Abstract
Some of the characteristics of cancer cells are high rates of cell proliferation, cell survival, and the ability to invade surrounding tissue. The cytoskeleton has an essential role in these processes. Dynamic changes in the cytoskeleton are necessary for cell motility and cancer cells are dependent on motility for invasion and metastasis. The signaling pathways behind the reshaping and migrating properties of the cytoskeleton in cancer cells involve a group of Ras-related small GTPases and their effectors, including the p21-activated kinases (Paks). Paks are a family of serine/threonine protein kinases comprised of six isoforms (Pak 1–6), all of which are direct targets of the small GTPases Rac and Cdc42. Besides their role in cytoskeletal dynamics, Paks have recently been shown to regulate various other cellular activities, including cell survival, mitosis, and transcription. Paks are overexpressed and/or hyperactivated in several human tumors and their role in cell transformation makes them attractive therapeutic targets. Pak-targeted therapeutics may efficiently inhibit certain types of tumors and efforts to identify selective Pak-inhibitors are underway.
Keywords: p21-activated kinase, Cell transformation, Cytoskeleton, Rac, Cdc42
1 Pak kinase family
The vast majority of cancers are controlled by oncogenes and their signaling pathways. The dissection of these pathways has led to a wealth of targets for therapeutic intervention and several new drugs that are already on the market to treat tumors. Some of the most successful drugs have been targeted against protein kinases. Protein kinases that have key roles in cell survival, cell proliferation, and cell migration, are important targets because of their roles in cancer cell growth and tumor invasion. One group of such kinases, the Pak kinases, show promise as a potential target.
1.1 Pak isoforms and structure
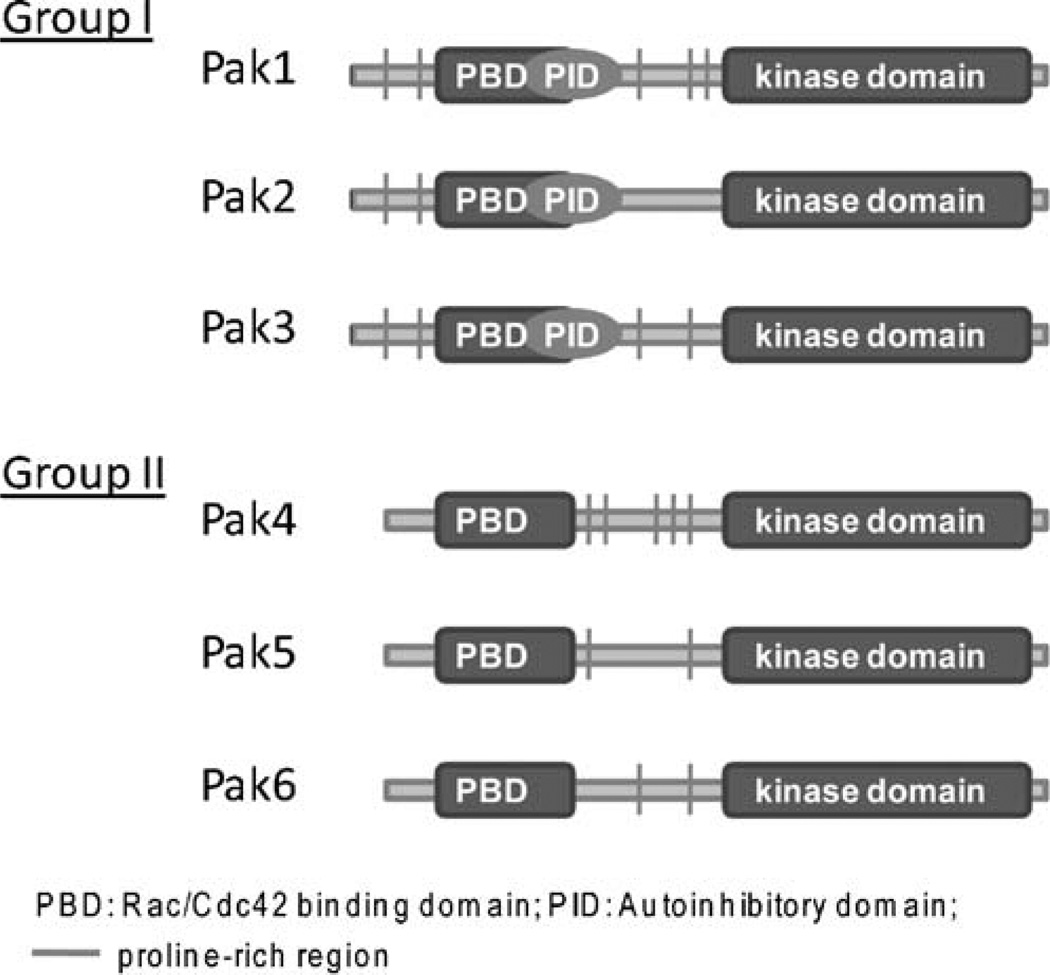
Pak kinases were first identified in screens for Rac and Cdc42 effectors and independently as a proteinase-activated kinase [1, 2]. They are widely conserved and found in yeast as well as Drosophila [3]. In mammals, six isoforms of Pak kinase have been found (Pak1–6), and these are subdivided into two groups (Fig. 1). Group I consists of Pak1 (αPak), Pak2 (γPak), and Pak3 (βPak), while Group II consists of Pak4, Pak5 and Pak6 [4].
Fig. 1.
Domain structure of Pak isoforms. The p21 (Rac/Cd42)-binding domain (PBD) and kinase domain are shown, as well as proline-rich putative SH3-binding motifs. Groups I Paks additionally contain an autoinhibitory domain (PID) that is overlapping with the PBD
All Paks are characterized by an N-terminal regulatory domain and a highly conserved C-terminal kinase domain. The kinase domains of Group I Paks are at least 93% homologous and about 54% homologous to members of the other group. The regulatory domains of all Paks consist of GTPase-binding domain (PBD) and several proline-rich regions that serve as docking sites for SH3 domain containing proteins. Group I Paks additionally possess an autoinhibitory domain (PID) overlapping with the PBD [5]. Group I Paks appear to form homodimers in cells, adopting a trans-inhibited conformation where the N-terminal PID of one molecule binds and inhibits the catalytic domain of the other [6]. Binding of activated Cdc42 or Rac to the PBD disrupts dimerization and activates Group I Paks, by releasing the PID-mediated inhibition and allowing autophosphorylation of the activation loop (Thr423 for Pak1). Phosphorylation of this site prevents refolding and consequent inhibition, even in absence of Rac and Cdc42. An acidic substitution at this site (T423E) renders Pak1 constitutively active. Autophosphorylation of other sites, mostly found in the N-terminus, also contributes to activation [7, 8].
Group II Paks do not possess an identifiable PID and have a higher basal kinase activity than Group I Paks. Although Group II Paks are still able to bind GTP-Rac and GTP-Cdc42, this does not enhance their kinase activities [9, 10]. However, binding of Rac and Cdc42 may regulate localization of Group II Paks and/or their interaction with other proteins.
1.2 Pak activation mechanisms
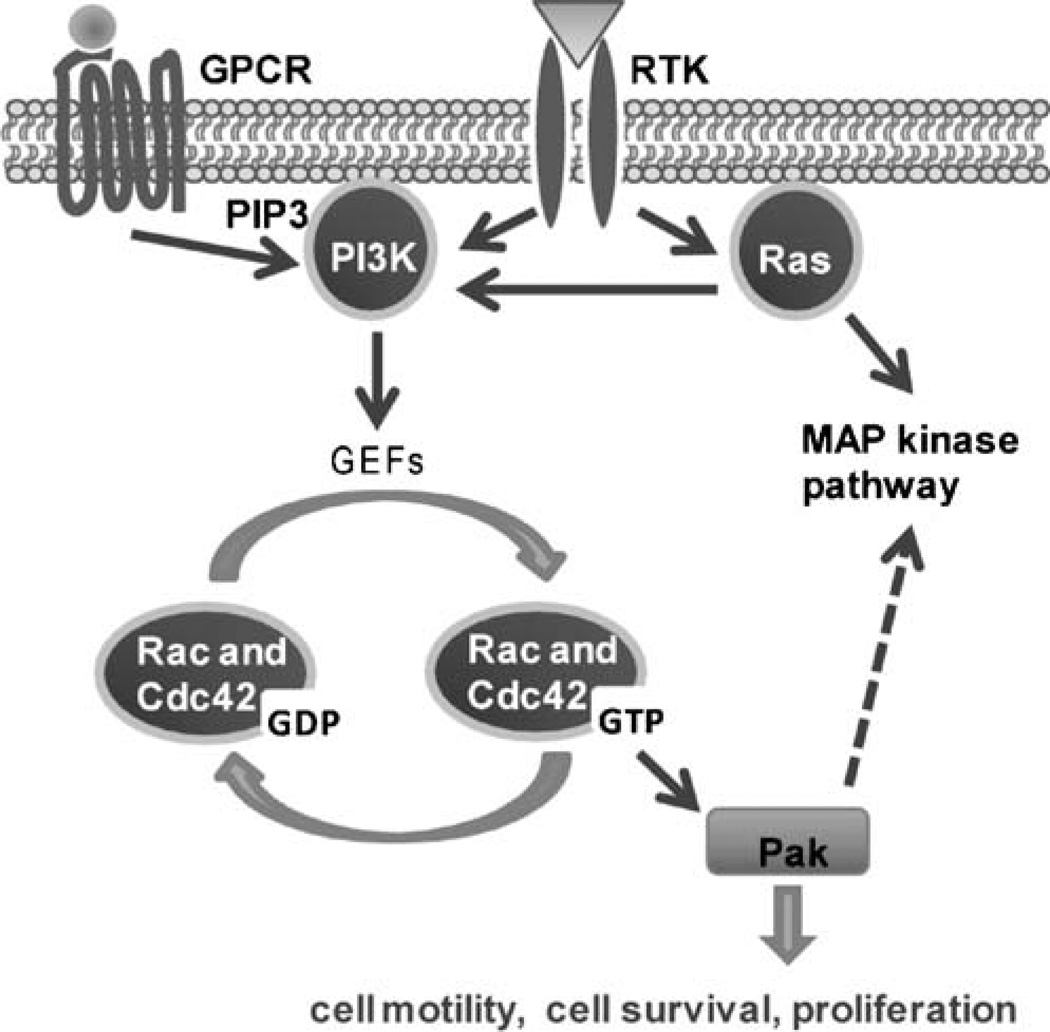
Signals from several growth factor receptor tyrosine kinases (e.g. insulin, EGF, PDGF, and VEGF receptors) and G protein-coupled receptors lead to activation of Pak [11–18]. These pathways generally activate Pak through sequential activation of PI-3 kinase (PI3K) and a guanine nucleotide exchange factor (GEF) from the Dbl family, which then activates the small GTPases Rac and Cdc42 (Fig. 2).
Fig. 2.
Schematic diagram of Pak activation by the small GTPases Rac and Cdc42. Signals from receptor tyrosine kinases, (e.g. insulin, EGF, PDGF, and VEGF receptors) and G protein-coupled receptors lead to activation of Pak via GTP-bound Rac and Cdc42. Activated Pak in turn initiates signaling cascades that culminate in the cellular response. In addition, activated Pak potentiates activation of the MAP kinase pathway. Of note, while activation of Pak via Rac and Cdc42 is well characterized, a number of GTPase-independent mechanisms for Pak activation have also been identified. GPCR, G protein-coupled receptors; RTK, receptor tyrosine kinase; PI3 K, phosphatidylinositol-3 kinase; PIP3, phosphatidylinositol (3, 4, 5) trisphosphate
While activation of Pak via Rac and Cdc42 is well characterized, a number of GTPase-independent mechanisms also modify Pak activity and function. For instance, the cell cycle regulated kinases Cdc2 and Cyclin-dependent kinase 5 (Cdk5) phosphorylate Pak1 at Thr212. Phosphorylation at this site alters the association of Pak1 with binding partners and/or substrates, affecting morphological changes such as postmitotic cell spreading in fibroblasts and microtubule dynamics in neurite outgrowth [19, 20]. PDK1 is able to activate Pak1 by direct phosphorylation of Thr423 [21], and the adaptor proteins Nck and Grb2 bind proline-rich regions near the N-terminus of Pak and can activate Pak by directing it to receptor tyrosine kinases at the cell membrane [22–24]. A family of guanine nucleotide exchange factors, collectively known as PIX or COOL, bind a different proline region on Pak to activate it through both a GTPase-dependent and a GTPase-independent mechanism [25, 26]. Paks can also be activated by sphingolipids [21]. Most of these GTPase-independent mechanisms have only been documented for Group I Paks.
In cancers, Pak activation frequently occurs via mutated Ras. Ras is one of the most commonly mutated oncogenes and activates the MAP kinase pathway as well as PI3 kinase (see Fig. 2) [27].
Several proteins have been identified, which negatively regulate Pak activity. Pak-interacting protein (PIP) abolishes kinase activity by binding to the regulatory domain of Pak, and inhibits Pak-mediated Jun kinase and NFkappaB signaling [28]. Nischarin interacts with the kinase domain of active Pak and inhibits the ability of Paks to phosphorylate substrates [29]. Nischarin may be important for local limitation of Pak activity in migrating cells. Merlin binds Pak1 through its PBD domain and is discussed in detail in Section 3.3 [30]. Partner of PIX1 and 2 (POPX1 and POPX2) are two phosphatases that dephosphorylate the Pak activation loop [31].
1.3 Pak substrate recognition
As for most protein kinases, there is some flexibility in the recognition sequences phosphorylated by Pak. One study used Pak2 and compared a limited number of peptides derived from the substrate KKRKSGL. This yielded a recognition sequence for Pak2 that is characterized by two basic amino acids in the −2 and −3 positions. For example, the peptide (K/R)RXS, in which the −2 position is an arginine, the −3 position is an arginine or a lysine is efficiently phosphorylated at the serine residue (X can be an acidic, basic, or neutral amino acid) [32]. A more recent study used a wider array of peptides and found that Pak1 and Pak2 preferred large hydrophobic residues in positions from +1 to +3, in addition to their preference for basic amino acids at the −2 and −3 positions [33]. While Pak1 and Pak2 shared nearly identical substrate specificities in this study, the substrate specificity of Pak4 was significantly different. Pak4 had a strong preference and for alanine at the +2 and serine at the +3 position. It should be noted that although there are differences in the preferred consensus sequences for Group I and Group II Paks, most known substrates are phosphorylated by both groups. Additionally, both groups strongly prefer serine over threonine as a phospho-acceptor site and do not phosphorylate tyrosine at all. Although the later study was able to identify a new Pak substrate by scanning databases, there are limitations to identifying substrates by sequence searches. The study found that none of the known Pak substrates fell into the top 2% of the predicted substrates, suggesting that other factors such as protein/protein interactions facilitate phosphorylation of what are otherwise less-ideal substrates.
2 Pak functions
Up to date, over 40 proteins have been identified as substrates for Paks (see Table 1), reflecting the significant roles the Pak kinase family plays in a range of biological activities. The most prominent functions are discussed below, and include stimulation of cell proliferation, cell survival, and cell motility. Deregulation of these cellular processes promotes carcinogenesis and Pak signaling can thus play a mechanistic role in cell transformation. Of note, Paks have also been implicated in other cellular processes that are relevant in tumorigenesis, such as angiogenesis [34], epithelial-mesenchymal transition [35], anchorage-independent growth [36, 37], and metabolism [38, 39].
Table 1.
Reported Pak substrates
| Process | Substrate | Sites | Isoform | Reference |
|---|---|---|---|---|
| Cytoskeleton remodelling | alpha-PIX | S488 | Pak1 | [33] |
| beta-PIX | S340, S525 (S497, S682)* | Pak1, Pak2 | [33, 105] | |
| Caldesmon | S657, S687 | Pak1, Pak3 | [106, 107, 108] | |
| CPI17 | T38 | Pak1 | [109] | |
| Desmin | Pak1 | [110] | ||
| Filamin A | S2152 | Pak1 | [111] | |
| GEF-H1 | S885 | Pak1 | [112] | |
| GIT1 | S517 | Pak1 | [43] | |
| LIM kinase | T508 | Pak1, Pak4 | [113, 114] | |
| MBS | T641 | Pak1 | [109] | |
| MLCK | S439, S991 | Pak1, Pak2 | [60, 115] | |
| NET1 | S152, S153 | Pak1 | [116] | |
| Op18/stathmin | S16 | Pak1 | [59] | |
| p41-ARC | T21 | Pak1 | [57] | |
| Rho GDI | S101, S174 | Pak1 | [117] | |
| R-MLC | S19 | Pak2 | [118, 119] | |
| TCoB | S65, S128 | Pak1 | [41] | |
| Vimentin | S25, S38, S50, S56, S65, S72 | Pak1 | [120–124] | |
| Cell growth | Abl1 | S637, S638 | Pak2 | [125, 126] |
| Aurora A | T288, S342 | Pak1 | [43] | |
| B-Raf | S446 | Pak1 | [127] | |
| c-Myc | T358, S373, T400 | Pak2 | [128] | |
| C-Raf1 | S338; S339 | Pak1, Pak2 Pak3 | [46, 50, 61, 129–131] | |
| ER alpha | S305 | Pak1 | [132, 133] | |
| Histone H3 | S10 | Pak1 | [40] | |
| MEK1 | S298 | Pak1 | [46, 134–137] | |
| MEKK1 | S67 | Pak1 | [138] | |
| Merlin | S518 | Pak1 | [95, 96, 100] | |
| MNK1 | S39 | Pak2 | [139] | |
| Plk1 | S49 | Pak1 | [44] | |
| Prolactin | S179 | Pak2 | [140] | |
| Cell survival | BAD | S111 (S136 through Raf-1) | Pak1, Pak2 | [61, 72, 141] |
| DLC1 | S88 | Pak1 | [68] | |
| FKHR | S256 | Pak1 | [67] | |
| Miscellaneous | CtBP1 | S158 | Pak1 | [142] |
| ESE1 | S207 | Pak1 | [143] | |
| G alpha z | S16 | Pak1 | [144] | |
| p47 phox | S303, S304, S320, S328 | Pak1 | [14, 145] | |
| p67 phox | Not mapped | Pak1 | [146] | |
| PGAM–B | S23, S118 | Pak1 | [38] | |
| PGM | T466 | Pak1 | [39] | |
| SHARP | S3486, T3568 | Pak1 | [147] | |
| SNAI1 | S246 | Pak1 | [35] | |
| STAT5a | S779 | Pak1 | [148] | |
| Syk | Not mapped | Pak2 | [149] | |
| Synapsin I | S603 | Pak1 | [150] | |
| Troponin I | S149 | Pak1 | [151] | |
| Pak auto-phosphorylation | Pak1 | S21, S57, S144, S149, S199, S204 | Pak1 | [8] |
| Pak2 | S19, S20, S55, S141, S165, S192, S197 | Pak2 | [7, 8] | |
| Pak3 | S50, S139 | Pak3 | [8] |
Abl1 Abelson murine leukemia viral oncogene homolog 1, BAD Bcl-2 antagonist of cell death, CPI17 17-kDa PKC-potentiated inhibitory protein of PP1, CtBP1 C-terminal-binding protein 1, DLC1 dynein light chain 1, ER estrogen receptor, ESE1 epithelium-specific Ets transcription factor 1, FKHR Forkhead box protein O1, G alpha z guanine nucleotide binding protein (G protein), alpha z, GEF-H1 guanine nucleotide exchange factor H1, GIT1 G protein-coupled receptor kinase-interactor 1, MBS myosin binding subunit of type 1 protein phosphatase, MEK1 mitogen-activated protein kinase kinase 1, MEKK1 mitogen-activated protein kinase kinase kinase 1, MLCK myosin light chain kinase, MNK1 MAP kinase interacting kinase 1, NET1 neuroepithelial cell transforming gene 1 (RhoA-specific guanine nucleotide exchange factor), P41-ARC actin-related protein 2/3 complex 41 kDa subunit, p47 phox neutrophil NADPH oxidase activator 1, p67 phox neutrophil NADPH oxidase factor 2, PGAM-B phosphoglyceratemutase-B, PGM phosphoglucomutase, PIX PAK-interacting exchange factor, Plk1 Polo-like kinase 1, Rho GDI Rho GDP dissociation inhibitor, R-MLC regulatory myosin light chain, SHARP SMART/HDAC1 associated repressor protein, SNAI1 snail 1 zinc finger protein, STAT5a signal transducer and activator of transcription 5A, Syk spleen tyrosine kinase, TCoB tubulin cofactor B;
beta-PIX phosphorylation sites for transcript B
2.1 Cell cycle and aneuploidy
Pak appears to have a critical role during cell cycle progression, its kinase activity peaks at mitosis entry and remains sustained during mitotic progression. Pak1 localizes to specific structures during mitosis, including chromosomes, centrosomes, mitotic spindles, and the contraction ring during cytokinesis [40]. Overexpression of activated Pak1 in MCF-7 breast cancer cells leads to abnormal centrosome number and spindle organization, and consequently to aneuploidy [37]. The genomic plasticity afforded by aneuploidy facilitates the loss of tumor-suppressor genes and the accumulation of oncogenes.
Mitotic spindle function is inextricably linked with microtubule dynamics. Tubulin cofactor B (TCoB), a cofactor in the assembly of alpha/beta-tubulin, was recently identified as an interacting substrate of Pak1 [41]. Pak1 phosphorylates TCoB on Ser65 and Ser128 and co-localizes with TCoB on newly polymerized microtubules and centrosomes. Coordinate deregulation of TCoB and Pak1 may contribute to the multiple-spindle phenotype seen in human breast cancer cells and other tumors.
In the early phase of mitosis Pak1 may have a role in chromosome condensation. Pak1 co-localizes with Histone H3 on condensing chromosomes and phosphorylates Histone H3 on Ser10, an event that is required for the initiation of chromosome condensation [40, 42]. Interestingly, Histone H3 appears to interact specifically with Pak1 but not Pak2 or Pak3.
The protein kinases Aurora-A and Polo-like kinase 1 (Plk1) are two other important regulators of mitotic events that are phosphorylated by Pak. As cells near the M phase, Pak1 is recruited to the centrosomes where it interacts with a GIT1-PIX complex. GIT1 and PIX are two Pak-binding proteins that also interact with Pak1 during focal adhesion turnover in cells. They are present at the centrosome throughout all phases of the cell cycle. Interaction with GIT1-PIX activates Pak1 independently of the small GTPases Cdc42 or Rac. Activated Pak subsequently activates Aurora-A via phosphorylation on Thr288 and Ser342 [43]. Maroto et al. showed that Pak1 regulates Plk1 activity by phosphorylation of Plk1 at Ser49 [44]. Pak1 and Plk1 co-localize on the spindle poles, the central spindle, and the midbody. Pak1-mediated phosphorylation of Plk1 is important in establishing a functional bipolar spindle.
Regulation of cyclin D1 expression may be another mechanism, by which Pak promotes cell cycle progression. Several studies have shown that constitutively active PAK1 induces transcription of cyclin D1 via activation of the transcription factor NFkappaB [12, 45].
The ability of Pak to regulate the MAP kinase pathway may also contribute to cell proliferation. Pak phosphorylates two mediators of the MAP kinase pathway, MEK1 and Raf1, at Ser298 and at Ser338, respectively [46–50]. While phosphorylation of these sites by Pak is not sufficient to activate Raf1 or MEK1, it significantly facilitates the activation of these kinases by their upstream activators Ras and Raf1, respectively.
2.2 Cell motility and cancer metastasis
Dynamic changes of the cytoskeleton are critical for normal cell motility, neurogenesis, and angiogenesis. During malignant transformation, the signaling pathways controlling these cytoskeletal dynamics are altered, and an increase in cell motility allows cancer cells to invade surrounding tissues and metastasize. Pak functions as a downstream effector of Rac in the regulation of the actin cytoskeleton and stimulates cell motility and invasion.
PDGF, insulin, and certain other cell stimuli cause the redistribution of Pak1 from the cytosol into cortical actin structures, such as lamellae at the leading edge, circular dorsal ruffles, and peripheral dorsal ruffles [13, 51]. In addition, Pak1 localizes to focal adhesions. Expression of a constitutively active form of Pak1 induces the rapid formation of lamellipodia, filopodia, and dorsal ruffles, as well as an in increase in focal adhesion turnover and the disassembly of stress fibers [52–55]. There are several reported substrates of Pak that are involved in cytoskeletal reorganization, including LIM kinase, myosin light chain kinase, merlin, filamin A, p41-ARC, and Op18/stathmin.
Pak1 activates Lim kinase (LIMK) by phosphorylating it on Thr508 in the kinase activation loop. Active LIMK then phosphorylates the actin binding protein cofilin and inhibits its activity. Active cofilin promotes actin filament cycling and retrograde flow, while inhibition of cofilin through Pak1/LIMK signaling promotes integrity of the actin filament network in the lamellipodium and cell protrusion efficiency [56]. Pak1 also phosphorylates the p41-ARC subunit of the ARP2/3, a protein complex controlling actin nucleation and branching. Phosphorylation of p41-ARC by Pak1 stimulates complex assembly at the cellular cortex of migrating cells and is required for both constitutive and growth factor-induced cell motility [57].
Leading edge microtubule dynamics also plays a role in cell motility. In the protruding edge of migrating cells, microtubules exhibit decreased catastrophe frequency and increased net growth. Local regulation of Op18/stathmin, a protein that inhibits tubulin polymerization, could account for this. Rac1/Pak1 signaling appears to negatively regulate Op18/stathmin [58, 59]. Expression of constitutively active Rac1 leads to phosphorylation of stathmin at Ser16, a site which downregulates its inhibitory activity on tubulin polymerization, and Pak1 readily phosphorylates stathmin at Ser16 in vitro.
A third component of the cytoskeleton that is involved in cytoskeletal dynamics is myosin. In nonmuscle cells, the activity of myosin II is regulated by phosphorylation of the myosin light chains. Phosphorylation of the light chains promotes the assembly of myosin into bipolar filaments that generate tension on the actin and bundle actin filaments into stress fibers. While protrusion at the leading edge is critical for cell movement, it is equally important to stimulate disassembly and turnover of focal adhesions and stress fibers. Myosin light chain kinase (MLCK), the kinase phosphorylating myosin light chains, is a substrate for Pak [60]. Phosphorylation of MLCK by Pak decreases its activity, which in turn results in decreased myosin light chain phosphorylation and a decrease in actin-myosin filament assembly. Pak’s ability to inhibit myosin light chain phosphorylation is likely to account for the disassembly of stress fibers and focal adhesions observed in cells overexpressing activated Pak.
2.3 Cell survival and apoptosis
Apoptosis, or programmed cell death, is a fundamental process in the development of multicellular organisms. Apoptosis enables an organism to eliminate unwanted or defective cells through an organized process of cellular disintegration. Apoptosis is also a prominent tumor-suppression mechanism and cancer cells require inactivation of pro-apoptotic pathways for tumor formation and progression. Pak activity has been shown to downregulate several important pro-apoptotic pathways.
Pak1 protects cells from intrinsic apoptotic signals via a Pak-Raf1-BAD pathway. Pak1, and as well Pak5, induce phosphorylation of Raf1 at Ser338 and stimulate translocation of a subpopulation of Raf1 to the mitochondria [61–63]. At the mitochondria, Raf-1 forms a protective complex with Bcl-2 and phosphorylates the pro-apoptotic protein BAD at Ser112. Bcl-2 is a proto-oncogene that maintains the integrity of the mitochondrial barrier if bound in protective complexes, whereas binding of Bcl-2 to the pro-apoptotic protein BAD induces release of pro-apoptotic factors from the mitochondria and leads to apoptosis. Phosphorylation of BAD at specific sites, including Ser112, renders it unable to bind Bcl-2. The phenotype of Raf-1 knock out cells supports a protective role of Raf-1 in apoptosis, as these cells have high rates of apoptosis while exhibiting normal proliferative rates and ERK activation [64].
Other protective signals transduced through Pak include stimulation of the transcription factor NFkappaB and inhibition of the pro-apoptotic transcription factor FKHR. NFkappaB has been shown to regulate genes involved in cell survival, proliferation, and angiogenesis. In tumor cells NFkappaB is commonly activated. Several studies have shown that Pak1 can activate NFkappaB but the exact mechanism is still elusive [12, 45, 65, 66]. FKHR promotes the expression of pro-apoptotic genes and is important for the execution of apoptosis. Pak1 appears to directly phosphorylate and inactivate FKHR by regulating its subcellular distribution [67]. Phosphorylated FKHR is maintained in the cytosol and is therefore unable to activate transcription of its target genes.
Pak1 also promotes cell survival by phosphorylating dynein light chain 1 (DLC1) and BimL [68]. BimL is a proapoptotic protein that inhibits Bcl-2 in a similar manner as BAD. Following apoptotic stimuli, DLC1-BimL dimers are released from the dynein motor complex, allowing BimL to interact with Bcl-2 at the mitochondria. Phosphorylation of BimL by Pak1 prevents it from binding and inhibiting Bcl-2.
Pak2 is unique among the Pak isoforms in that it has both pro- and anti-apoptotic functions. During the late events of apoptosis, Pak2 is cleaved by caspase-3, which removes the N-terminal regulatory domain. This generates a constitutively active 34 kDa Pak2 kinase fragment [69–71]. This fragment is subsequently myristoylated and accumulates at plasma membrane ruffles and internal membranes, where it promotes cell death. Interestingly, the Pak2 fragment promotes cell death without compromising mitochondrial integrity. Instead, changes in the cytoskeleton mediated by the Pak2 kinase fragment may trigger cell death due to mechanical stress. Of note, while the 34 kDa Pak2 fragment promotes cell death, the full length activated Pak2 protects cells through mechanisms similar to those seen with Pak1 [72].
2.4 Isoform-specific functions of Pak
Although many identified Pak substrates are phosphorylated to a similar extent by all Pak isoforms, differences in preferred consensus sequences have been found (see Section 1.3. for Pak substrate recognition), and there is evidence that substrate specificities differ under physiological circumstances [73].
Individual Pak isoforms show differences in tissue distribution and subcellular localization, which may in part account for individual substrate specificities. In mice, Pak2 and Pak4 are expressed ubiquitously, whereas Pak1, Pak3, Pak5, and Pak6 have more restricted tissue-specific expression patterns. Pak1, Pak3 and Pak5 are all highly expressed in neuronal tissues [10, 74, 75]. Pak6 is highly expressed in prostate and appears to have a unique role in hormone signaling, as it binds to the androgen receptor and represses androgen receptor-mediated transcription [76]. A substantial portion of Pak5 localizes to the mitochondria, mediating Raf-1 translocation and BAD phosphorylation in survival signaling [63].
Targeted deletions of Pak isoforms in mice have further elucidated biological processes controlled by individual Pak isoforms. Individual Pak1, Pak3, and Pak5 null mice are viable, whereas Pak2 or Pak4 gene deletion results in embryonic lethality [74, 77, 78]. In humans, loss-of-function mutations in the Pak3 gene have been identified, which were associated with X-linked nonsyndromic mental retardation [79, 80]. In accordance with this, mice lacking Pak3 exhibit abnormalities in synaptic plasticity and cognition [74]. Analysis of Pak4-null embryos revealed abnormalities in the heart and nervous system, indicating an essential role for Pak4 in the development of the heart and neural tube, as well as in differentiation and migration of neurons. Mice lacking Pak5 appear not to have any defects and there may be functional redundancy between Pak5 and other Pak kinases. A recent study by Coniglio et al. used siRNAs in breast carcinoma cells to identify functional differences between Pak1 and Pak2. In this study, both Pak1 and Pak2 contributed to promote cell motility but had distinct roles in the regulation of cofilin, myosin light chain, lamellipodia formation, and focal adhesions.
3 Pak functions in human cancers
Accumulating evidence implicates Pak kinases in oncogenic growth. In cultured cells, ectopic expression of a constitutively active form of Pak induces many phenotypic hallmarks of transformation, such as increased cell motility, anchorage-independent growth, and resistance to apoptosis. Transgenic mice that overexpress a constitutively active Pak1 under a beta-lactoglobulin promoter develop malignant mammary gland tumors, although with a relatively long latency period and low penetrance [81]. Inhibition of Pak on the other hand, efficiently blocks the ability of oncogenic Ras to cause cell transformation [36, 47, 82].
3.1 Pak expression and activation in cancer
Overexpression and/or hyperactivation of Pak family members have been detected in various human tumors. Most frequently, the Pak1 isoform is overexpressed but other Pak family members have also been found overexpressed in specific cancers (Table 2). Pak4, for example, is overexpressed in 75% of the NCI 60 cell line panel and a dominant negative mutant will block anchorage-independent growth of a colon cancer cell line [36].
Table 2.
Cancers with altered expression of Pak family members
| Cancer type |
Pak isoform |
Type of alterations | References |
|---|---|---|---|
| Brain | Pak1 | Increased phospho-Pak1 in cytoplasm. | [152] |
| Esophagus | Pak4 | Protein overexpression. | [153] |
| Breast | Pak1, Pak4 | Protein overexpression and increased nuclear localization; Gene amplification (11q13–>q14 amplicon). | [45, 83, 85, 92, 153] |
| Liver | Pak1 | Protein and gene overexpression. | [154] |
| Kidney | Pak1 | Protein overexpression and increased activity. | [155] |
| Pancreas | Pak4 | Gene amplification (19q13 amplicon), protein overexpression. | [156] |
| Colon | Pak1, Pak4 | Protein overexpression. Pak4 gene amplification (19q13 amplicon) and 2 somatic mutations. | [86, 153, 157] |
| Bladder | Pak1 | Gene amplification (11q13–>q14 amplicon). | [158] |
| Ovarian | Pak1 | Protein overexpression and gene amplification (11q13–>q14 amplicon). | [84, 159, 160] |
| Prostate | Pak6 | Protein overexpression. | [161] |
| T-cell lymphoma | Pak1 | Gene amplification. | [162] |
Several distinct molecular mechanisms have been identified that cause aberrant Pak signaling in cancer, including gene amplification and alteration of upstream regulators. Both Pak1 and Pak4 are localized to genomic regions, which are frequently amplified in cancer cells. The Pak1 gene is localized within the 11q13 region, and 11q13. q14 amplifications involving the Pak1 locus have been recently reported in bladder, ovary, and breast cancer [83–85]. Pak4 localizes to another amplicon, 19q13.2, and Pak4 gene amplification has been found in colorectal and pancreatic cancers [86, 87].
Pak gene amplifications are not frequent enough to account as the only molecular mechanism leading to Pak overexpression/hyperactivation in cancer. A recent report identified a novel mechanism for the overexpression of Pak1 through microRNA downregulation. Reddy et al. found that the levels of endogenous microRNA miR-7 inversely correlated with Pak1 expression in a variety of cancer cell lines [88]. Moreover, transfection of miR-7 downregulated Pak1 expression in breast cancer cells, and suppressed motility and invasiveness of these cells.
There is little evidence for cancer cells having activating mutations in Pak genes. In a colorectal tumor sample a mutation was identified in the Pak4 kinase domain (E329 K) but whether it affected kinase activity was not investigated [86]. There are no reports of Pak involvement in lung cancers; however, a mouse model for Ras-induced lung cancers is highly sensitive to Rac inhibition, suggesting that lung cancers may also be dependent on Pak [89]. Activation of the Ras pathway could be a general mechanism of Pak activation in cancers.
There are two types of cancers, Neurofibromatosis and breast cancers, in which the biological consequences of increased Pak signaling have been explored extensively. These are discussed below.
3.2 Pak in breast cancer
In breast cancer, deregulation of Pak1 is well documented and correlates with increased invasiveness and survival of these cancer cells. More than 50% of human breast cancers display overexpression and/or hyperactivation of Pak1 [45]. The molecular mechanisms by which Pak1 promotes mammary epithelial cell transformation have been extensively studied in 3-dimensional culture model systems.
A recent study examined Pak1 activity in a pre-malignant progression series of MCF10A mammary epithelial cell variants. Pak1 expression levels increased in correlation with the progression stages in this series, indicating a role for Pak1 in the early stages of cell transformation [90]. Activation of the transcription factor NFkappaB appears to be a prominent mechanism by which Pak1 regulates survival of breast cancer cells. Friedland et al. showed a functional link between the resistance of mammary epithelial cells to apoptosis in 3-dimensional cultures and Pak1-mediated activation of NFkappaB [66]. Notably, NFkappaB also promotes cell proliferation via cyclin D1 transcription in breast cancer cells [45]. Phosphoylation of the pro-apoptotic proteins BAD and FKHR, and phosphorylation of DLC1 are other mechanisms by which Pak1 may promote breast cancer cell survival.
Pak substrates that control different aspects of cytoskeletal dynamics, such as LIM kinase, p41-ARC, filamin A, Op18/stathmin, and TCoB, are likely to promote the invasiveness of breast cancer cells (see chapter 2 for discussion of Pak substrates). In addition, a recent study showed that the multimodular protein Scrib positively regulates activation of Pak1 and participates in lamellipodia formation at the leading edge of migratory breast cancer cells [91].
Holm et al. showed a mechanistic link between increased nuclear levels of active Pak1 and tamoxifen resistance in breast cancer [92]. Approximately 70% of all breast cancers express estrogen receptor, and tamoxifen is a selective antiestrogen, which is widely used for treatment of this group of breast cancers. Pak1 is one of many kinases that phosphorylate estrogen receptor alpha (ERalpha). Deregulated activation of Pak1 produces multiple or inappropriate phosphorylation of ERalpha, creating a promiscuous receptor that is resistant to tamoxifen treatment and activates growth mechanisms in absence of estrogen. The link between Pak1 and ERalpha raises the possibility that tamoxifen resistance might be prevented or reversed by Pak1 inhibition.
3.3 Neurofibromatosis
Neurofibromatosis types 1 and 2 (NF1 and NF2) are dominantly inherited autosomal diseases caused by the loss-of-function mutations of the tumor suppressor genes NF1 and NF2, respectively. NF1 is a common disease, having a birth incidence of about 1 in 3,000, while NF2 is a relatively rare disorder with an incidence of about 1 in 25,000. Neurofibromatosis patients are predisposed to the development of multiple tumors of the central and peripheral nervous system. Schwann cells, the cells that comprise the myelin sheath around nerves, are predominantly affected in both tumors. Patients carry heterozygous mutations in either the NF1 or NF2 gene but their tumors typically display loss of the residual wild-type allele, conforming to the classic two hit Knudsen paradigm seen with most tumor suppressors. Although NF1 and NF2 are genetically and clinically distinct diseases, loss of each gene product leads to abnormal activation of Pak1, albeit through different mechanisms. Experimental results suggest that Pak1 is important for the malignant growth in both types of neurofibromatosis.
The mechanism of Pak1 activation through NF1 proceeds through the Ras pathway. The product of the NF1 gene is a cytoplasmic protein called Neurofibromin. It is widely expressed across a range of tissues but with high concentrations in the nervous system. Neurofibromin is a GTPase activating protein (GAP) and acts by accelerating the intrinsic GTPase activity of Ras. Consequently, loss of Neurofibromin is associated with increased levels of activated GTP-bound Ras, which activates oncogenic pathways, including the MAP kinase cascade and PI3 kinase. Downstream signals of PI3 kinase activate Pak via Rac and Cdc42. Dominant negative Pak mutants were shown to be potent inhibitors of Ras transformation in both rat Schwann cells and a malignant peripheral nerve sheath tumor (MPNST or neurofibrosarcoma) cell line from an NF1 patient [93].
While NF1 activates Pak through effector pathways, NF2 interacts directly with Pak1. The NF2 gene product is a cytoskeleton-associated tumor suppressor named Merlin (also called Schwannomin). Merlin is structurally related to the moesin/ezrin/radixin proteins, which link the actin cytoskeleton to cell surface glycoproteins that control growth and cellular remodeling. Merlin is widely expressed in Schwann cells, meningeal cells, peripheral nerves, and the lens. In non-neoplastic cells, merlin mediates contact-dependent growth inhibition. The growth suppressive function of Merlin depends on its phosphorylation status at Ser518 [94]. Under growth restrictive conditions, Merlin is unphosphorylated and inhibits cell proliferation, while under growth permissive conditions, Merlin is phosphorylated. Both cAMP-dependent protein kinase A (PKA) and Pak1 are able to phosphorylate Merlin at Ser518 and thereby inhibit its growth suppressive activity [95–97]. Phosphorylation of Merlin at Ser518 was also demonstrated by Pak2 and Pak6, however they may not be highly expressed in Schwann cells and therefore are of uncertain relevance to NF2 [96].
While Pak phosphorylates and inhibits Merlin, there is also an important inhibitory feedback mechanism from Merlin to Pak. Merlin associates with inactive Pak and prevents its activation, perhaps by competing with Rac [30, 98]. Phosphorylation at Ser518 induces a conformation change in Merlin and consequently disrupts interaction with Pak1, allowing Pak1 to be activated. Thus, in NF2 patients, loss of Merlin is associated with abnormal Pak1 activity, which also leads to elevated levels of Rac as well as pronounced cell ruffling [99, 100]. In cell culture experiments, the Pak1 inhibitors CEP-1347 and WR-PAK18 were able to inhibit the growth of merlin-deficient tumor cells, but not merlin-positive cells [98].
Together, these studies suggest that Pak1 is a major driver underlying Schwann cell transformation and an attractive target for therapeutics.
4 Pak as drug target
Protein kinases currently constitute a major focus for drug discovery with most major pharmaceutical companies developing inhibitors. Small molecular weight inhibitors typically target the highly conserved ATP-binding pockets of the kinase domain and compete with ATP binding. Because of similarities in the active sites of many kinases, specificity issues are common for inhibitors targeting the ATP-binding pocket, and cross-reactivity may cause unwanted toxicities. However, for several kinases this approach has been successful and in recent years a number of protein kinase inhibitors have successfully been taken through clinical trials to enter clinical practice. Sorafenib (Nexavar®), imatinib mesylate (Gleevec®), temsirolimus (Torisel®), erlotinib (Tarceva®), sunitinib (Sutent®), and gefitinib (Iressa®) are examples of such small molecule kinase inhibitors. The targets for these drugs include Raf-1, Abl, mTOR, and the receptor tyrosine kinases EGFR and VEGFR.
The importance of Pak in cell and animal models of tumorigenesis and metastasis provides a rationale for the development of Pak inhibitors as anti-cancer therapeutics. Several compounds have been identified that can act as ATP-competitive Pak inhibitors but they lack selectivity and inhibit too many other kinases to be of therapeutic use. The most potent are CEP-1347, a derivative of staurosporine, and OSU- 03012, a derivative of the Cox-2 inhibitor celecoxib [101, 102]. Nevertheless, ATP competitive compounds with enough specificity to be useful may eventually be developed.
Another approach has been to use non-ATP competitive inhibitors. Most of these derive from Pak itself. Recombinant pepide fragments of the Pak autoinhibitory domain (Pak PID) efficiently inhibit the activity of endogenous Pak protein. However, the need to introduce the autoinhibitory peptide into cells makes this approach less suitable for therapeutic use. Moreover, a recent study reported unintended side-effects with the use of the Pak-PID fragment [103]. Isolated Pak-PID induced cell cycle arrest and inhibition of cyclin D1 and D2 expression, independently of Pak1 kinase activity. This indicates that the PID may bind to and inhibit other targets and experiments using this fragment should be interpreted with caution.
The tight regulation of Group I Paks by autoinhibition presents an opportunity to develop allosteric inhibitors. Allostery refers to the phenomenon whereby the binding of a small molecule to one site regulates the activity of another site on the protein. This approach of targeting allosteric transition states during kinase activation has led to the identification of very selective inhibitors for Akt isoforms and other kinases, as the inhibitor binding site is usually more unique than the ATP-binding pocket. An elegant proof of principle for allosteric inhibition of Pak was recently shown by Peterson and his colleagues [104]. Peterson’s group identified a small molecule inhibitor that they named IPA-3, by screening for inhibitors that prevented Cdc42 activation of Pak. While all three Group I Paks were inhibited by IPA-3, Group II Paks were insensitive to the compound. This is to be expected, as Group II Paks are not regulated by an autoinhibitory domain. IPA-3 targets the distinct autoregulatory mechanism used by the Group I Paks and does not inhibit Pak after it is already activated. Although IPA-3 is not stable or potent enough to be used in vivo, it is a promising lead to develop effective inhibitors for Group I Paks.
Acknowledgments
J.F. is supported by a grant from the NIH (GM48241) and B.D. is supported by the Novartis Foundation and the Swiss National Foundation (PBBSA-120519).
Contributor Information
Bettina Dummler, Department of Pharmacology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, USA.
Kazufumi Ohshiro, Department of Biochemistry and Molecular Biology, The George Washington University School of Medicine, Washington, D.C. 20037, USA.
Rakesh Kumar, Department of Biochemistry and Molecular Biology, The George Washington University School of Medicine, Washington, D.C. 20037, USA.
Jeffrey Field, Email: jfield@upenn.edu, Department of Pharmacology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, USA.
References
- 1.Manser E, et al. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 2.Tahara SM, Traugh JA. Cyclic Nucleotide-independent protein kinases from rabbit reticulocytes. Identification and characterization of a protein kinase activated by proteolysis. Journal of Biological Chemistry. 1981;256(22):11558–11564. [PubMed] [Google Scholar]
- 3.Bokoch GM. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 4.Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. International Journal of Ciochemistry and Cell Biology. 2002;34(7):713–717. doi: 10.1016/s1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Z, et al. A conserved Negative Regulatory Region in aPAK: inhibition of PAK Kinases Reveals Their Morphological Roles Downstream of Cdc42 and Rac1. Molecular and Cell Biology. 1998;18(4):2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei M, et al. Structure of PAK1 in an Autoinhibited Conformation Reveals a Multistage Activation Switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 7.Gatti A, et al. Multisite autophosphorylation of p21-activated protein kinase g-Pak as a function of Activation. Journal of Biological Chemistry. 1999;274:8022–8028. doi: 10.1074/jbc.274.12.8022. [DOI] [PubMed] [Google Scholar]
- 8.Chong C, et al. The mechanism of PAK activation. Journal of Biological Chemistry. 2001;276:17347–17353. doi: 10.1074/jbc.M009316200. [DOI] [PubMed] [Google Scholar]
- 9.Abo A, et al. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. European Molecular Biology Organization Journal. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dan C, et al. PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Molecular and Cellular Biology. 2002;22(2):567–577. doi: 10.1128/MCB.22.2.567-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagheri-Yarmond RB, et al. Vascular endothelial growth factor upregulation via p21-activated kinase-1 signaling regulates heregulin-b1-mediated angiogenesis. Journal of Biological Chemistry. 2000;275:39451–39457. doi: 10.1074/jbc.M006150200. [DOI] [PubMed] [Google Scholar]
- 12.Dadke D, et al. Activation of p21-activated kinase 1-nuclear factor kappaB signaling by Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor during cellular transformation. Cancer Research. 2003;63(24):8837–8847. [PubMed] [Google Scholar]
- 13.Dharmawardhane S, et al. Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. Journal of Cell Biology. 1997;138(6):1265–1278. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knaus UG, et al. Regulation of human leukocyte p21-activated kinases through G protein-coupled receptors. Science. 1995;269(5221):221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- 15.Menard RE, Mattingly RR. Cell surface receptors activate p21-activated kinase 1 via multiple Ras and PI3-kinase-dependent pathways. Cell Signal. 2003;15(12):1099–1109. doi: 10.1016/s0898-6568(03)00087-1. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz U, et al. Lysophosphatidic Acid Stimulates p21-Activated Kinase in Vascular Smooth Muscle Cells. Biochemical and Biophysical Research Communication. 2002;291(3):687–691. doi: 10.1006/bbrc.2002.6493. [DOI] [PubMed] [Google Scholar]
- 17.Teo M, Manser E, Lim L. Identification and molecular cloning of a p21cdc42/rac1-activated serine/threonine kinase that is rapidly activated by thrombin in platelets. Journal of Biological Chemistry. 1995;270:26690–26697. doi: 10.1074/jbc.270.44.26690. [DOI] [PubMed] [Google Scholar]
- 18.Tsakiridis T, et al. Insulin activates a p21-activated kinase in muscle cells via phosphatidylinositol 3-kinase. Journal of Biological Chemistry. 1996;271:19664–19667. doi: 10.1074/jbc.271.33.19664. [DOI] [PubMed] [Google Scholar]
- 19.Banerjee M, et al. Pak1 phosphorylation on T212 affects microtubules in cells undergoing mitosis. Current Biology. 2002;12:1233–1239. doi: 10.1016/s0960-9822(02)00956-9. [DOI] [PubMed] [Google Scholar]
- 20.Thiel DA, et al. Cell cycle regulated phosphorylation of p21-activated kinase 1. Current Biology. 2002;12:1227–1232. doi: 10.1016/s0960-9822(02)00931-4. [DOI] [PubMed] [Google Scholar]
- 21.King CC, et al. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1) Journal of Biological Chemistry. 2000;275(52):41201–41209. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- 22.Bokoch GM, et al. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) Journal of Biological Chemistry. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 23.Galisteo ML, et al. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. Journal of Biological Chemistry. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 24.Lu W, et al. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Current Biology. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 25.Manser E, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Molecular Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 26.Bagrodia S, et al. A Tyrosine-phosphorylated Protein That Binds to an Important Regulatory Region on the Cool Family of p21-activated Kinase-binding Proteins. Journal of Biological Chemistry. 1999;274:22393–22400. doi: 10.1074/jbc.274.32.22393. [DOI] [PubMed] [Google Scholar]
- 27.Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–238. doi: 10.1016/s0092-8674(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 28.Xia C, et al. Regulation of the p21-activated kinase (PAK) by a human Gbeta -like WD-repeat protein, hPIP1. Proc Natl Acad Sci U S A. 2001;98(11):6174–6179. doi: 10.1073/pnas.101137298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alahari SK, Reddig PJ, Juliano RL. The integrin-binding protein Nischarin regulates cell migration by inhibiting PAK. European Molecular Biology Organization Journal. 2004;23(14):2777–2788. doi: 10.1038/sj.emboj.7600291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kissil JL, et al. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Molecular Cell. 2003;12(4):841–849. doi: 10.1016/s1097-2765(03)00382-4. [DOI] [PubMed] [Google Scholar]
- 31.Koh CG, et al. The p21-activated kinase PAK is negatively regulated by POPX1 and POPX2, a pair of serine/ threonine phosphatases of the PP2C family. Current Biology. 2002;12(4):317–321. doi: 10.1016/s0960-9822(02)00652-8. [DOI] [PubMed] [Google Scholar]
- 32.Tuazon PT, et al. Determinants for Substrate Phosphorylation by p21-Activated Protein Kinase (g-PAK) Biochemistry. 1997;36:16059–16064. doi: 10.1021/bi9717845. [DOI] [PubMed] [Google Scholar]
- 33.Rennefahrt UEE, et al. Specificity Profiling of Pak Kinases Allows Identification of Novel Phosphorylation Site. Journal of Biological Chemistry. 2007;282(21):15667–15678. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- 34.Kiosses WB, et al. A role for p21—activated kinase in endothelial cell migration. Journal of Cell Biology. 1999;147(4):831–844. doi: 10.1083/jcb.147.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, et al. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail’s subcellular localization and functions. Cancer Research. 2005;65(8):3179–3384. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- 36.Callow MG, et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. Journal of Biological Chemistry. 2002;277(1):550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 37.Vadlamudi RK, et al. Regulatable Expression of p21-activated Kinase-1 Promotes Anchorage-independent Growth and Abnormal Organization of Mitotic Spindles in Human Epithelial Breast Cancer Cells. Journal of Biological Chemistry. 2000;275:36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 38.Shalom-Barak T, Knaus UGA. p21-activated kinase-controlled metabolic switch up-regulates phagocyte NADPH oxidase. Journal of Biological Chemistry. 2002;277(43):40659–40665. doi: 10.1074/jbc.M206650200. [DOI] [PubMed] [Google Scholar]
- 39.Gururaj A, et al. Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene. 2004;3(49):8118–8127. doi: 10.1038/sj.onc.1207969. [DOI] [PubMed] [Google Scholar]
- 40.Li F, et al. p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. European Molecular Biology Organization Reports. 2002;3(8):767–773. doi: 10.1093/embo-reports/kvf157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vadlamudi RK, et al. p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Molecular and Cellular Biology. 2005;25(9):3726–3736. doi: 10.1128/MCB.25.9.3726-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Molecular Cell. 2000;5(6):905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhao ZS, et al. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Molecular Cell. 2005;20(2):237–249. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 44.Maroto B, et al. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27(36):4900–4908. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- 45.Balasenthil S, et al. P21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. Journal of Biological Chemistry. 2003 doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 46.Beeser A, et al. Role of group A p21-activated kinases in activation of extracellular-regulated kinase by growth factors. Journal of Biological Chemistry. 2005;80:36609–36615. doi: 10.1074/jbc.M502306200. [DOI] [PubMed] [Google Scholar]
- 47.Tang Y, et al. Kinase-deficient Pak1 mutants inhibit Ras transformation of Rat-1 fibroblasts. Molecular and Cell Biology. 1997;17:4454–4464. doi: 10.1128/mcb.17.8.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frost JA, et al. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. European Molecular Biology Organization Journal. 1997;16(21):6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran NH, Frost JA. Phosphorylation of Raf-1 by p21-activated kinase 1 and Src regulates Raf-1 autoinhibition. Journal of Biological Chemistry. 2003;278(13):11221–11226. doi: 10.1074/jbc.M210318200. [DOI] [PubMed] [Google Scholar]
- 50.King A, et al. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396:180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- 51.Sells MA, Pfaff A, Chernoff J. Temporal and Spatial Distribution of Activated Pak1 in Fibroblasts. Journal of Cell Biology. 2000;151:1449–1457. doi: 10.1083/jcb.151.7.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sells MA, et al. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Current Biology. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 53.Manser E, et al. Expression of constitutively active a- Pak reveals effects of the kinase on actin and focal complexes. Molecular and Cell Biology. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sells M, Boyd JT, Chernoff J. p21-Activated Kinase 1 (Pak1) regulates Cell Motility in Mammalian Fibroblasts. Journal Cell Biology. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frost JA, et al. Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. Journal of Biological Chemistry. 1998;273(43):28191–28198. doi: 10.1074/jbc.273.43.28191. [DOI] [PubMed] [Google Scholar]
- 56.Delorme V, et al. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev Cell. 2007;13(5):646–662. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vadlamudi RK, et al. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. European Molecular Biology Organization Reports. 2004;5(2):154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. Journal of Biological Chemistry. 2004;279(7):6196–6203. doi: 10.1074/jbc.M307261200. [DOI] [PubMed] [Google Scholar]
- 59.Daub H, et al. Rac/Cdc42 and p65PAK regulate the microtubule-destabilizing protein stathmin through phosphorylation at serine 16. Journal of Biological Chemistry. 2001;276(3):1677–1680. doi: 10.1074/jbc.C000635200. [DOI] [PubMed] [Google Scholar]
- 60.Sanders LC, et al. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 1999;283(5410):2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- 61.Jin A, et al. PAK1-dependent phosphorylation of RAF-1 regulates its mitochondrial localization, phosphorylation of BAD, and BCL-2 association. Journal of Biological Chemistry. 2005;280:24698–24705. doi: 10.1074/jbc.M413374200. [DOI] [PubMed] [Google Scholar]
- 62.Wu X, et al. p21 activated kinase 5 activates Raf-1 and targets it to mitochondria. Journal of Cellular Biochemistry. 2008;105(1):167–175. doi: 10.1002/jcb.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cotteret S, et al. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Molecular and Cellular Biology. 2003;23(16):5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huser M, et al. MEK kinase activity is not necessary for Raf-1 function. European Molecular Biology Organization Journal. 2001;20(8):1940–1951. doi: 10.1093/emboj/20.8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frost JA, et al. Stimulation of NFkB Activity by Multiple Signaling Pathways Requires PAK1. Journal of Biological Chemistry. 2000;275:19693–19699. doi: 10.1074/jbc.M909860199. [DOI] [PubMed] [Google Scholar]
- 66.Friedland JC, et al. {alpha}6{beta}4 integrin activates Rac-dependent p21-activated kinase 1 to drive NF-{kappa}B-dependent resistance to apoptosis in 3D mammary acini. Journal of Cell Science. 2007 doi: 10.1242/jcs.03484. jcs.03484. [DOI] [PubMed] [Google Scholar]
- 67.Mazumdar A, Kumar R. Estrogen regulation of Pak1 and FKHR pathways in breast cancer cells. FEBS Lett. 2003;535(1–3):6–10. doi: 10.1016/s0014-5793(02)03846-2. [DOI] [PubMed] [Google Scholar]
- 68.Vadlamudi RK, et al. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 2004;5(6):575–585. doi: 10.1016/j.ccr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 69.Lee N, et al. Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc. Natl. Acad. Sci. USA. 1997;94:13642–13647. doi: 10.1073/pnas.94.25.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rudel T, Bokoch GM. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of Pak2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 71.Walter BN, et al. Cleavage and activation of p21-activated protein kinase -PAK by CPP32 (Caspase 3). Effects of Autophosphorylation on Activity. Journal of Biological Chemistry. 1998;273:28733–28739. doi: 10.1074/jbc.273.44.28733. [DOI] [PubMed] [Google Scholar]
- 72.Jakobi R, Moertl E, Koeppel MA. p21-activated protein kinase g-PAK suppresses programmed cell death of BALB3T3 fibroblasts. Journal of Biological Chemistry. 2001;276:16624–16634. doi: 10.1074/jbc.M007753200. [DOI] [PubMed] [Google Scholar]
- 73.Eswaran J, et al. UnPAKing the class differences among p21-activated kinases. Trends Biochem Sci. 2008 doi: 10.1016/j.tibs.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Meng J, et al. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. Journal of Neuroscience. 2005;25(28):6641–6650. doi: 10.1523/JNEUROSCI.0028-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandey A, et al. Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene. 2002;21(24):3939–3948. doi: 10.1038/sj.onc.1205478. [DOI] [PubMed] [Google Scholar]
- 76.Yang F, et al. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. Journal of Biological Chemistry. 2001;276(18):15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 77.Qu J, et al. PAK4 kinase is essential for embryonic viability and for proper neuronal development. Molecular and Cellular Biology. 2003;23(20):7122–7133. doi: 10.1128/MCB.23.20.7122-7133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li X, Minden A. Targeted disruption of the gene for the PAK5 kinase in mice. Molecular and Cellular Biology. 2003;23(20):7134–7142. doi: 10.1128/MCB.23.20.7134-7142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Allen KM, et al. PAK3 mutation in nonsyndromic X-linked mental retardation. Nature Genetics. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 80.Bienvenu T, et al. Missense mutation in PAK3, R67C, causes X-linked nonspecific mental retardation. American Journal of Medical Genetics. 2000;93(4):294–298. doi: 10.1002/1096-8628(20000814)93:4<294::aid-ajmg8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 81.Wang RA, et al. PAK1 hyperactivation is sufficient for mammary gland tumor formation. Oncogene. 2005 doi: 10.1038/sj.onc.1209309. [DOI] [PubMed] [Google Scholar]
- 82.Osada S, et al. A domain containing the Cdc42/Rac interactive binding (CRIB) region of p65PAK inhibits transcriptional activation and cell transformation mediated by the Ras-Rac pathway. Febs letters. 1997;404:227–233. doi: 10.1016/s0014-5793(97)00139-7. [DOI] [PubMed] [Google Scholar]
- 83.Bekri S, et al. Detailed map of a region commonly amplified at 11q13->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79(1–2):125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 84.Brown LA, et al. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47(6):481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 85.Bostner J, et al. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene. 2007;26(49):6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- 86.Parsons DW, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436(7052):792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 87.Chen S, et al. Copy number alterations in pancreatic cancer identify recurrent Pak4 amplification. Cancer Biol. Ther. 2008;X:XXX. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reddy SD, et al. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Research. 2008;68(20):8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kissil JL, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67(17):8089–8094. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 90.Li Q, et al. p21-Activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia. 2008;10(4):314–329. doi: 10.1593/neo.07970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nola S, et al. Scrib regulates PAK activity during the cell migration process. Human Molecular Genetics. 2008;17(22):3552–3565. doi: 10.1093/hmg/ddn248. [DOI] [PubMed] [Google Scholar]
- 92.Holm C, et al. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. Journal of the National Cancer Institute. 2006;98(10):671–680. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 93.Tang Y, et al. A role for Pak protein kinases in Schwann cell transformation. Proc. Natl. Acad. Sci. USA. 1998;95:5139–5144. doi: 10.1073/pnas.95.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Surace EI, Haipek CA, Gutmann DH. Effect of merlin phosphorylation on neurofibromatosis 2 (NF2) gene function. Oncogene. 2004;23(2):580–587. doi: 10.1038/sj.onc.1207142. [DOI] [PubMed] [Google Scholar]
- 95.Kissil JL, et al. Merlin phosphorylation by p21-activated kinase 2 and effects of phosphorylation on merlin localization. Journal of Biological Chemistry. 2002;277(12):10394–10399. doi: 10.1074/jbc.M200083200. [DOI] [PubMed] [Google Scholar]
- 96.Xiao G-H, et al. p21-activated Kinase Links Rac/ Cdc42 Signaling to Merlin. Journal Biological Chemistry. 2002;277:883–886. doi: 10.1074/jbc.C100553200. [DOI] [PubMed] [Google Scholar]
- 97.Alfthan K, et al. Cyclic AMP-dependent Protein Kinase Phosphorylates Merlin at Serine 518 Independently of p21-activated Kinase and Promotes Merlin-Ezrin Heterodimerization. Journal of Biological Chemistry. 2004;279(18):18559–18566. doi: 10.1074/jbc.M313916200. [DOI] [PubMed] [Google Scholar]
- 98.Hirokawa Y, et al. A clue to the therapy of neurofibromatosis type 2: NF2/merlin is a PAK1 inhibitor. Cancer Journal. 2004;10(1):20–26. doi: 10.1097/00130404-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 99.Pelton PD, et al. Ruffling membrane, stress fiber, cell spreading and proliferation abnormalities in human Schwannoma cells. Oncogene. 1998;17:2195–2209. doi: 10.1038/sj.onc.1202141. [DOI] [PubMed] [Google Scholar]
- 100.Shaw RJ, et al. The Nf2 tumor suppressor, Merlin, functions in Rac-dependent signalling. Developmental Cell. 2001;1:63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 101.Porchia LM, et al. 2-Amino-N-{4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]-phe nyl} Acet-amide (OSU-03012), a Celecoxib Derivative, Directly Targets p21-Activated Kinase. Molecular Pharmacology. 2007;72(5):1124–1131. doi: 10.1124/mol.107.037556. [DOI] [PubMed] [Google Scholar]
- 102.Nheu TV, et al. The K252a derivatives, inhibitors for the PAK/MLK kinase family selectively block the growth of RAS transformants. Cancer Journal. 2002;8(4):328–336. doi: 10.1097/00130404-200207000-00009. [DOI] [PubMed] [Google Scholar]
- 103.Thullberg M, et al. The kinase-inhibitory domain of p21-activated kinase 1 (PAK1) inhibits cell cycle progression independent of PAK1 kinase activity. Oncogene. 2007;26(12):1820–1828. doi: 10.1038/sj.onc.1209983. [DOI] [PubMed] [Google Scholar]
- 104.Deacon SW, et al. An Isoform-Selective, Small- Molecule Inhibitor Targets the Autoregulatory Mechanism of p21-Activated Kinase. Chemistry & Biology. 2008;5(4):322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shin EY, et al. Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. Journal of Biological Chemistry. 2002;277(46):44417–44430. doi: 10.1074/jbc.M203754200. [DOI] [PubMed] [Google Scholar]
- 106.Foster DB, et al. Phosphorylation of caldesmon by p21-activated kinase. Implications for the Ca(2+) sensitivity of smooth muscle contraction. Journal of Biological Chemistry. 2000;275(3):1959–1965. doi: 10.1074/jbc.275.3.1959. [DOI] [PubMed] [Google Scholar]
- 107.McFawn PK, et al. Calcium-independent contraction and sensitization of airway smooth muscle by p21-activated protein kinase. American journal of physiology. Lung cellular and molecular physiology. 2003;284(5):L863–L870. doi: 10.1152/ajplung.00068.2002. [DOI] [PubMed] [Google Scholar]
- 108.Van Eyk JE, et al. Different molecular mechanisms for Rho family GTPase-dependent, Ca2+-independent contraction of smooth muscle. Journal of Biological Chemistry. 1998;273(36):23433–23439. doi: 10.1074/jbc.273.36.23433. [DOI] [PubMed] [Google Scholar]
- 109.Takizawa N, Koga Y, Ikebe M. Phosphorylation of CPI17 and myosin binding subunit of type 1 protein phosphatase by p21-activated kinase. Biochem Biophys Res Commun. 2002;297(4):773–778. doi: 10.1016/s0006-291x(02)02302-1. [DOI] [PubMed] [Google Scholar]
- 110.Ohtakara K, et al. p21-activated kinase PAK phosphorylates desmin at sites different from those for Rho-associated kinase. Biochem Biophys Res Commun. 2000;272(3):712–716. doi: 10.1006/bbrc.2000.2854. [DOI] [PubMed] [Google Scholar]
- 111.Vadlamudi RK, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nature Cell Biology. 2002;4(9):681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 112.Zenke FT, et al. p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. Journal of Biological Chemistry. 2004;279(18):18392–18400. doi: 10.1074/jbc.M400084200. [DOI] [PubMed] [Google Scholar]
- 113.Edwards DC, et al. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nature Cell Biology. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 114.Dan C, et al. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. Journal of Biological Chemistry. 2001;276(34):32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- 115.Goeckeler ZM, et al. Phosphorylation of myosin light chain kinase by p21-activated kinase PAK2. Journal of Biological Chemistry. 2000;275(24):18366–18374. doi: 10.1074/jbc.M001339200. [DOI] [PubMed] [Google Scholar]
- 116.Alberts AS, et al. PAK1 negatively regulates the activity of the Rho exchange factor NET1. Journal of Biological Chemistry. 2005;280(13):12152–12161. doi: 10.1074/jbc.M405073200. [DOI] [PubMed] [Google Scholar]
- 117.DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Molecular Cell. 2004;15(1):117–127. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 118.Chew TL, et al. Phosphorylation of non-muscle myosin II regulatory light chain by p21-activated kinase (gamma-PAK) Journal of Muscle Research and Cell Motility. 1998;19(8):839–854. doi: 10.1023/a:1005417926585. [DOI] [PubMed] [Google Scholar]
- 119.Ramos E, Wysolmerski RB, Masaracchia RA. Myosin phosphorylation by human cdc42-dependent S6/H4 kinase/gammaPAK from placenta and lymphoid cells. Recept Signal Transduct. 1997;7(2):99–110. [PubMed] [Google Scholar]
- 120.Goto H, et al. Phosphorylation and reorganization of vimentin by p21-activated kinase (PAK) Genes Cells. 2002;7(2):91–97. doi: 10.1046/j.1356-9597.2001.00504.x. [DOI] [PubMed] [Google Scholar]
- 121.Li QF, et al. Critical role of vimentin phosphorylation at Ser-56 by p21-activated kinase in vimentin cytoskeleton signaling. 2006;281(45):34716–34724. doi: 10.1074/jbc.M607715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang DD, Bai Y, Gunst SJ. Silencing of p21-activated kinase attenuates vimentin phosphorylation on Ser-56 and reorientation of the vimentin network during stimulation of smooth muscle cells by 5-hydroxytryptamine. Biochemical Journal. 2005;388(Pt 3):773–783. doi: 10.1042/BJ20050065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang R, et al. Dissociation of Crk-associated substrate from the vimentin network is regulated by p21-activated kinase on ACh activation of airway smooth muscle. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2007;292(1):L240–L248. doi: 10.1152/ajplung.00199.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chan W, et al. Vimentin intermediate filament reorganization by Cdc42: involvement of PAK and p70 S6 kinase. European Journal of Cell Biology. 2002;81(12):692–701. doi: 10.1078/0171-9335-00281. [DOI] [PubMed] [Google Scholar]
- 125.Roig J, et al. Functional interaction between c-Abl and the p21-activated protein kinase g-PAK. PNAS. 2000;97(26):14346–14351. doi: 10.1073/pnas.97.26.14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jung JH, et al. Phosphorylation of c-Abl by protein kinase Pak2 regulates differential binding of ABI2 and CRK. Biochemistry. 2008;47(3):1094–1104. doi: 10.1021/bi701533j. [DOI] [PubMed] [Google Scholar]
- 127.Tran NH, Wu X, Frost JA. B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. Journal of Biological Chemistry. 2005;280(16):16244–16253. doi: 10.1074/jbc.M501185200. [DOI] [PubMed] [Google Scholar]
- 128.Huang Z, Traugh JA, Bishop JM. Negative control of the Myc protein by the stress-responsive kinase Pak2. Molecular and Cellular Biology. 2004;24(4):1582–1594. doi: 10.1128/MCB.24.4.1582-1594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Edin ML, Juliano RL. Raf-1 serine 338 phosphorylation plays a key role in adhesion-dependent activation of extracellular signal-regulated kinase by epidermal growth factor. Molecular and Cellular Biology. 2005;25(11):4466–4475. doi: 10.1128/MCB.25.11.4466-4475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chaudhary A, et al. Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Current Biology. 2000;10(9):551–554. doi: 10.1016/s0960-9822(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 131.Zang M, Hayne C, Luo Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. Journal of Biological Chemistry. 2002;277(6):4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- 132.Wang RA, et al. P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. European Molecular Biology Organization Journal. 2002;21(20):5437–5447. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rayala SK, et al. P21-activated kinase 1 regulation of estrogen receptor-alpha activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Research. 2006;66(3):1694–1701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- 134.Frost JA, et al. Actions of Rho family small G proteins and p21 activated protein kinases on Mitogen-Activated Protein Kinase family members. Molecular and Cellular Biology. 1996 Jul;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Slack-Davis JK, et al. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. Journal Cell Biology. 2003;162(2):281–291. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eblen ST, et al. Mitogen-activated protein kinase feedback phosphorylation regulates MEK1 complex formation and activation during cellular adhesion. Molecular and Cellular Biology. 2004;24(6):2308–2317. doi: 10.1128/MCB.24.6.2308-2317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Coles LC, Shaw PE. PAK1 primes MEK1 for phosphorylation by Raf-1 kinase during cross-cascade activation of the ERK pathway. Oncogene. 2002;21(14):2236–2244. doi: 10.1038/sj.onc.1205302. [DOI] [PubMed] [Google Scholar]
- 138.Gallagher ED, et al. Binding of JNK/SAPK to MEKK1 is regulated by phosphorylation. Journal of Biological Chemistry. 2002;277(48):45785–45792. doi: 10.1074/jbc.M207702200. [DOI] [PubMed] [Google Scholar]
- 139.Orton KC, et al. Phosphorylation of Mnk1 by caspase-activated Pak2/gamma-PAK inhibits phosphorylation and interaction of eIF4G with Mnk. Journal of Biological Chemistry. 2004;279(37):38649–38657. doi: 10.1074/jbc.M407337200. [DOI] [PubMed] [Google Scholar]
- 140.Tuazon PT, et al. p21-activated protein kinase gamma- PAK in pituitary secretory granules phosphorylates prolactin. FEBS Lett. 2002;515(1–3):84–88. doi: 10.1016/s0014-5793(02)02444-4. [DOI] [PubMed] [Google Scholar]
- 141.Tang Y, et al. The Akt proto-oncogene links Ras to Pak and cell survival signals. Journal of Biological Chemistry. 2000;275:9106–9109. doi: 10.1074/jbc.275.13.9106. [DOI] [PubMed] [Google Scholar]
- 142.Liberali P, et al. The closure of Pak1-dependent macro-pinosomes requires the phosphorylation of CtBP1/BARS. European Molecular Biology Organization Journal. 2008;27(7):970–981. doi: 10.1038/emboj.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Manavathi B, Rayala SK, Kumar R. Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. Journal of Biological Chemistry. 2007;282(27):19820–19830. doi: 10.1074/jbc.M702309200. [DOI] [PubMed] [Google Scholar]
- 144.Wang J, et al. Reciprocal Signaling between Heterotrimeric G Proteins and the p21-stimulated Protein Kinase. Journal of Biological Chemistry. 1999;274:31641–31647. doi: 10.1074/jbc.274.44.31641. [DOI] [PubMed] [Google Scholar]
- 145.Martyn KD, et al. p21-activated kinase (Pak) regulates NADPH oxidase activation in human neutrophils. Blood. 2005;106(12):3962–3969. doi: 10.1182/blood-2005-03-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ahmed S, et al. Cryptic Rac-binding and p21(Cdc42Hs/Rac)-activated kinase phosphorylation sites of NADPH oxidase component p67(phox) Journal of Biological Chemistry. 1998;273(25):15693–15701. doi: 10.1074/jbc.273.25.15693. [DOI] [PubMed] [Google Scholar]
- 147.Vadlamudi RK, et al. An essential role of Pak1 phosphorylation of SHARP in Notch signaling. Oncogene. 2005;24(28):4591–4596. doi: 10.1038/sj.onc.1208672. [DOI] [PubMed] [Google Scholar]
- 148.Wang RA, et al. Essential functions of p21-activated kinase 1 in morphogenesis and differentiation of mammary glands. Journal of Cell Biology. 2003;161(3):583–592. doi: 10.1083/jcb.200212066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miah SM, et al. Activation of Syk protein tyrosine kinase in response to osmotic stress requires interaction with p21-activated protein kinase Pak2/gamma-PAK. Molecular and Cellular Biology. 2004;24(1):71–83. doi: 10.1128/MCB.24.1.71-83.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sakurada K, et al. Synapsin I is phosphorylated at Ser603 by p21-activated kinases (PAKs) in vitro and in PC12 cells stimulated with bradykinin. Journal of Biological Chemistry. 2002;277(47):45473–45479. doi: 10.1074/jbc.M206673200. [DOI] [PubMed] [Google Scholar]
- 151.Buscemi N, et al. p21-activated kinase increases the calcium sensitivity of rat triton-skinned cardiac muscle fiber bundles via a mechanism potentially involving novel phosphorylation of troponin I. Circulation Research. 2002;91(6):509–516. doi: 10.1161/01.res.0000035246.27856.53. [DOI] [PubMed] [Google Scholar]
- 152.Aoki H, et al. Phosphorylated Pak1 level in the cytoplasm correlates with shorter survival time in patients with glioblastoma. Clinical Cancer Research. 2007;13(22 Pt 1):6603–6609. doi: 10.1158/1078-0432.CCR-07-0145. [DOI] [PubMed] [Google Scholar]
- 153.Liu Y, et al. The pak4 protein kinase plays a key role in cell survival and tumorigenesis in athymic mice. Mol Cancer Res. 2008;6(7):1215–1224. doi: 10.1158/1541-7786.MCR-08-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ching YP, et al. P21-activated protein kinase is overexpressed in hepatocellular carcinoma and enhances cancer metastasis involving c-Jun NH2-terminal kinase activation and paxillin phosphorylation. Cancer Research. 2007;67(8):3601–3608. doi: 10.1158/0008-5472.CAN-06-3994. [DOI] [PubMed] [Google Scholar]
- 155.O’Sullivan GC, et al. Modulation of p21-activated kinase 1 alters the behavior of renal cell carcinoma. International Journal of Cancer. 2007;121(9):1930–1940. doi: 10.1002/ijc.22893. [DOI] [PubMed] [Google Scholar]
- 156.Mahlamaki EH, et al. High-resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia. 2004;6(5):432–439. doi: 10.1593/neo.04130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carter JH, et al. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clinical Cancer Research. 2004;10(10):3448–3456. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- 158.Ito M, et al. P21-activated kinase 1: a new molecular marker for intravesical recurrence after transurethral resection of bladder cancer. Journal of Urology. 2007;178(3 Pt 1):1073–1079. doi: 10.1016/j.juro.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 159.Schraml P, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. American Journal of Pathology. 2003;163(3):985–992. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Davidson B, Shih IM, Wang TL. Different clinical roles for p21-activated kinase-1 in primary and recurrent ovarian carcinoma. Hum Pathol. 2008 doi: 10.1016/j.humpath.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 161.Kaur R, et al. Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate. 2008;68(14):1510–1516. doi: 10.1002/pros.20787. [DOI] [PubMed] [Google Scholar]
- 162.Mao X, et al. Genomic alterations in blastic natural killer/extranodal natural killer-like T cell lymphoma with cutaneous involvement. Journal of Investigative Dermatology. 2003;121(3):618–627. doi: 10.1046/j.1523-1747.2003.12406.x. [DOI] [PubMed] [Google Scholar]