Summary
The mechanisms of transcriptional regulation underlying human primordial germ cell (PGC) differentiation are largely unknown. The transcriptional repressor Prdm1/Blimp-1 is known to play a critical role in controlling germ cell specification in mice. Here, we show that PRDM1 is expressed in developing human gonads and contributes to the determination of germline versus neural fate in early development. We show that knockdown of PRDM1 in human embryonic stem cells (hESCs) impairs germline potential and upregulates neural genes. Conversely, ectopic expression of PRDM1 in hESCs promotes the generation of cells that exhibit phenotypic and transcriptomic features of early PGCs. Furthermore, PRDM1 suppresses transcription of SOX2. Overexpression of SOX2 in hESCs under conditions favoring germline differentiation skews cell fate from the germline to the neural lineage. Collectively, our results demonstrate that PRDM1 serves as a molecular switch to modulate the divergence of neural or germline fates through repression of SOX2 during human development.
Graphical Abstract
Highlights
-
•
PRDM1 serves as a molecular switch that determines neural or germline fate in human
-
•
PRDM1 is expressed in early germ cells of developing human fetal gonads
-
•
PRDM1 is expressed in hESC-derived germ cells and directs germline differentiation
-
•
Germline differentiation in hESC requires PRDM1-mediated suppression of SOX2
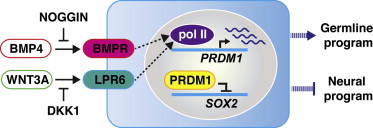
In this study, Lin and colleagues investigate the function of PRDM1 in germline differentiation of human embryonic stem cells. They find that BMP4- and WNT3A-mediated upregulation of PRDM1 transcriptionally suppresses SOX2, thereby inducing the activation of germline fate and preventing neural program.
Introduction
Primordial germ cells (PGCs) are the founders of germ cells that give rise to eggs and sperm as the end products (Surani, 2007). In mice, extraembryonic tissues direct a small number of pluripotent epiblast cells to express interferon-induced transmembrane protein 3 (Ifitm3) as PGC precursors by providing bone morphogenetic protein 4 (BMP4) growth factor (Saitou et al., 2002). WNT3A is required for the responsiveness of epiblast cells to BMP4 (Ohinata et al., 2009). Following BMP4 signaling, approximately 40 cells in the posterior-proximal extraembryonic mesoderm begin to express Stella, which permits the specification of PGCs. During PGC specification from the pluripotent epiblast cells, it is crucial to repress the somatic program (Saitou et al., 2002). Thereafter, PGCs migrate along the hindgut into the genital ridges concomitantly with an extensive genome-wide epigenetic reprogramming, including alteration of histone modification and erasure of imprinted loci (Hajkova et al., 2002). Another key event during mouse germline specification is the expression of pluripotency-associated genes such as Pou5f1 (Oct3/4), Sox2, and Nanog (Yabuta et al., 2006). However, human PGCs appear to express only OCT4 and NANOG, and not SOX2 (Perrett et al., 2008), indicating the possibility of interspecies differences in germline development and an unidentified mode of action of pluripotency-associated genes in germline commitment.
The transcriptional repressor PRDM1, also known as B lymphocyte-induced maturation protein-1 (Blimp-1), was identified as the key regulator of the differentiation of mature B lymphocytes into antibody-producing plasma cells (Shaffer et al., 2002). It is also expressed and required for mouse embryonic development as well as for the differentiation of many adult cell lineages (Bikoff et al., 2009). It is of importance that, in mouse, some Prdm1-expressing cells can be traced in the inner cell mass (ICM) and used to predict the outgrowth of PGCs (Chu et al., 2011), in which the sustained expression of Prdm1 permits the generation of PGCs at embryonic day 6.25 (E6.25) in proximal posterior epiblast cells (Ohinata et al., 2005). It has been noted that Prdm1 is crucial for the specification of PGCs in early mouse development because mice lacking Prdm1 produce rare PGCs that are unable to migrate (Ohinata et al., 2005; Vincent et al., 2005). Prdm1 temporally associates with an arginine-specific histone methyltransferase, Prmt5, to establish epigenetic changes during mouse germ cell development (Ancelin et al., 2006). Whether PRDM1 is expressed and plays a role in human germline specification remains elusive.
Embryonic stem cells (ESCs) provide a valuable tool to elucidate the molecular mechanisms underlying the developmental path of cellular lineages, particularly with regard to human development. PGCs can be derived from pluripotent mouse ESCs or human ESCs (hESCs) (Chuang et al., 2012; Geijsen et al., 2004; Hübner et al., 2003). Furthermore, hESCs or human induced pluripotent stem cells (hiPSCs) were recently directed into adult-type postmeiotic spermatogenic cells with largely improved frequencies (Easley et al., 2012). Due to ethical issues involving the fact that the precursors of human PGCs are practically inaccessible in vivo, as they colonize between 5 and 8 weeks of gestation (Clark, 2007; Freeman, 2003), hESCs have become an important tool for producing potential PGCs in vitro. Therefore, it is important to develop improved methods to isolate and generate human PGCs or functional gametes from hESCs. For instance, manipulation of gene expression in hESCs, such as overexpression of deleted in azoospermia-like (DAZL) or silencing DAZL, affects the formation of cells expressing VASA (Kee et al., 2009), the postmigratory PGC marker (Castrillon et al., 2000).
Herein, we examine whether PRDM1 is expressed by human PGCs and involved in human germline differentiation. We show that PRDM1 is expressed in the second trimester of human embryonic ovary and testis development. Additionally, using hESCs as the differentiating platform, we show that PRDM1 is necessary and sufficient for the formation of hESC-derived germline cells, which may be attributed to the role of PRDM1, at least partly, in the suppression of SOX2.
Results
PRDM1 Is Expressed in Early Human Fetal Gonads
We first examined the expression of PRDM1 in various human embryonic organs. Figure 1A shows that PRDM1 was predominantly expressed in human fetal testis collected from an 18-week-old abortus, as demonstrated by immunohistochemistry (IHC) staining with anti-PRDM1 antibody. However, PRDM1 immunopositive signals, which were stained in nuclei, were not detected in human fetal liver, lung, heart, and kidney (Figure 1A), supporting previous findings obtained in mice (Chang et al., 2002). Of note, in fetal testis, PRDM1 is expressed in enclosed seminiferous cords (Figure 1A).
Figure 1.
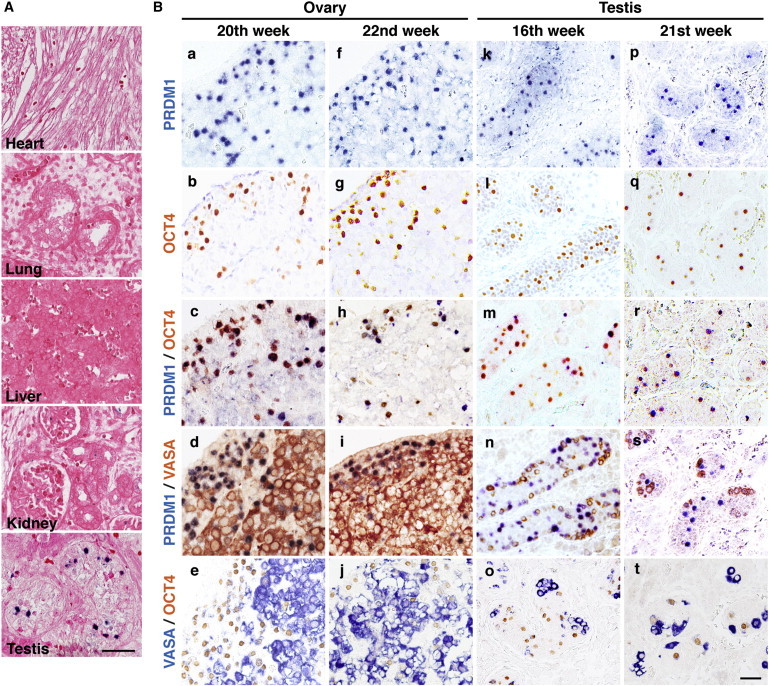
PRDM1 Is Expressed in Human Fetal Gonads
(A) IHC staining of the indicated human embryonic tissues isolated from an 18-week-old male abortus shows the expression of PRDM1 in fetal testis. Sections were also counterstained with eosin. Scale bar, 100 μm.
(B) IHC staining of human fetal gonads isolated from two female and two male abortus at the indicated age with anti-PRDM1, anti-OCT4, and anti-VASA antibodies. Scale bar, 30 μm.
Next, to verify the cell lineage expressing PRDM1 in fetal gonads, we collected embryonic germline tissues from 16- to 22-week-old human fetal gonads to analyze the expression of OCT4, VASA, and PRDM1 by IHC staining. Here, OCT4 was used as the indicator for PGCs (Kehler et al., 2004). We found that PRDM1+ cells appeared to be round and small, and located in the cortex of the ovary (Figure 1B, a and f). OCT4 immunopositive signals, which were present in nuclei, were also detected in the ovarian cortex (Figure 1B, b and g). In fetal testis, the PRDM1+ and OCT4+ cells were enclosed in the seminiferous cords (Figure 1B, k-l and p-q). Indeed, double stains of PRDM1 and OCT4 revealed that a large proportion of PRDM1+ cells overlapped with OCT4+ cells in either fetal ovary or testis (Figure 1B, c, h, m, and r). In contrast, the majority of PRDM1+ cells or OCT4+ cells were not coexpressed with cells stained strongly by anti-VASA antibody (Figure 1B, d-e, i-j, n-o, and s-t). Because VASA is the hallmark of postmigrating germ cells (Castrillon et al., 2000), we suspected that PRDM1 might be involved in early human PGC commitment.
PRDM1 Is Expressed in hESC-Derived Germ Cells
Next, we used hESCs as an in vitro model to examine the expression of PRDM1 in human germline differentiation. We used two hESC lines: H9 hESCs and previously established NTU1 hESCs, which possess the potential to spontaneously differentiate into germ cells (Chen et al., 2007). H9 hESCs were plated for spontaneous differentiation, and cells were harvested for quantitative RT-PCR (qRT-PCR) and immunofluorescence staining with antibodies against PRDM1 and various markers associated with germline lineages. PRDM1 mRNA was readily induced following spontaneous differentiation of H9 hESCs and showed induction kinetics to similar to those of NANOS3, which is important for maintaining PGCs (Julaton and Reijo Pera, 2011). The induction of PRDM1 also preceded the expression of VASA and synaptonemal complex protein 3 (SCP3), a marker for meiosis (Moens and Spyropoulos, 1995; Figure 2A). Immunofluorescence staining showed that PRDM1 began to express at day 5 and was primarily coexpressed with STELLA and NANOS3, which is important for guiding PGC migration in vivo (Tanaka et al., 2005; Figure 2B). The level of PRDM1 was enriched at day 10, at which time the majority of PRDM1+ cells were also OCT4+ (Figure 2B and Figure S1A available online). However, VASA+ cells became prominent by day 20 (Figure S1B) and excluded the expression of PRDM1 (Figure 2B). Likewise, following spontaneous differentiation of NTU1 hESCs, PRDM1 and NANOS3 mRNA was significantly increased before upregulation of VASA and SCP3 occurred (Figure 2C). Immunofluorescence staining showed that PRDM1 was coexpressed with OCT4 at days 5 and 10 (Figures 2D and S1A), and with STELLA and NANOS3 at day 5, but the majority of VASA+ cells did not coexpress with PRDM1 at day 20 (Figure 2D). The significant induction of PRDM1 before the induction of VASA and SCP3 was also observed in NTU1 hESCs plated for spontaneous differentiation by two other protocols (Figures S1C and S1D).
Figure 2.
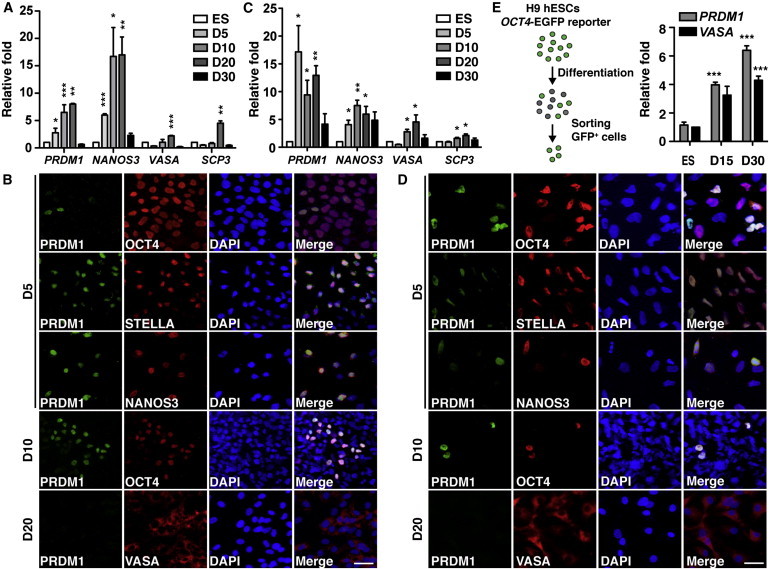
PRDM1 Is Expressed in Germline Lineage Spontaneously Differentiated from hESCs
(A) qRT-PCR for the indicated mRNA isolated from H9 hESCs plated for spontaneous differentiation on the indicated days.
(B) Immunofluorescence staining of H9 hESCs plated for spontaneous differentiation with indicated antibodies on indicated days. DAPI was used as the nuclear counterstain. Scale bar, 30 μm.
(C) qRT-PCR for the indicated mRNA isolated from NTU1 hESCs plated for differentiation by adherent culture on the indicated days.
(D) Immunofluorescence staining of NTU1 hESCs with the indicated antibodies and DAPI at 5, 10, and 20 days after differentiation by adherent culture. Scale bar, 30 μm.
(E) qRT-PCR for the levels of PRDM1 and VASA mRNA in sorted EGFP+ cells produced by H9:OCT4-EGFP plated for differentiation on the indicated days. The experimental flowchart is illustrated. Results in (A), (C), and (E) are the mean ± SEM of three independent experiments.
See also Figure S1.
In parallel, we validated the expression of PRDM1 in enriched germline cells derived from differentiating hESCs using a stable hES transfectant that carried an OCT4-EGFP transgene reporter, H9:OCT4-EGFP (Chuang et al., 2012). The reporter hESCs were plated for spontaneous differentiation and then the EGFP+ cells representing the progeny of germline lineage at days 15 and 30 were sorted (Figure S1E). Indeed, in comparison with the undifferentiated hESCs, sorted EGFP+ H9:OCT4-EGFP cells expressed higher levels of VASA and PRDM1 mRNA (Figure 2E), indicating that PRDM1 is enriched in hESC-derived germ cells. Consistently, certain OCT4+ (EGFP+) cells were also PRDM1+/STELLA+ or PRDM1+/NANOS3+ (Figure S1F).
Taken together, these results indicate that PRDM1 expression is indicative of early germline lineage derived from differentiating hESCs.
PRDM1 Is Required for BMP4- and WNT3A-Induced Germ Cell Differentiation
To facilitate germ cell formation in culture, H9 hESCs were treated with BMP4 and WNT3A (Chuang et al., 2012; Ohinata et al., 2009). Immunofluorescence staining showed that a subset of cells expressed PRDM1 5 days after exposure to BMP4 and WNT3A, whereas undifferentiated hESCs did not express PRDM1, and a large proportion of PRDM1+ cells were also OCT4+ cells (Figures 3A and S2A). At day 20, some cells expressed VASA (Figure S2B), but they did not coexpress with PRDM1 (Figure 3A). Consistently, like the early induction of NANOS3, PRDM1 mRNA was induced most pronouncedly at day 5 following treatment with BMP4 and WNT3A (Figure 3B). Similarly, the induction of PRDM1 mRNA occurred prior to the induction of VASA and SCP3 (Figure 3B). It is noted that treatment with NOGGIN and/or dickkopf-1 (DKK1), the inhibitors of BMPs and canonical Wnt signaling, respectively, suppressed the induction of PRDM1 and NANOS3 mRNA at day 5 (Figure 3C), suggesting that the induction of PRDM1 depends on BMP4 and WNT3A signaling in germline development in vitro.
Figure 3.
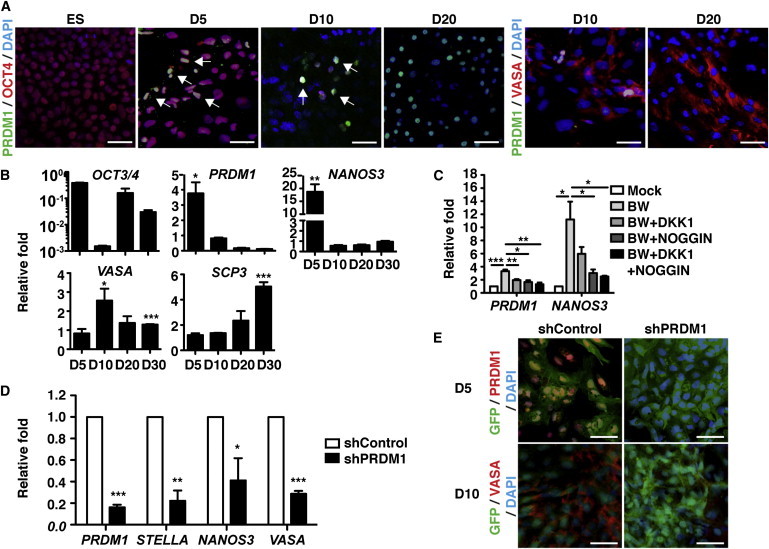
PRDM1 Is Required for BMP4- and WNT3A-Induced Germ Cell Formation in hESCs
(A) Immunofluorescence staining of H9 hESCs treated with BMP4 (100 ng/mL) and WNT3A (50 ng/mL) for the indicated days using DAPI and the indicated antibodies. Scale bar, 30 μm. Arrows indicate PRDM1+/OCT4+ cells.
(B) qRT-PCR analysis showed the expression of the indicated mRNA in H9 hESCs exposed to BMP4 and WNT3A treatment for the indicated days. Relative fold was compared with spontaneously differentiated cells without BMP4 and WNT3A treatment.
(C) qRT-PCR analysis showed the expression of the indicated mRNA in H9 hESCs treated with BMP4 (B) and WNT3A (W) in the presence or absence of NOGGIN (250 ng/mL) and DKK1 (500 ng/mL) for 5 days. Relative fold was compared with the levels of mRNA indicated in cells without treatment with BMP4 and WNT3A (mock).
(D) qRT-PCR for the indicated mRNA isolated from BMP4- and WNT3A-treated shControl or shPRDM1-expressing H9 cells on day 10. Relative fold was compared with the shControl group.
(E) Immunofluorescence staining of DAPI, PRDM1, and VASA in control and PRDM1-KD (GFP+) cells 5 days and 10 days after BMP4 and WNT3A treatment. Scale bar, 30 μm. The values in (B)−(D) are represented as the mean ± SEM of three independent experiments.
See also Figure S2.
To determine whether the induction of PRDM1 is crucial for germline formation, we transduced H9 hESCs with lentiviral vectors producing small hairpin RNA (shRNA) against PRDM1, or control shRNA (shControl). The induction of PRDM1 mRNA was impaired in PRDM1 knockdown (PRDM1-KD) cells after treatment with BMP4 and WNT3A (Figure 3D). It is noted that, compared with shControl-expressing cells, the mRNA levels of STELLA, NANOS3, VASA, and SCP3 were reduced in PRDM1-KD cells (Figures 3D and S2C). Likewise, immunofluorescence staining showed that shControl-transduced (GFP+) H9 hESCs expressed PRDM1 and VASA in response to BMP4 and WNT3A signaling at days 5 and 10, respectively, whereas the expression of both proteins was dampened in PRDM1-KD cells (Figures 3E and S2D). These data show that PRDM1 is necessary for BMP4- and WNT3A-induced germ cell fate in vitro.
PRDM1 Promotes Germline Differentiation of hESCs
Given that PRDM1 is important for germline fate driven by BMP4 and WNT3A treatment in hESCs, we wondered whether the BMP4- and WNT3A-induced germ cell formation could be further enhanced by overexpression of PRDM1. H9 hESCs were transduced with lentiviral vectors encoding GFP-PRDM1 or GFP and then treated with BMP4 and WNT3A. As shown in Figures 4A and S2C, a small portion (4.5%) of GFP control vector-transduced H9 cells expressed VASA 12 days after treatment with BMP4 and WNT3A. To our surprise, combined treatment with BMP4/WNT3A and GFP-PRDM1 expression synergistically enhanced the expression of VASA (Figures 4A and S2E), suggesting that ectopic PRDM1 expression can further promote the effect of BMP4 and WNT3A. It is noteworthy that treatment with BMP4 and WNT3A along with PRDM1 overexpression generated more cells expressing punctate and elongated SCP3 (Figures 4B and 4C). Supporting this finding, fluorescence in situ hybridization (FISH) analysis with probes for chromosomes 16 and X on GFP-PRDM1 or GFP-expressing and BMP4/WNT3A-treated cells revealed that ectopic expression of GFP-PRDM1 resulted in a small percentage of cells (0.67%) having only a single staining signal for both probes (haploid), whereas the chromosome contents of GFP-expressing cells were all diploid (Figure 4D). In combination, these results indicate that ectopic expression of PRDM1 together with the exposure to BMP4 and WNT3A facilitated the generation of germ cells with meiotic properties.
Figure 4.
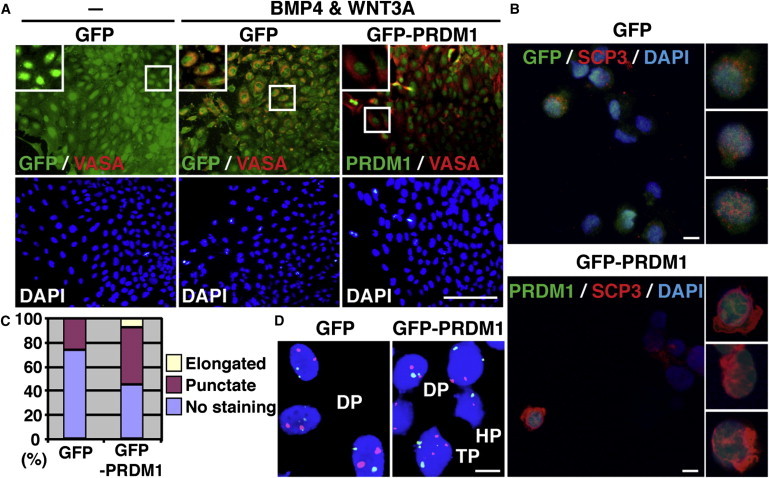
Ectopic Expression of PRDM1 in hESCs Enhances BMP4- and WNT3A-Induced Germline Differentiation
(A) Immunofluorescence staining of VASA shows that in comparison with GFP-expressing cells, GFP-PRDM1-expressing cells generated enhanced VASA signals after exposure to BMP4 or WNT3A for 2 weeks. The images shown in the selected boxes are enlarged in the upper-left corner of each panel. Scale bar, 100 μm.
(B) Immunofluorescence staining shows the formation of SCP3+ cells in GFP-PRDM1-expressing H9 cells treated with BMP4 and WNT3A for 2 weeks. Scale bar, 10 μm.
(C) Percentage of cells showing punctate, elongated, or negative stains for SCP3 within 300 GFP+ or 300 GFP-PRDM1+ H9 cells treated with BMP4 and WNT3A on day 14.
(D) FISH results for chromosome 16 (green) and X (red) in GFP-PRDM1- or GFP-expressing cells as described in (B). DP, diploid; HP, haploid; TP, tetraploid. Scale bar, 10 μm.
See also Figure S2.
Because Prdm1-expressing cells in the ICM are predisposed to a PGC fate in mice (Chu et al., 2011), we wondered whether overexpression of PRDM1 in hESCs is sufficient to enhance the outburst of germ cells. Lentiviral vectors producing GFP-PRDM1 fusion protein or GFP were used to transduce H9 hESCs that were maintained in ES media and cocultured with feeder cells. Of note, in contrast to GFP, forced expression of GFP-PRDM1 in H9 hESCs resulted in a subset of cells expressing STELLA, NANOS3, or VASA at week 2 (Figure 5A), suggesting that enforced PRDM1 expression in hESCs upregulated these markers. In parallel, GFP+ or GFP-PRDM1+ cells were harvested and subjected to analysis of expression of several germ cell markers by qRT-PCR. Accordingly, the mRNA levels of several germline markers, such as SSEA1, NANOS3, and VASA, were increased, whereas DNMT3b mRNA was reduced, in GFP-PRDM1-expressing cells compared with GFP-expressing cells (Figure 5B).
Figure 5.
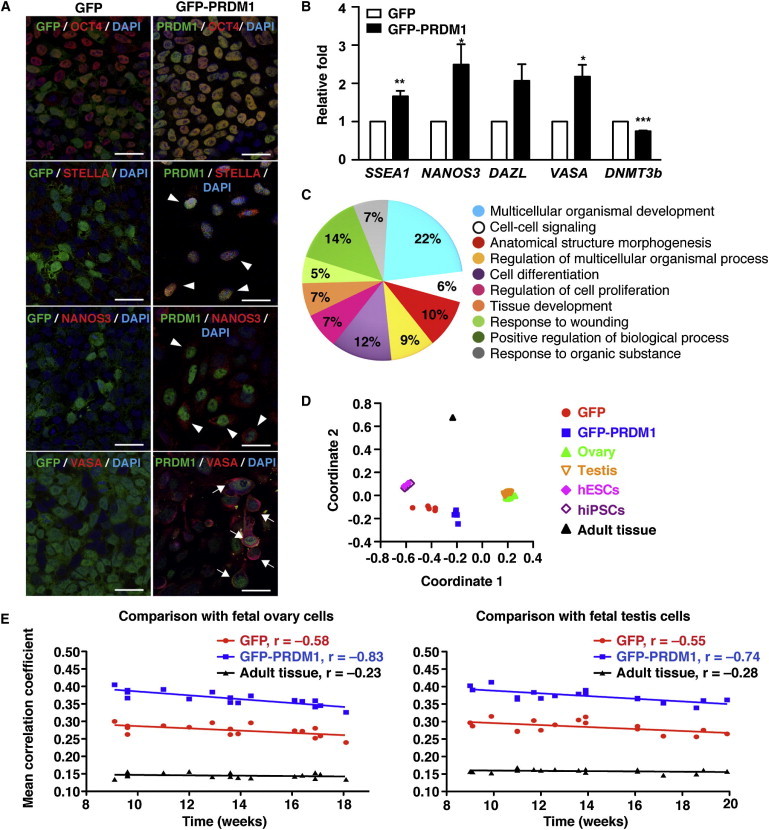
Ectopic Expression of PRDM1 in hESCs Enhances Embryonic Germ Cell Differentiation
(A) Immunofluorescence staining shows that ectopic expression of PRDM1 coexpressed with OCT4+ cells, and produced STELLA+ and NANOS3+ (indicated by the arrowheads), and VASA+ (indicated by the arrows) cells in H9 hESCs. GFP-PRDM1- or GFP-expressing H9 colonies were picked under the fluorescence microscope 7 days after lentiviral transduction and the GFP+ cells were seeded onto a new feeder layer, followed by the immunofluorescence staining and DAPI staining at week 2. Scale bar, 30 μm.
(B) qRT-PCR for the indicated mRNA using cDNA from isolated GFP+ H9 cells that expressed either GFP-PRDM1 or GFP for 2 weeks as described in (A). Results are the mean ± SEM from three independent experiments.
(C) GO analysis showed a significant enrichment of genes related to differentiation and development following PRDM1 expression in H9 cells. The list shows the top ten enriched GO terms based on the p value. The percentage represents the relative numbers of genes belonging to each category within the top ten enriched GO terms of 1,071 preferentially expressed genes.
(D) Multidimensional scaling plot of the 51 global gene-expression profiles of selected stem cells and tissue samples. Distances between data points approximate dissimilarity (1 − correlation coefficient) of their expression profiles.
(E) The average correlation coefficients between the expression profiles of H9 hESCs transduced with GFP- or GFP-PRDM1-expressing vector and fetal ovary (left) or testis (right) tissues at the indicated developmental time points.
To further establish whether hESCs expressing ectopic GFP-PRDM1 displayed a gene-expression signature similar to that observed in the germline lineages, we compared the transcription profiles of H9 hESCs transduced with GFP-PRDM1 and GFP control vectors on days 7 and 17 with data sets derived from hESCs and hiPSCs (Huang et al., 2011), human fetal testis and ovary tissues (Houmard et al., 2009), and differentiated tissues of human adults (Su et al., 2004). We found that 1,071 genes were selectively expressed in GFP-PRDM1 vector-transduced cells (Table S1). Gene Ontology (GO) analysis of these differentially expressed genes revealed that the top ten enriched GO categories were majorly involved in the processes of development, differentiation, proliferation, and signaling (Figure 5C; Table S2). We then projected the 1,071-dimensional transcription profiles of 216 experiments into a two-dimensional plane using nonclassical multidimensional scaling (MDS). Strikingly, the results in Figure 5D indicate that the developmental specification from hESCs toward germ cells following PRDM1 expression is reflected by the global gene-expression patterns. The data points from left to right correspond to the experiments with hESCs and hiPSCs, hESCs transduced with GFP-PRDM1, and fetal gonads. This trend is also pronounced by the average correlation coefficients between transcription from our experiments and three other studies in Figure 5D (Table S3).
We then analyzed the average correlation coefficients of global gene-expression profiles between GFP- and GFP-PRDM1-transduced H9 cells and fetal ovary or testis tissues along with developmental time points. In both ovary and testis, correlation coefficients with GFP-PRDM1-transduced cells unequivocally decreased with time. In contrast, the correlation coefficients with GFP-transduced cells exhibited only moderate dependencies with time. Furthermore, the correlation coefficients with the average expression profiles of differentiated adult tissues exhibited weak dependencies with time (Figure 5E). To ensure the similarities in global expression profiles between GFP-PRDM1 cells and gonad samples were attributed to germ cells, we calculated the expression profile correlation coefficients between four adult testis substructures in the data set of adult tissues (Su et al., 2004), and PRDM1-treated and control H9 hESCs (Table S3). GFP-PRDM1-transduced cells had the highest average correlation coefficient to germ cells (0.176), followed by Leydig cells (0.138), seminiferous tubules (0.131), and interstitial cells (0.119). These results suggest that the global gene-expression profiles of GFP-PRDM1-transduced hESCs resemble those of the early stage of germ cells.
PRDM1 Directly Suppresses SOX2 Expression
We then examined whether the expression of pluripotent genes was affected after ectopic expression of GFP-PRDM1 in H9 hESCs. Interestingly, OCT3/4 and NANOG mRNA levels were only marginally affected, but SOX2 mRNA levels were significantly reduced in GFP-PRDM1-expressing cells, when compared with GFP-expressing cells (Figure 6A). This finding was further verified by immunofluorescence staining, which showed that GFP-PRDM1+ cells displayed strikingly less SOX2 compared with GFP+ cells, but the majority of PRDM1+ cells remained to coexpress with OCT4 and NANOG, 7 days after lentiviral transduction (Figure 6B). To further clarify this observation, we transduced three human embryonal carcinoma (EC) lines (PA-1, NTERA-2 and NCCIT cells) with GFP- or GFP-PRDM1-expressing lentiviral vectors. Consistently, SOX2 mRNA and protein were dramatically repressed by exogenous PRDM1 in all three EC lines, whereas the expression of OCT4 and NANOG was only marginally influenced (Figure S3), suggesting that SOX2 is likely suppressed by PRDM1. Indeed, seven potential PRDM1-binding sites between the genomic region of −4.4 Kb and transcriptional start site of human SOX2 gene were found, according to the 12 bp consensus binding site, GTAGTGAAAGTG, determined previously (Kuo and Calame, 2004; Figure 6C). Chromatin immunoprecipitation (ChIP) assay using chromatins prepared from NCCIT EC cells expressing GFP-PRDM1 or GFP and anti-PRDM1 antibody demonstrated that PRDM1 bound to some of these predicted sites (Figure 6D). The CIITA gene was used as the positive control locus (Su et al., 2009), and the 3′ UTR of SOX2 and CIITA loci was used as the negative control (Figure 6D). Moreover, we constructed luciferase reporters fused with the 5′ regulatory region, located between −4.4 Kb and +268 of human SOX2 containing all seven potential PRDM1-binding sites (SOX2-Luc) or other various lengths of the 5′ flanking region of SOX2 (Figure 6E), in order to dissect the major PRDM1-binding site(s) that can contribute to the suppression of SOX2. Figure 6E shows that PRDM1 suppressed the luciferase activity regulated by the −4.4 Kb 5′ regulatory region of human SOX2 (a). The luciferase reporter construct deleting sites 1, 2, and 3 of the SOX2 5′ flanking region (b) was suppressed significantly less by PRDM1, whereas deletion of an additional region containing site 4 (c) did not reveal further repression (Figure 6E). Further deletion of the SOX2 5′ flanking region composed of sites 1–5 (d) rendered the luciferase activity refractory to PRDM1 suppression (Figure 6E). Lastly, luciferase activity driven by the −1.5 Kb 5′ regulatory region of SOX2, which lacks either sites 1–6 or all seven potential PRDM1-binding sites (e and f, respectively), were resistant to PRDM1 suppression (Figure 6E). To further validate these findings, we mutated sites 1, 2, and/or 3 in construct (a) by site-directed mutagenesis. We found that when sites 1/2 and 1/2/3 were simultaneously, but not individually, mutated, PRDM1-mediated repression of the SOX2 promoter activity was reduced (Figure 6F). These data suggested that sites 1, 2, and 3 (primarily sites 1 and 2) on the SOX2 5′ flanking region are involved in PRDM1-mediated transcriptional repression. Together, these data show that germline specification in hESCs triggered by the expression of PRDM1 is linked with the suppression of SOX2.
Figure 6.
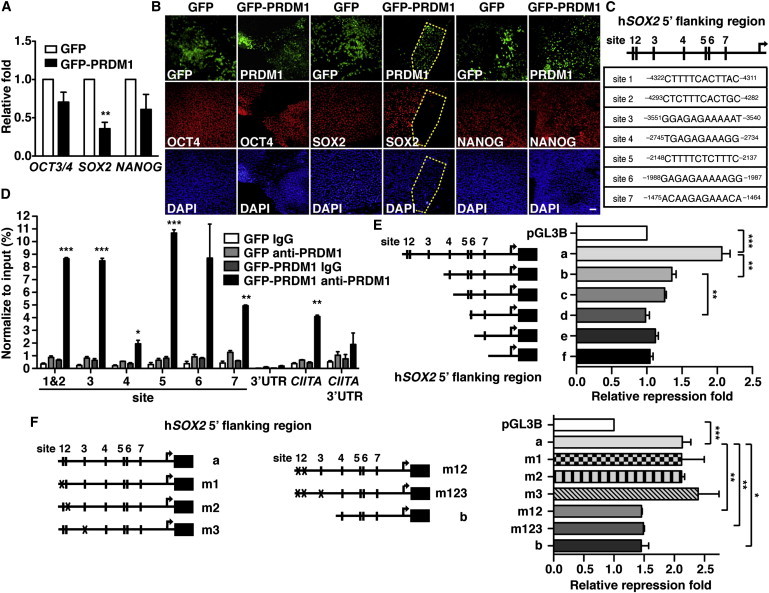
Enforced PRDM1 Expression Represses SOX2
(A) qRT-PCR shows the relative mRNA levels of OCT3/4, SOX2, and NANOG in H9 cells 7 days after transduction with GFP-PRDM1- or GFP-expressing lentiviral vectors.
(B) Immunofluorescence staining of OCT4, SOX2, or NANOG in H9 cells expressing GFP-PRDM1 or GFP for 7 days. Scale bar, 100 μm.
(C) Schematic representation of seven potential PRDM1-binding sites on the SOX2 5′ flanking region. The transcriptional start site (+1) is indicated.
(D) ChIP showed the binding of PRDM1 in the SOX2 5′ flanking region in NCCIT EC cells expressing GFP-PRDM1. NCCIT EC cells were transduced with either GFP-PRDM1 or GFP producing lentiviral vectors. Two days later, ChIP analysis was performed. Immunoglobulin G (IgG) was used as isotype control antibody. The 3′ UTR of SOX2 and CIITA gene loci and CIITA promoter III were used as the negative control and positive loci for PRDM1 binding, respectively. Results (mean ± SEM) are triplicate data for one representative experiment of three independent experiments.
(E and F) The luciferase reporter assay showed that multiple PRDM1-binding sites on the SOX2 5′ flanking region are involved in PRDM1-mediated suppression. Relative luciferase activity was measured using lysates from NCCIT EC cells transfected with PRDM1 expression or control vector, equal molar of the indicated luciferase reporters driven by various lengths of SOX2 5′ flanking region (E) or SOX2 5′ flanking region containing various PRDM1-binding-site mutants (m) (F) together with thymidine kinase promoter-Renilla luciferase reporter plasmid (RL-tk). Luciferase activity assay was analyzed 48 hr later. Results in (A), (E), and (F) are the mean ± SEM of three independent experiments.
See also Figure S3.
Suppression of SOX2 Is Required for Germline Differentiation In Vitro
In addition to ESCs, SOX2 is expressed in neuroectoderm and is a functional marker of neural lineages (Graham et al., 2003). Since PRDM1 suppresses SOX2, we wondered whether BMP4- and WNT3A-mediated induction of PRDM1, which favors germline differentiation, participates in the suppression of neural differentiation. Indeed, BMP4 and WNT3A treatment of H9 hESCs resulted in the downregulation of PAX6, the neural marker (Zhang et al., 2010; Figure S4), as well as SOX2 (Figure 7A). We thus analyzed the expression of several neural markers (Hou et al., 2013), including SOX1, LHX2, NESTIN, TUJ1, and MAP2, in PRDM1-KD H9 hESCs treated with BMP4 and WNT3A. We found that PRDM1-KD led to increased SOX2 mRNA and protein (Figures 7A and S5A), consistent with the notion that PRDM1 directly represses SOX2. Interestingly, the mRNA levels of several neural genes, such as SOX1, PAX6, NESTIN, and MAP2, but not LHX2, were all elevated in PRDM1-KD cells at various time points (Figure 7A). Immunofluorescence staining results also showed that PRDM1-KD GFP+ cells exhibited stronger signals for the neural markers SOX1, NESTIN, TUJ1, and MAP2 after treatment with BMP4 and WNT3A, as compared with shControl-transduced GFP+ cells (Figure 7B). These data suggested that BMP4- and WNT3A-induced PRDM1 not only drives germ cell differentiation but also prohibits the neural program.
Figure 7.
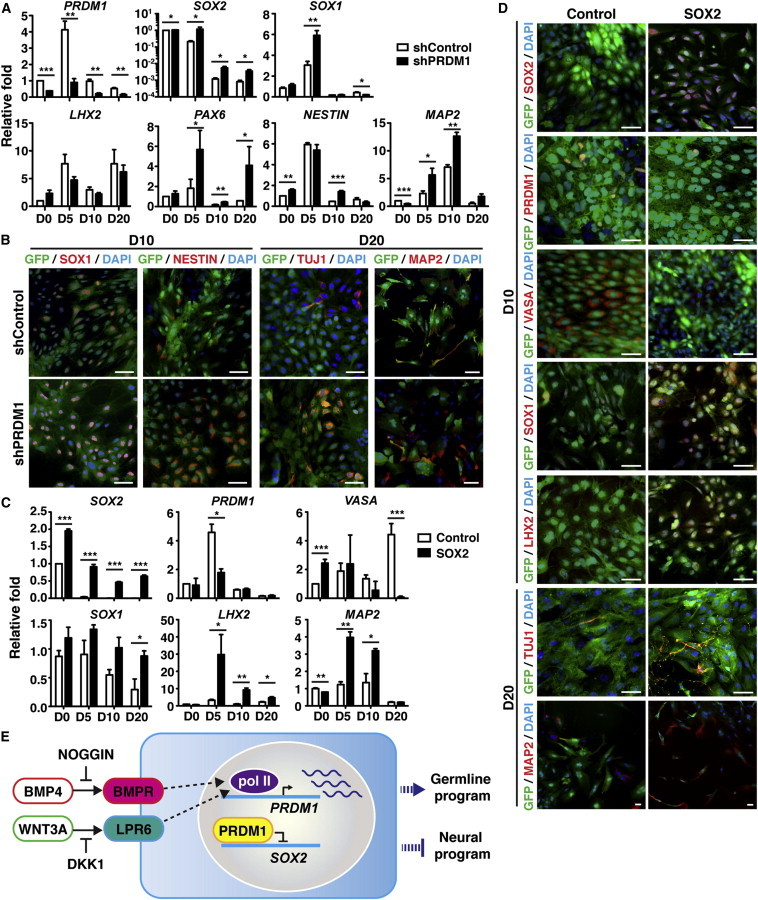
The PRDM1/SOX2 Regulatory Axis Controls Germ Cell versus Neural Cell Fate in BMP4- and WNT3A-Treated hESCs
(A) shControl or shPRDM1 lentiviral vectors were transduced into BMP4- and WNT3A-treated H9 cells. Alteration of mRNA expression was quantified by qRT-PCR. Relative values were calculated by comparing the mRNA levels of the indicated genes in PRDM1-KD cells at the indicated days with shControl-transduced cells on day 0.
(B) Immunofluorescence staining indicated that shPRDM1-transduced, but not shControl-transduced, cells showed enhanced SOX1, NESTIN, TUJ1, and MAP2 at the indicated days after BMP4 and WNT3A treatment. Scale bar, 30 μm.
(C) Sorted GFP+ H9 cells that were expressing control GFP alone or SOX2 and GFP simultaneously were treated with BMP4 and WNT3A, followed by qRT-PCR analysis to detect the indicated genes at various days. The fold change was plotted relative to the values obtained from the control vector-transduced cells on day 0.
(D) Immunofluorescence staining showed the expression of exogenous SOX2, reduced expression of PRDM1 and VASA, and increased expression of SOX1, LHX2, TUJ1, and MAP2 as compared with control GFP+ cells at the indicated days after BMP4 and WNT3A treatment. Scale bar, 30 μm. Results in (A) and (C) are the mean ± SEM of three independent experiments.
(E) Working model of the function of PRDM1 in germline differentiation by hESCs. BMP4- and WNT3A-mediated upregulation of PRDM1 transcriptionally suppresses SOX2, thereby inducing the activation of germline fate and preventing the neural program. Dashed lines indicate that the detailed signaling pathways leading to the induction of PRDM1 have not been characterized.
See also Figures S4 and S5.
Given that downregulation of SOX2 is linked with human germ cell differentiation, we wondered whether ectopic expression of SOX2 in hESCs blocks BMP4- and WNT3A-mediated germline differentiation. H9 hESCs stably expressing control GFP or SOX2 and GFP simultaneously were established for this purpose (Figures S5B–S5E). Four weeks after ectopic expression of SOX2, H9 hESCs cultured in ES media showed slightly increased numbers of SOX1+, LHX2+, PAX6+, and NESTIN+ cells as compared with control GFP-expressing H9 hESCs (Figure S5E). Endogenous SOX2 was rapidly decreased in control GFP+ cells at day 5 after BMP4 and WNT3A treatment, whereas the expression of exogenous SOX2 was sustained during differentiation (Figure 7C). It is noted that in the presence of exogenous SOX2, induction of PRDM1 at day 5 and VASA at day 20 after BMP4 and WNT3A treatment was perturbed as compared with control GFP-expressing cells (Figure 7C). qRT-PCR or immunofluorescence staining showed that the expression levels of neural markers, including SOX1, LHX2, TUJ1, and MAP2, were elevated in SOX2-overexpressing cells (Figures 7C and 7D). These data suggest that SOX2 impedes germ cell differentiation induced by BMP4 and WNT3A, and redirects the germline-committed hESCs toward a neural fate.
Discussion
Here, we provide evidence showing the expression of PRDM1 in human fetal germline tissues. Although it has been reported that Prdm1 is absent in zebrafish PGCs (Ng et al., 2006), the expression of PRDM1 in the germ cell lineage appears to be evolutionarily conserved between mouse and human. In the mouse germline, Prdm1 expression was found in several developmental stages: the ICM, proximal epiblast PCG precursors, and migrating PGCs along the hindgut and primitive gonadal ridge (Chang et al., 2002; Chu et al., 2011; Ohinata et al., 2005). Although in this study we were unable to demonstrate whether PRDM1 is expressed in human PGC precursors, as their formation occurs between 5 and 8 weeks of gestation (Clark, 2007; Freeman, 2003), we showed that PRDM1 is expressed in the second trimester of fetal gonads. Given that PRDM1 is coexpressed with OCT4, but not with the migratory germ cell marker VASA, in the developing human fetal testis or ovary of second-trimester abortus, it is plausible that the PRDM1-expressing cells in human fetal gonads are newly immigrating PGCs that can further differentiate into late germline progenitors/germ cells. Supporting this finding, it has been reported that OCT4 is expressed in a subpopulation of germ cells where the PGCs reside in testis cords in the second trimester of human testis development (Gaskell et al., 2004). Consistent with the in vivo observation, our in vitro differentiation studies demonstrated that PRDM1 is coexpressed with other germ cell markers in differentiating hESCs in early differentiation stages (days 5 and 10), but is not coexpressed with VASA in the late stage (day 20), although VASA has been detected in the fetal germline population as early as 7 weeks of gestation (Castrillon et al., 2000). This could be due to the heterogeneous nature of early germline development, and cells with different expression patterns of germline markers may represent a different subpopulation of germline progenitors that give rise to a larger population of VASA-expressing cells at later developmental stages.
The functional role of BMP/Smad and Wnt/β-catenin signaling pathways in early germ cell formation has been studied extensively in mice (Ohinata et al., 2009). It was demonstrated that only a subset of WNT3A-expressing epiblast cells of E5.5–E6.25 embryos respond to BMPs secreted from their surrounding extraembryonic cells, thereby acquiring the competence of early germ cells (Ohinata et al., 2009). Similar to these findings in mice, we previously demonstrated that BMP4 and WNT3A treatment significantly enhances the potential of germline differentiation by hESCs (Chuang et al., 2012). These observations led us to investigate the importance of BMP and/or Wnt signaling for PRDM1 induction in germline differentiation of hESCs. Indeed, we showed that BMP4 and WNT3A significantly increased the expression of germline-related marker genes as well as PRDM1, whereas blocking the activation of either BMP or Wnt signaling with their antagonists significantly reduced the expression of PRDM1. Furthermore, the germline potential was evidently impaired in PRDM1-KD hESCs after BMP4 and WNT3A treatment, suggesting that PRDM1 is most probably a crucial downstream effector of germline inducers such as BMP and Wnt signaling molecules. Thus, these results suggest an evolutionarily conserved BMP4/PRDM1 regulatory axis between human and mouse in controlling germ cell specification. Although we demonstrated BMP4-dependent PRDM1 induction, the mechanism by which BMP signaling activates PRDM1 is still elusive. It was previously suggested that, in mouse, BMP signals may upregulate Prdm1 via BMP-dependent activation of Smad1 and Smad5 (Ohinata et al., 2009). Further studies are necessary to determine whether SMAD proteins directly regulate PRDM1 during human germline differentiation.
During mouse PGC specification, Prdm1 served as a key transcriptional regulator responsible for repression of the somatic program and establishment of germline characteristics, including upregulation of pluripotency-associated genes such as Oct3/4, Nanog, and Sox2, in the PGC precursors (Kurimoto et al., 2008). We showed that ectopic expression of PRDM1 in hESCs downregulated SOX2 expression, but had no effect on the expression of OCT4 and NANOG. Further, combined ChIP and reporter assay analyses showed that PRDM1 directly targets the promoter of SOX2 and represses its transcription. However, these results also reveal a discrepancy between human and mouse in terms of the PRDM1-mediated germline specification. Sox2 is expressed in mouse germ cells (Saitou, 2009), whereas it is absent from human PGCs and gonocytes during the early stage of human development (Perrett et al., 2008). The functional significance of the absence of SOX2 from early human germ cells remains unclear. We show here that overexpression of SOX2 in hESCs under the condition favoring germline differentiation reduced the production of VASA+ cells, suggesting that SOX2 may perturb the expression of germline-associated genes. In hESCs, the SOX family proteins SOX2 and SOX3 are redundant and repress mesendoderm differentiation (Wang et al., 2012). It is thus plausible that in human, other SOX proteins may provide substitutive functions that similarly mimic the action of SOX2 in mouse for modulating germline formation. Supporting this notion, SOX17 was found to be coexpressed with OCT4 in early human germ cells (de Jong et al., 2008). This observation further suggests that SOX17 may play a role in conjunction with other factors, such as OCT4, in the generation of human germ cells.
The functional roles of SOX2 in human pluripotency maintenance and lineage differentiation were recently reported (Wang et al., 2012). SOX2 is not essential for hESC self-renewal. However, overexpression of SOX2 results in the promotion of neural ectodermal differentiation and the suppression of definitive endodermal differentiation in hESCs under in vitro differentiation (Wang et al., 2012). In agreement with these findings, we found that the neural program, including the expression of SOX1, LHX2, and NESTIN, was upregulated in hESCs overexpressing SOX2 (Figure S5E). Furthermore, our results demonstrated that the germline program was suppressed under the same condition, suggesting an opposing role of SOX2 in directing human neural and germ cell fate. Thus, it is tempting to speculate that PRDM1 may serve as a molecular switch to modulate the induction of neural or germline fates by turning off SOX2 during early human development (Figure 7E). The BMP4/PRDM1/SOX2 regulatory axis identified in this study may also provide a mechanistic clue to elucidate the molecular mechanisms underlying the suppression of neural development by BMP4 signaling (Hemmati-Brivanlou and Melton, 1997).
The study of germline development from hESCs has been hampered by the lack of proper protocols for efficient germline induction and/or selection from differentiating hESCs. Previous approaches, including combined growth factor/cytokine stimulation (Chuang et al., 2012) and human fetal gonadal cell coculture (Park et al., 2009), have shown promising effects on germline induction of hESCs and/or hiPSCs. Furthermore, advances in generating transgenic hESCs/hiPSCs harboring germline reporters, such as VASA (Kee et al., 2006) and OCT4 (Chuang et al., 2012), have also enabled us to enrich the potential germ cell populations from differentiating PSCs. Nevertheless, the generation of germ cells from hESCs/hiPSCs by the aforementioned protocols is still not satisfactory. In this study, we provide multiple lines of evidence demonstrating that ectopic expression of PRDM1 in hESCs is sufficient to promote the germline program, resulting in the generation of increased numbers of VASA-expressing PGCs during in vitro differentiation. In addition to promoting germ cell marker expression, we detected a small number of cells capable of expressing the meiotic marker SCP3 after ectopic expression of PRDM1. Furthermore, the decreased expression of DNMT3b in the differentiated cells overexpressing PRDM1 suggests that PRDM1 may contribute to somatic-to-germline epigenomic reprogramming by promoting the erasure of DNA methylation. However, we did not observe a significantly increased number of cells exhibiting the ability to complete meiosis as judged by haploidy of two chromosome FISH probes. This suggests that, in contrast to the function of previously reported factors, such as DAZ and BOULE (Kee et al., 2006), PRDM1 is not fully competent to promote the later stages of meiosis and development of haploid gametes. These results suggest that PRDM1 acts as an early rather than late germline effector. Our studies thus provide an induction system that will be beneficial for initiating early gem cell specification and subsequent germ cell generation by hESCs/hiPSCs.
Experimental Procedures
Collection of Human Fetal Samples
Human fetal tissues were obtained from women undergoing an induced termination of pregnancy, with the approval of the Human Subject Research Ethics Committee of Academia Sinica and of National Taiwan University Hospital. The weeks of gestational age were estimated by ultrasound and by measurement of the foot length. Fetal samples obtained from abortus between weeks 16 and 22 of gestational age, without morphological abnormality, were immediately fixed in 10% buffered formalin and then processed for IHC staining.
Culture and Differentiation of hESCs
The conditions used to maintain human EC cell lines are described in Supplemental Experimental Procedures. NTU1 hESCs were described previously (Chen et al., 2007). They were cultured on mitotically inactivated mouse embryonic fibroblast (MEF) feeder layers in primate ESC culture medium (ReproCELL) and 5 ng/mL human recombinant basic fibroblast growth factor (bFGF; Invitrogen). H9 hESCs (NIH code WA09, 46XX) were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (1:1) supplemented with 20% KnockOut serum replacement (Invitrogen) and 5 ng/mL bFGF (Invitrogen) on the MEF feeders. ESCs were subcultured by mechanical scraping and then transferred to fresh MEF feeder cells in ESC media weekly. During differentiation, the hESC split colonies were transferred to dishes coated with 10% Pluronic F-127 (Sigma) for suspension culture/embryoid body formation for 3 days. They were then replated on dishes coated with 0.1% gelatin (Sigma) for adherent culture, and incubated in differentiation media (DMEM from Invitrogen supplemented with 15% fetal bovine serum, 1% nonessential amino acid, 1% L-glutamine, and 0.5% antibiotics). In some experiments, adherent cultures were added with 100 ng/mL BMP4 (R&D Systems) and 50 ng/mL WNT3A (R&D Systems) in differentiation media. Cells were harvested for analysis at the indicated days within the range of not less or more than 36 hr as described in the text. The generation and maintenance of the OCT4-EGFP transgene reporter H9:OCT4-EGFP were described in a previous report (Chuang et al., 2012). H9:OCT4-EGFP cells were spontaneously differentiated on the indicated days. For qRT-PCR analysis, EGFP+ cells were sorted by FACS Aria (BD Biosciences). For immunofluorescence staining, EGFP+ cells were enriched by manual scraping and then replated on gelatin-coated coverslips.
IHC and Immunofluorescence Staining
Paraffin-embedded samples were sectioned at 3 μm. The tissue sections were then mounted on glass slides at 60°C for 10 min, followed by deparaffinization and rehydration as previously described (Angelin-Duclos et al., 2000). Detailed procedures are described in Supplemental Experimental Procedures. The procedures for immunofluorescence staining were performed essentially as described previously (Su et al., 2009) and the detailed protocol is described in Supplemental Experimental Procedures.
Meiotic Spreads, Meiotic Marker Staining, and FISH
Detailed protocols are provided in Supplemental Experimental Procedures.
RT-PCR, qRT-PCR, and Immunoblotting
Detailed protocols are provided in Supplemental Experimental Procedures.
Generation of Lentiviral Vector and Transduction
The lentiviral vectors used in this study, the procedure for preparing lentiviral vectors, and transduction are described in Supplemental Experimental Procedures.
ChIP, Transfection, and Luciferase Reporter Assay
Detailed procedures are described in Supplemental Experimental Procedures.
Microarray, Comparison with External Gene-Expression Microarray Data, and GO Analysis
Detailed procedures of microarray analysis and bioinformatic analysis to compare the gene-expression profiles of hESCs transduced with GFP-PRDM1 and GFP vectors with other tissues are described in Supplemental Experimental Procedures.
Statistics
All of the quantitative analyses were performed by Prism (GraphPad statistics software) and presented as the mean value ± SEM. The statistical significance was determined by two-tailed Student’s t tests; p < 0.05 was considered significant (∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05).
Acknowledgments
We thank Li-Wen Lo for technical assistance with confocal imaging. This work was supported by Academia Sinica (AS-99-CDA-L12 to K.-I L.) and the National Science Council, Taiwan (100-2628-B-001-015-MY4 to K.-I L., and 101-2321-B-001-019 and 100-2314-B-001-001-MY3 to H.-C.K.).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Hung-Chih Kuo, Email: kuohuch@gate.sinica.edu.tw.
Kuo-I Lin, Email: kuoilin@gate.sinica.edu.tw.
Accession Numbers
The accession number for the microarray data reported in this paper is GSE53283.
Supplemental Information
References
- Ancelin K., Lange U.C., Hajkova P., Schneider R., Bannister A.J., Kouzarides T., Surani M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Angelin-Duclos C., Cattoretti G., Lin K.I., Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J. Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- Bikoff E.K., Morgan M.A., Robertson E.J. An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet. Dev. 2009;19:379–385. doi: 10.1016/j.gde.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Castrillon D.H., Quade B.J., Wang T.Y., Quigley C., Crum C.P. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl. Acad. Sci. USA. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.H., Cattoretti G., Calame K.L. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech. Dev. 2002;117:305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Chen H.F., Kuo H.C., Chien C.L., Shun C.T., Yao Y.L., Ip P.L., Chuang C.Y., Wang C.C., Yang Y.S., Ho H.N. Derivation, characterization and differentiation of human embryonic stem cells: comparing serum-containing versus serum-free media and evidence of germ cell differentiation. Hum. Reprod. 2007;22:567–577. doi: 10.1093/humrep/del412. [DOI] [PubMed] [Google Scholar]
- Chu L.F., Surani M.A., Jaenisch R., Zwaka T.P. Blimp1 expression predicts embryonic stem cell development in vitro. Curr. Biol. 2011;21:1759–1765. doi: 10.1016/j.cub.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.Y., Lin K.I., Hsiao M., Stone L., Chen H.F., Huang Y.H., Lin S.P., Ho H.N., Kuo H.C. Meiotic competent human germ cell-like cells derived from human embryonic stem cells induced by BMP4/WNT3A signaling and OCT4/EpCAM (epithelial cell adhesion molecule) selection. J. Biol. Chem. 2012;287:14389–14401. doi: 10.1074/jbc.M111.338434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.T. Establishment and differentiation of human embryonic stem cell derived germ cells. Soc. Reprod. Fertil. Suppl. 2007;63:77–86. [PubMed] [Google Scholar]
- de Jong J., Stoop H., Gillis A.J., van Gurp R.J., van de Geijn G.J., Boer Md., Hersmus R., Saunders P.T., Anderson R.A., Oosterhuis J.W., Looijenga L.H. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J. Pathol. 2008;215:21–30. doi: 10.1002/path.2332. [DOI] [PubMed] [Google Scholar]
- Easley C.A.t., Phillips B.T., McGuire M.M., Barringer J.M., Valli H., Hermann B.P., Simerly C.R., Rajkovic A., Miki T., Orwig K.E. Direct Differentiation of Human Pluripotent Stem Cells into Haploid Spermatogenic Cells. Cell reports. 2012;2:440–446. doi: 10.1016/j.celrep.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B. The active migration of germ cells in the embryos of mice and men is a myth. Reproduction. 2003;125:635–643. doi: 10.1530/rep.0.1250635. [DOI] [PubMed] [Google Scholar]
- Gaskell T.L., Esnal A., Robinson L.L., Anderson R.A., Saunders P.T. Immunohistochemical profiling of germ cells within the human fetal testis: identification of three subpopulations. Biol. Reprod. 2004;71:2012–2021. doi: 10.1095/biolreprod.104.028381. [DOI] [PubMed] [Google Scholar]
- Geijsen N., Horoschak M., Kim K., Gribnau J., Eggan K., Daley G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A., Melton D. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell. 1997;88:13–17. doi: 10.1016/s0092-8674(00)81853-x. [DOI] [PubMed] [Google Scholar]
- Hou P.S., Chuang C.Y., Kao C.F., Chou S.J., Stone L., Ho H.N., Chien C.L., Kuo H.C. LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 2013;41:7753–7770. doi: 10.1093/nar/gkt567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houmard B., Small C., Yang L., Naluai-Cecchini T., Cheng E., Hassold T., Griswold M. Global gene expression in the human fetal testis and ovary. Biol. Reprod. 2009;81:438–443. doi: 10.1095/biolreprod.108.075747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.P., Chen P.H., Hwu W.L., Chuang C.Y., Chien Y.H., Stone L., Chien C.L., Li L.T., Chiang S.C., Chen H.F. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum. Mol. Genet. 2011;20:4851–4864. doi: 10.1093/hmg/ddr424. [DOI] [PubMed] [Google Scholar]
- Hübner K., Fuhrmann G., Christenson L.K., Kehler J., Reinbold R., De La Fuente R., Wood J., Strauss J.F., 3rd, Boiani M., Schöler H.R. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- Julaton V.T., Reijo Pera R.A. NANOS3 function in human germ cell development. Hum. Mol. Genet. 2011;20:2238–2250. doi: 10.1093/hmg/ddr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K., Gonsalves J.M., Clark A.T., Pera R.A. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006;15:831–837. doi: 10.1089/scd.2006.15.831. [DOI] [PubMed] [Google Scholar]
- Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomelí H., Nagy A., McLaughlin K.J., Schöler H.R., Tomilin A. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.C., Calame K.L. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J. Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P.B., Spyropoulos B. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma. 1995;104:175–182. doi: 10.1007/BF00352182. [DOI] [PubMed] [Google Scholar]
- Ng T., Yu F., Roy S. A homologue of the vertebrate SET domain and zinc finger protein Blimp-1 regulates terminal differentiation of the tracheal system in the Drosophila embryo. Dev. Genes Evol. 2006;216:243–252. doi: 10.1007/s00427-005-0044-5. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O’Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Park T.S., Galic Z., Conway A.E., Lindgren A., van Handel B.J., Magnusson M., Richter L., Teitell M.A., Mikkola H.K., Lowry W.E. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett R.M., Turnpenny L., Eckert J.J., O’Shea M., Sonne S.B., Cameron I.T., Wilson D.I., Rajpert-De Meyts E., Hanley N.A. The early human germ cell lineage does not express SOX2 during in vivo development or upon in vitro culture. Biol. Reprod. 2008;78:852–858. doi: 10.1095/biolreprod.107.066175. [DOI] [PubMed] [Google Scholar]
- Saitou M. Germ cell specification in mice. Curr. Opin. Genet. Dev. 2009;19:386–395. doi: 10.1016/j.gde.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., Staudt L.M. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.T., Ying H.Y., Chiu Y.K., Lin F.R., Chen M.Y., Lin K.I. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell. Biol. 2009;29:1421–1431. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M.A. Germ cells: the eternal link between generations. C. R. Biol. 2007;330:474–478. doi: 10.1016/j.crvi.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Tanaka S.S., Yamaguchi Y.L., Tsoi B., Lickert H., Tam P.P. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell. 2005;9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Vincent S.D., Dunn N.R., Sciammas R., Shapiro-Shalef M., Davis M.M., Calame K., Bikoff E.K., Robertson E.J. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Yabuta Y., Kurimoto K., Ohinata Y., Seki Y., Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol. Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang C.T., Chen J., Pankratz M.T., Xi J., Li J., Yang Y., Lavaute T.M., Li X.J., Ayala M. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


