D, Maass G, Thiebe R, Philippsen P, Zachau HG 1973. The conformational transitions in yeast tRNAPhe as studied with tRNAPhe fragments. Eur J Biochem 36: 76–88 [DOI] [PubMed] [Google Scholar]
- Romani AM 2007. Magnesium homeostasis in mammalian cells. Front Biosci 12: 308–331 [DOI] [PubMed] [Google Scholar]
- Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SG, Pande V, Doniach S, Herschlag D, et al. 2002. Rapid compaction during RNA folding. Proc Natl Acad Sci 99: 4266–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, Uhlenbeck OC 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci 85: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattin BD, Zhao W, Travers K, Chu S, Herschlag D 2008. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc 130: 6085–6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA 1998. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279: 1940–1943 [DOI] [PubMed] [Google Scholar]
- Serra MJ, Baird JD, Dale T, Fey BL, Retatagos K, Westhof E 2002. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA 8: 307–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiman R, Draper DE 2000. Stabilization of RNA tertiary structure by monovalent cations. J Mol Biol 302: 79–91 [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Bevilacqua PC 2009. Thinking inside the box: Designing, implementing, and interpreting thermodynamic cycles to dissect cooperativity in RNA and DNA folding. Methods Enzymol 455: 365–393 [DOI] [PubMed] [Google Scholar]
- Sokoloski JE, Dombrowski SE, Bevilacqua PC 2011. Thermodynamics of ligand binding to a heterogeneous RNA population in the malachite green aptamer. Biochemistry 51: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink CH, Chaires JB 1998. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 38: 496–508 [DOI] [PubMed] [Google Scholar]
- Spink CH, Garbett N, Chaires JB 2007. Enthalpies of DNA melting in the presence of osmolytes. Biophys Chem 126: 176–185 [DOI] [PubMed] [Google Scholar]
- Stein A, Crothers DM 1976a. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry 15: 160–168 [DOI] [PubMed] [Google Scholar]
- Stein A, Crothers DM 1976b. Equilibrium binding of magnesium(II) by Escherichia coli tRNAfMet. Biochemistry 15: 157–160 [DOI] [PubMed] [Google Scholar]
- Strulson CA, Molden RC, Keating CD, Bevilacqua PC 2012. RNA catalysis through compartmentalization. Nat Chem 4: 941–946 [DOI] [PubMed] [Google Scholar]
- Strulson CA, Yennawar NH, Rambo RP, Bevilacqua PC 2013. Molecular crowding favors reactivity of a human ribozyme under physiological ionic conditions. Biochemistry 52: 8187–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM 2002. Productive folding to the native state by a group II intron ribozyme. J Mol Biol 315: 297–310 [DOI] [PubMed] [Google Scholar]
- Takamoto K, He Q, Morris S, Chance MR, Brenowitz M 2012. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nat Struct Mol Biol 9: 928–933 [DOI] [PubMed] [Google Scholar]
- Thirumalai D 1998. Native secondary structure formation in RNA may be a slave to tertiary folding. Proc Natl Acad Sci 95: 11506–11508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Lee N, Woodson SA, Klimov DK 2001. Early events in RNA folding. Annu Rev Phys Chem 52: 751–762 [DOI] [PubMed] [Google Scholar]
- Truong DM, Sidote DJ, Russell R, Lambowitz AM 2013. Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core. Proc Natl Acad Sci 110: E3800–E3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DH, Bevilacqua PC 1993. Thermodynamic considerations for evolution by RNA. In The RNA world (ed. Gesteland RF, Atkins JF), pp. 447–464 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Weikl TR, Palassini M, Dill KA 2004. Cooperativity in two-state protein folding kinetics. Protein Sci 13: 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman EE 2011. “Identification and biophysical characterization of factors influencing the thermostablility of biological RNAs.” Master thesis, Pennsylvania State University, University Park, PA [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc 127: 4659–4667 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA 2010. Compact intermediates in RNA folding. Annu Rev Biophys 39: 61–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Tinoco I 1998. RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci 95: 11555–11560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SK, Soll DG, Crothers DM 1972. Properties of a dimer of tRNA I Tyr 1 (Escherichia coli). Biochemistry 11: 2311–2320 [DOI] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR 1994. Kinetic intermediates in RNA folding. Science 265: 918–924 [DOI] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP 2008. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys 37: 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]