APE1 is recruited to the transcription initiation site of the SIRT1 promoter during early cell response to oxidative stress. This reveals the importance of BER enzyme involvement in controlling specific gene expression at the transcriptional level.
Abstract
Apurinic/apyrimidinic endonuclease 1 (APE1) is a multifunctional protein contributing to genome stability via repair of DNA lesions via the base excision repair pathway. It also plays a role in gene expression regulation and RNA metabolism. Another, poorly characterized function is its ability to bind to negative calcium responsive elements (nCaRE) of some gene promoters. The presence of many functional nCaRE sequences regulating gene transcription can be envisioned, given their conservation within ALU repeats. To look for functional nCaRE sequences within the human genome, we performed bioinformatic analyses and identified 57 genes potentially regulated by APE1. We focused on sirtuin-1 (SIRT1) deacetylase due to its involvement in cell stress, including senescence, apoptosis, and tumorigenesis, and its role in the deacetylation of APE1 after genotoxic stress. The human SIRT1 promoter presents two nCaRE elements stably bound by APE1 through its N-terminus. We demonstrate that APE1 is part of a multiprotein complex including hOGG1, Ku70, and RNA Pol II, which is recruited on SIRT1 promoter to regulate SIRT1 gene functions during early response to oxidative stress. These findings provide new insights into the role of nCaRE sequences in the transcriptional regulation of mammalian genes.
INTRODUCTION
Apurinic/apyrimidinic endonuclease 1 (APE1), also known as redox effector factor-1 (Ref-1), is a multifunctional and essential protein in mammals. It plays a vital role during cellular response to oxidative stress (Fung and Demple, 2005) and contributes to the maintenance of genome integrity (Tell et al., 2005, 2009, 2010a). As an AP endonuclease, APE1 is involved in the base excision repair (BER) pathway, which deals with DNA damage induced by oxidative and alkylating agents, including chemotherapeutic agents (Chen and Stubbe, 2005). APE1 also has transcriptional regulatory activity, modulating gene expression through a redox-based coactivating function on several transcription factors involved in cancer promotion and progression (Huang and Adamson, 1993; Tell et al., 1998; Gaiddon et al., 1999). These two major APE1 activities are independent and located in distinct protein domains. The N-terminal portion of the protein is devoted to the transcriptional coactivating function, and the C-terminal domain exerts the endonuclease activity on DNA abasic sites (Xanthoudakis et al., 1996; Tell et al., 2005). The latter domain is highly conserved, whereas the N-terminal region presents wider variability among different organisms, being more conserved in mammals, thus suggesting recent acquisition during evolution (Georgiadis et al., 2008; Fantini et al., 2010; Poletto et al., 2013). Through its N-terminal portion, APE1 also interacts with different proteins involved in ribosome biogenesis, pre-mRNA maturation/splicing, and ribonucleotide catabolism. These observations highlight an unexpected role of APE1 in RNA metabolism (Vascotto et al., 2009b; Tell et al., 2010b), as also shown by two independent studies that demonstrated the ability of APE1 to cleave abasic RNA in vitro and in vivo (Berquist et al., 2008; Barnes et al., 2009). Accordingly, APE1 has been proposed to be a main factor in the abasic RNA cleansing process, suggesting that some of the activities of APE1 in gene expression may involve posttranscriptional mechanisms (Tell et al., 2010b).
Another interesting, yet poorly characterized aspect of APE1 transcriptional activity is its role as a component of a trans-acting complex that acts as a Ca2+-dependent repressor of the parathyroid hormone (PTH) gene by binding the negative calcium responsive elements (nCaRE) in its promoter region (Okazaki et al., 1991). In particular, an increase in extracellular Ca2+ concentration inhibits PTH expression through a mechanism involving APE1 binding to two nCaRE elements, nCaRE-A and nCaRE-B (Yamamoto et al., 1989). This observation was further extended to the promoter region of renin (Fuchs et al., 2003), Bax (Bhattacharyya et al., 2009), and APE1 itself (Izumi et al., 1996). This last case represents the first example of such a negative regulatory mechanism for a DNA repair enzyme. Other experiments demonstrated that APE1 requires additional factors, such as heterogeneous ribonucleoprotein L (Kuninger et al., 2002), Ku antigen (Chung et al., 1996), and PARP-1 (Bhattacharyya et al., 2009), to stably bind to nCaRE elements.
nCaRE-B sequences are located within ALU repeats (McHaffie and Ralston, 1995; Shankar et al., 2004). Therefore, given that ALU elements are transposable elements that occupy at least 1/10 of the expressed human genome, many other functional nCaRE-B sequences could exist and play a role in the transcriptional regulation of genes. However, information is lacking on 1) an accurate numbering, 2) the identity of genes containing these sequences within their promoter, and 3) the active biological function of these elements. Thus the quest is for functional nCaRE-B sequences in the human genome to identify new potential genes whose expression may be regulated by APE1 through nCaRE binding.
The present work is devoted to this issue and focuses on the characterization of the molecular mechanisms responsible for APE1 binding to nCaRE-B sequences on sirtuin-1 (SIRT1) promoter. Bioinformatic analyses of human gene expression data obtained upon APE1 knockdown in cells (Vascotto et al., 2009a) show the presence of multiple nCaRE-B sequences in genes deregulated upon APE1 silencing and conserved in the mouse genome. Among these, we focus on the two nCaRE sequences present within the promoter region of the human deacetylase SIRT1 gene and their role in regulating the corresponding gene transcription. SIRT1 is a deacetylase participating in cell growth, adaptation to caloric restriction, apoptosis, and tumorigenesis (Gorospe and de Cabo, 2008; Kim and Um, 2008), as well as in cell response to genotoxic agents through the deacetylation of APE1 (Yamamori et al., 2010). Together the data show the importance of APE1 during transcriptional initiation in positively promoting transcription of genes under genotoxic conditions.
RESULTS
Bioinformatic search for nCaRE sequence–containing genes reveals the SIRT1 gene as a novel candidate target of APE1 regulation
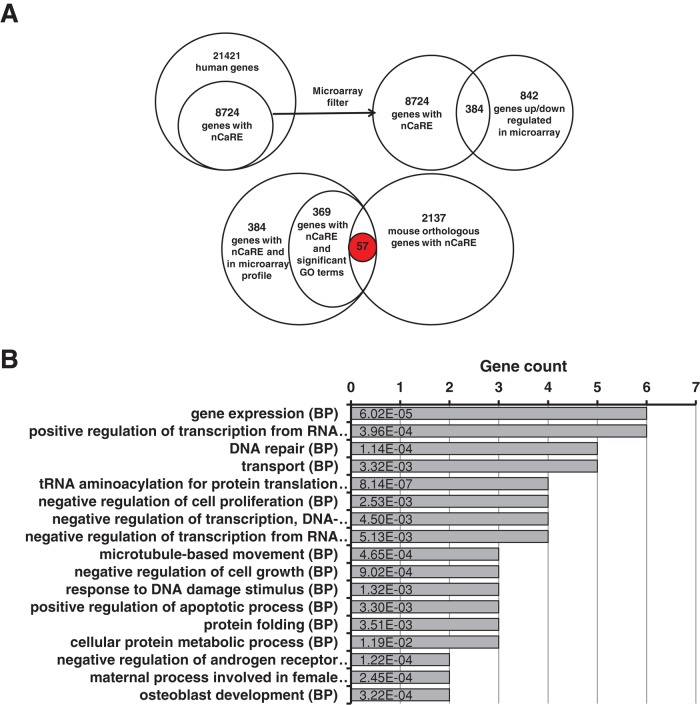
Bioinformatic analysis of the systematic retrieval of functional nCaRE-B sequences in the human genome was carried out by filtering biological data obtained from the gene expression profile of HeLa cells knocked down for APE1 (Vascotto et al., 2009a). Here we develop a method that integrates different approaches to the problem on a whole-genome scale while minimizing the number of false positives. To this purpose, classic DNA pattern-matching studies are integrated with independent information on gene regulation. We use three main sources for data filtering: 1) functional annotation data collected via the Gene Ontology (GO) project; 2) gene expression data derived from the microarray profile of APE1-knockdown HeLa cells (Vascotto et al., 2009a); and 3) human–mouse gene sequence comparisons. In fact, both expression data and functional annotation database provide a wealth of information about coregulation. This is of particular interest, since coregulated genes likely share similar transcriptional regulatory mechanisms. Comparison with orthologous gene promoters highlights sequences retained during evolution, whose conservation suggests their potential functionality. As a final result, we obtain a set of genes that pass the mentioned filters and are considered bona fide candidates coregulated through nCaRE-B sequences (Supplemental Figure S1).
We collected the 6000–base pair upstream regions of all human and mouse protein-coding genes, which we then analyzed for the presence of nCaRE-B elements using Glsearch for local alignment (Pearson, 2000). We found 8724 human genes to contain one or more nCaRE-B matches within their promoters; 2173 matches were retrieved in the mouse genome. We then cross-checked candidate genes with gene expression data obtained from APE1-knockdown cells (Vascotto et al., 2009a), verifying coexpression between genes carrying nCaRE-B elements. Through this analysis, we identified 384 common genes in the two data sets (Figure 1A). Then we considered the GO annotations of these 384 genes, searching for statistically significant common annotations. We observed strong overrepresentation of terms related to RNA processing and metabolism, in accordance with our previous studies (Vascotto et al., 2009a). All the significant associations between genes and GO terms are reported in Supplemental Table S1. Finally, we applied the phylogenetic footprinting filter, which evaluates whether a significant fraction of the selected genes, as obtained through the GO filter, share homologous genes containing nCaRE-B-related sequences in the upstream region with respect to the mouse gene data set. As a final result, we extracted 57 genes that may be considered bona fide candidates bearing the putative nCaRE-B sequences within their regulatory elements (Supplemental Table S2). We performed a functional enrichment analysis to determine whether the 57 genes found are involved in common biological processes. This showed that candidate genes were associated with processes related to gene expression, activation, or increment of the extent of transcription from an RNA polymerase II–driven promoter (Figure 1B and Supplemental Table S3). Among these 57 genes, several are involved with DNA repair process and cellular response to external stimuli and DNA damage: for example, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a member 4 (SMARCA4), sirtuin 1 (SIRT1), valosin-containing protein (VCP), multiple endocrine neoplasia I (MEN1), structural maintenance of chromosomes protein 6 (SMC6), early growth response 1 (EGR-1), and APE1 itself. We paid particular attention to SIRT1, a NAD-dependent histone deacetylase belonging to class III of the sirtuin family, based on the recent demonstration of a functional involvement of this enzyme in the deacetylation of some K residues in the N-terminal region of APE1 (Yamamori et al., 2010; Lirussi et al., 2012). The latter information and the data derived from our bioinformatic analysis led us to hypothesize the existence of an autoregulatory loop between APE1 and SIRT1.
FIGURE 1:
Bioinformatic research on nCaRE sequences. (A) Results obtained from the application of the different filters. Top, data derived from alignment research on nCaRE sequences on human gene promoters and subsequent cross-checking with microarray data. Bottom, final results from combined GO and phylogenetic footprinting analyses. (B) Functional enrichment analysis of the 57 putative genes regulated by APE1 performed according to their biological process annotations. For simplicity, only the most representative functional categories are reported. The number of genes for each category is provided on the horizontal axis, together with the list of the first 17 co-occurrence terms. Statistical significance for each category is shown within each bar.
APE1 binds the nCaRE-B sequences present in the human SIRT1 promoter
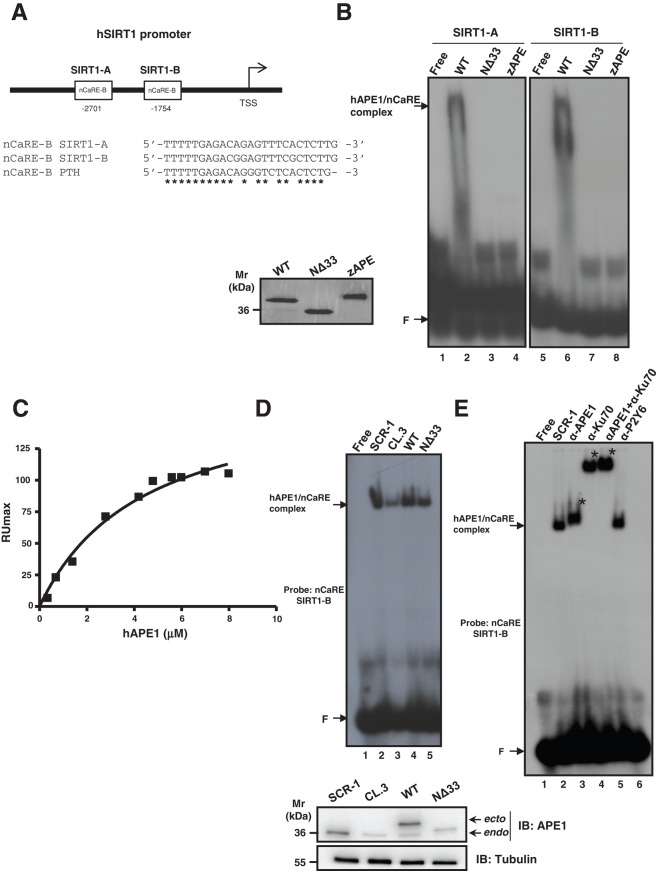
To prove the functional relevance of the nCaRE-B sequences identified in the human SIRT1 promoter, we first tested the ability of APE1 to specifically bind these elements in vitro. Thus we performed electrophoretic mobility shift assay (EMSA) analyses using different APE1 recombinant protein forms and two double-stranded (ds) oligonucleotides containing the SIRT1 nCaRE-B elements corresponding to the sequences present at −2701 (SIRT1-A) and −1754 base pairs (SIRT1-B) from the transcription start site (TSS), respectively (Figure 2A). Full-length human wild-type (WT) APE1 (APE1WT), the N-terminal APE1-deletion mutant (APE1NΔ33), and the orthologous zebrafish APE1 (zAPE1) expressed in Escherichia coli were used for this purpose. As clearly demonstrated by EMSA analyses, only the APE1WT protein was able to stably bind to both the SIRT1 nCaRE-B sequences (Figure 2B, lanes 2 and 6), whereas a complete absence of retarded complex was observed in the case of the truncated APE1NΔ33 form (Figure 2B, lanes 3 and 7). These findings show the importance of the first 33 amino acids at the APE1 N-terminus for proper binding of the protein to nCaRE-B sequences. Similar poor DNA-binding activity was apparent in the case of zAPE1 (Figure 2B, lanes 4 and 8), which bears a nonrelated N-terminal domain (Fantini et al., 2010). Together these results suggest that the phylogenetically evolved N-terminal domain of the protein is essential for stable interaction between APE1 and the nCaRE-B sequences. It is conceivable that K residues present within this region and acquired during evolution in mammals (Georgiadis et al., 2008; Fantini et al., 2010; Poletto et al., 2013) play a major role in protein binding to these DNA elements.
FIGURE 2:
APE1 is part of a nuclear protein complex that binds to the SIRT1 nCaRE sequence through its N-terminal domain. (A) Schematic representation (top) and multiple sequence alignment (bottom) of two nCaRE-B sequences found on the human SIRT1 gene promoter (SIRT1-A and SIRT1-B) with the nCaRE sequence found on the human PTH promoter (Okazaki et al., 1991). (B) EMSA analysis of nCaRE SIRT1-A and SIRT1-B sequence challenged with 10 pmol of purified APE1WT, APE1NΔ33, and zAPE1 proteins. Left, Coomassie staining of the purified recombinant proteins. (C) SPR analysis of the human APE1 (hAPE1)–nCaRE interaction. Recombinant hAPE1 and biotinylated nCaRE SIRT1-B (Table 1) were used as analyte and ligand, respectively. Plot of RUmax from each binding vs. hAPE1 concentrations (0.5–8 μM); data were fitted by nonlinear regression analysis. (D) Top, EMSA analysis of nCaRE SIRT1-B incubated with HeLa nuclear extract of different clones: control clone, APE1SCR-1 (lane 2), clone silenced for APE1, APE1CL.3 (lane 3), and clones reconstituted with APE1WT (lane 4) or APE1NΔ33 (lane 5). Bottom, Western blot analysis of APE1 protein in HeLa nuclear cell extracts. (E) EMSA analysis of nCaRE SIRT1-B with HeLa nuclear extract from APE1SCR-1 clone alone (lane 2) or preincubated with monoclonal antibody against APE1 (lane 3) or/and with an antibody against Ku-70 (lanes 4 and 5). Lane 6 corresponds to APE1SCR-1 nuclear extract incubated with a nonspecific antibody (α-P2Y6). Free indicates probe alone; F shows the position of the free oligonucleotide probe. Specific APE1/nCaRE interaction is indicated by the arrow. Asterisk indicates supershift.
We then estimated the affinity of APE1 for SIRT1 nCaRE-B sequences through surface plasmon resonance (SPR) analysis (Figure 2C). Biotinylated versions of the nCaRE-B (SIRT1-B) or polyT sequences were immobilized onto a streptavidin chip for use as ligands in SPR experiments. APE1WT and APE1NΔ33 were then analyzed for their DNA-binding properties. When testing APE1WT as analyte, KD = 3.90 ± 0.08 μM was measured (Poletto et al., 2013); corresponding kinetic parameters are shown in Table 1. Conversely, when using APE1NΔ33, we did not observe any SPR signal variation (unpublished data), in accordance with EMSA experiments (Figure 2B). This result confirmed that the protein region 1–33 is essential for stable interaction of APE1 with these DNA elements. As a DNA-repair enzyme, APE1 has intrinsic ability to bind to DNA in a sequence-independent manner. Moreover, independent observations clearly point to a role of the nucleic acid secondary structure in positively modulating this activity (Poletto et al., 2013). Here we evaluated the protein capacity to bind to a single-stranded 24-mer oligo-dT (here called polyT). A KD = 308 ± 3 μM was measured in the case of APE1WT, whereas no binding was observed for APE1NΔ33 (Table 1 and Supplemental Figure S2), in agreement with EMSA analysis. These results indicate that APE1 poorly recognizes a nonstructured oligonucleotide formed by a stretch of thymidines, confirming that this protein may bind to DNA with different affinities and the oligo-dT sequence may be used in EMSA analysis as a nonspecific competitor.
TABLE 1:
Dissociation constant and kinetic parameter values as determined for APE1 by SPR analysis.
| Ligand | Ka (ms × 105) | Kd (1/s) | KD (μM) |
|---|---|---|---|
| SIRT1-B nCaRE | 0.270 | 0.105 | 3.90 ± 0.08 |
| SIRT1-B mutated | 0.0198 | 0.236 | 119 ± 8 |
| polyT | 0.004 | 0.112 | 308 ± 3 |
Our EMSA and SPR analyses on APE1 binding activity to the SIRT1 nCaRE-B sequences showed low affinity for recombinant APE1 alone. Therefore we tested whether additional factors present in nuclear extracts of the cells may increase protein-binding affinity to its oligonucleotide target. To this aim, further EMSA analyses, performed using HeLa cell nuclear fractions, confirmed that nuclear activity able to specifically bind nCaRE-B sequences was indeed present (Figure 2D). The high-affinity complex measured, even using much lower amounts of APE1 (0.63 pmol, as estimated from Lirussi et al., 2012) with respect to that obtained with the recombinant purified protein alone (10 pmol; Figure 2B), suggested that additional factors are required for efficient APE1 binding to SIRT1 nCaRE-B sequences. To demonstrate the presence of APE1 in the retarded complex observed in Figure 2D, we used nuclear extracts obtained from a HeLa line (CL.3) for which endogenous APE1 protein expression was previously knocked down through stable short hairpin RNA (shRNA) transfection (Vascotto et al., 2009a). As is apparent from lane 3 in Figure 2D, an overall reduction of the intensity (almost 70% with respect to lane 2) of the retarded nCaRE-bound complex was observed, similar to what occurred for the clone expressing APE1NΔ33 (lane 5; almost 50% with respect to lane 2). Nuclear extracts obtained from HeLa cells reconstituted with ectopic APE1WT (lane 4) showed the same amount of bound complex as the control cell clone expressing a scrambled shRNA vector (indicated as SCR-1; recovery of almost 80% with respect to lane 2). Preincubation of the nuclear extract from the SCR-1 clone with an anti-APE1 antibody resulted in the formation of a supershifted complex (Figure 2E, lane 3). This complex was absent when a nonrelated antibody was used (lane 6), clearly demonstrating that APE1, present in the nuclear cell extract, is involved in the recognition of the nCaRE-B elements of the SIRT1 promoter. The reduced intensity of the retarded complex band upon APE1 silencing or in the reconstituted cells expressing APE1NΔ33 protein (Figure 2D), as well as the lower apparent electrophoretic mobility of the protein–DNA retarded complex when using nuclear extracts in place of the recombinant purified protein (Figure 2B), suggests that APE1 may be part of a multiprotein complex. Because Ku70 antigen protein was already demonstrated to bind nCaRE-A sequences in complex with APE1 (Chung et al., 1996), we incubated the HeLa nuclear extract with an antibody recognizing the Ku70 antigen. We then subjected the reaction to EMSA analysis, showing the formation of a supershifted complex (Figure 2E, lane 4). The concurrent presence of APE1 and Ku70 in the same retarded complex was confirmed by performing simultaneous preincubation with antibodies specific for these proteins (Figure 2E, lane 5). The presence of Ku70 in the complex with APE1 was also corroborated by additional EMSA analysis performed with the purified recombinant protein attesting that, when present alone, Ku70 is not able to stably bind to the SIRT1 nCaRE-B sequence per se. On the contrary, when concomitantly incubated with APE1, Ku70 significantly enhances APE1 DNA-binding activity to SIRT1 nCaRE-B element (Supplemental Figure S3, compare lanes 2–5 and 8–10). Unpublished data demonstrated that this stimulatory activity on APE1 binding required interaction through the APE1 33N-terminal domain. Overall these data demonstrate that APE1 must be part of a multiprotein complex containing Ku70 to elicit its high-affinity binding potential with regard to the nCaRE-B sequences present on the SIRT1 promoter.
Topology of the APE1-nCaRE complex
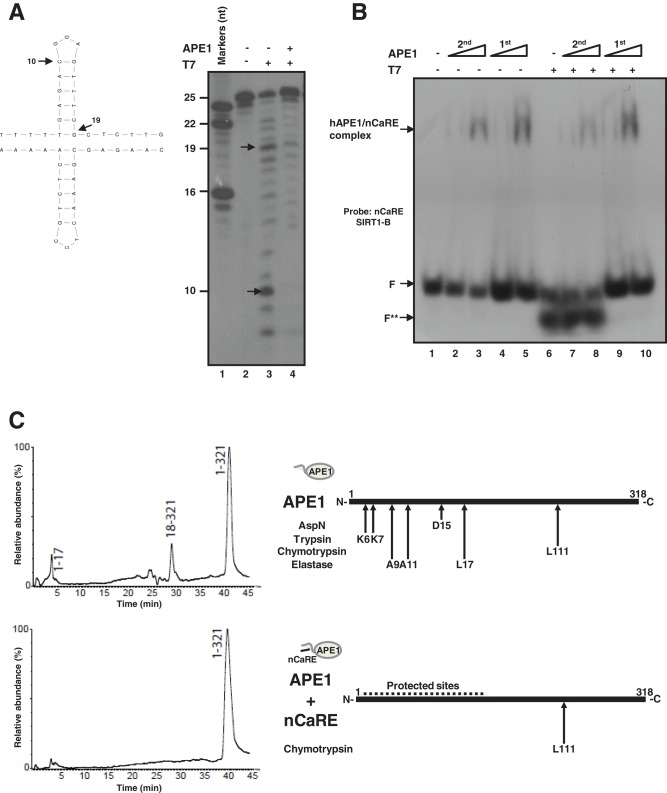
nCaRE-B element consists of a palindromic sequence that can fold into self-complementary hairpins (Figure 3A, left). In silico analyses with the mfold program suggested that these elements could fold into cruciform-like structures. To evaluate whether SIRT1 nCaRE-B sequences can fold into cruciform duplexes and specifically assess whether APE1 binding to this element may depend on such secondary structures, we performed footprinting analyses by using the T7 endonuclease I (Figure 3A, right). This protein is a structure-sensitive enzyme that specifically recognizes conformationally branched DNA and Holliday structures or junctions (Parkinson and Lilley, 1997; Déclais et al., 2003; Fan et al., 2006). Footprinting data support the hypothesis of the existence of a secondary structure for the nCaRE-B sequences (Figure 3A and Supplemental Figure S4). In particular, our experiments show the predominant formation of two bands (respectively 10 and 19 nucleotides in length) in the digested samples, which correspond to a cutting site close to the predicted loop or immediately adjacent to the predicted stem (Figure 3A, arrows). Of interest, preincubation of the SIRT1 nCaRE-B oligonucleotide with APE1 impairs T7 endonuclease digestion (Figure 3A, lane 4). Consistent with these observations, EMSA analysis demonstrated that digestion of the SIRT1 nCaRE-B oligonucleotide with T7 endonuclease affects APE1 binding to this element, but, conversely, when APE1 was first incubated with the nCaRE-B probe and then digested with T7 endonuclease, the binding was not affected. These data suggest a protective role of APE1 with regard to T7 endonuclease action, strongly supporting the hypothesis that these proteins may compete for the same binding site on the nCaRE-B sequences (Figure 3B).
FIGURE 3:
APE1 recognizes structured nCaRE sequences through its N-terminal domain. (A) Left, predicted cruciform structure of nCaRE SIRT1-B ds oligonucleotide. Arrows indicate the cleavage site of T7 endonuclease I and the length of the products. Right, 5′-32P-end-labeled nCaRE was preincubated (lane 4) or not (lane 3) with APE1 recombinant protein and then subject to T7 endonuclease digestion. (B) EMSA analysis of APE1 binding to nCaRE sequence after digestion with T7 endonuclease (lanes 7 and 8) or upon preincubation with APE1 and subsequent digestion with T7 endonuclease (lanes 9 and 10). Lane 1 is the probe alone; APE1 incubation with the probe was performed temporally before (1st) or after (2nd) T7 digestion; F shows the position of the free oligonucleotide probe. F** indicates the T7 endonuclease–digested probe. Specific APE1/nCaRE interaction is indicated by the arrow. (C) Schematic representation of the amino acids within the N-terminal domain of APE1 and involved in nCaRE oligonucleotide binding. Left, proteolytic maps obtained after incubation of recombinant APE1 alone (top) or recombinant APE1 complexed with SIRT1 nCaRE-B oligonucleotide (bottom) with endoprotease AspN. Experiments were performed on a recombinant APE1 form bearing three additional amino acids at the protein N-terminus with respect to the native counterpart. Peptides identified by mass spectrometry analysis are indicated at the top of the corresponding chromatographic peaks. Right, proteolytic sites identified in native APE1 alone (top) and in APE1 complexed with SIRT1 nCaRE-B oligonucleotide (bottom); summary of results from independent experiments performed by using different proteases. See the Supplemental Information for experimental details and Supplemental Figure S4 and Supplemental Table S4 for complete data.
The first 33 N-terminal amino acids of APE1 are required for protein binding to the nCaRE sequences (Figure 2, A and D). To further investigate the role of this protein portion on APE1’s ability to bind to DNA elements, we performed combined limited proteolysis-mass spectrometry experiments on recombinant APE1 protein in either the absence or the presence of its target nCaRE oligonucleotide (Figure 3C, left). In particular, parallel experiments with a specific proteolytic enzyme (used at a defined APE1/protease weight/weight ratio) were carried out on a time-course basis on 1) isolated APE1, 2) APE1 complexed with SIRT1 nCaRE-B oligonucleotide, and 3) APE1 complexed with PTH nCaRE-B oligonucleotide (Okazaki et al., 1992; Supplemental Figure S5 and Supplemental Table S4). This last was used as control. Resulting differential peptide maps provided information on amino acid(s) eventually protected from proteolytic attack as result of their presence at the protein–DNA complex interface (Figure 3C, left). The data were interpreted according to the concept of “molecular shielding” of amino acids from proteolytic attack (Scaloni et al., 1998, 1999; Renzone et al., 2007) and were rationalized on the basis of x-ray crystallographic APE1 structures (Gorman et al., 1997; Beernink et al., 2001). To maximize information resulting from limited proteolysis experiments, we used different proteolytic enzymes in separate assays.
Figure 3C (right) summarizes the results of the limited proteolysis experiments, as obtained by using different proteases on recombinant APE1 alone (Figure 3C, top) or after its complex formation with the SIRT1 nCaRE substrate (Figure 3C, bottom; see Supplemental Figure S5 and Supplemental Table S4 for experimental details). General considerations concerning the native protein are as follows. Preferential hydrolyzed peptide bonds on isolated APE1 gathered into a specific region of the protein, namely the most exposed segment, the unstructured N-terminal domain, which contained six proteolytic sites (K6, K7, A9, A11, D15, and L17). An additional site was within the globular APE1 domain (L111). Of interest, no other cleavage sites were detected in other protein regions, although exposed on the molecular surface (Gorman et al., 1997; Beernink et al., 2001). After complex formation with nCaRE-B oligonucleotides, a marked protective effect was observed, as demonstrated by the large decrease in the number of proteolytic sites present in the N-terminal region (from six to one), thus confirming the involvement of this APE1 portion in binding to these DNA elements.
Transcriptional regulation of SIRT1 expression by APE1 protein
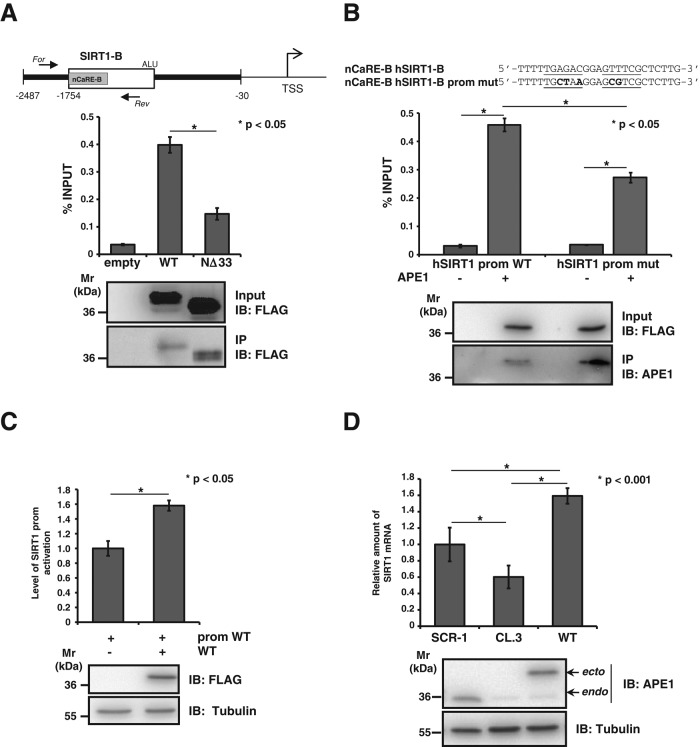
To determine whether the observations obtained in vitro regarding APE1 binding to SIRT1 nCaRE-B sequences have any relevance in vivo, we examined the APE1 occupancy of the nCaRE-B sequence in the SIRT1 promoter through chromatin immunoprecipitation (ChIP) analyses. To this purpose, we used HeLa cells cotransfected with a human SIRT1 promoter–containing plasmid (Yamamori et al., 2010) and FLAG-tagged APE1WT- or APE1NΔ33-expressing plasmids. The amount of immunoprecipitated SIRT1 promoter was significantly enriched in APE1WT-transfected cells compared with that obtained from control cells transfected with the empty vector alone (Figure 4A). As expected, APE1NΔ33-transfected cells had a remarkable reduction, although not completely so, in the amount of immunoprecipitated SIRT1 promoter. A similar degree of reduction was observed when ChIP analysis was performed by using a SIRT1 promoter bearing a mutated sequence within the nCaRE-B motif (Figure 4B), whose reduced specificity was previously assessed through SPR analysis (Table 1 and Supplemental Figure S2). These experiments revealed that the mutation introduced in the nCaRE-B sequence significantly reduced the APE1 binding to DNA, lowering the affinity of the complex by 30-fold (KD = 119 ± 3 μM). Competitive EMSA analyses were in agreement with these findings. In fact, addition of the unlabeled nCaRE ds oligonucleotide resulted in almost complete elimination of bound complex formation. On the other hand, competition with the unlabeled mutant nCaRE oligo (nCaRE-mut) caused only a slight reduction of the nCaRE-binding complex, in agreement with ChIP and SPR data, supporting the notion of sequence-dependent binding specificity (unpublished data). These data were also confirmed with the endogenous APE1 promoter via ChIP followed by high-throughput sequencing (ChIP-Seq) analyses (unpublished data), further demonstrating the physiological relevance of these findings. Together these results confirm our in vitro observations (Figure 2) and support the hypothesis that, under basal conditions, APE1 is associated with the nCaRE-B sequence within the SIRT1 promoter also in vivo, possibly as part of a multiprotein complex.
FIGURE 4:
APE1 positively regulates SIRT1 expression at the promoter level. (A). Top, schematic representation of the human SIRT1 promoter used for HeLa transfection. For and Rev arrows indicated the reverse transcription-PCR primer designed for the quantification of the human SIRT1 nCaRE sequence bound to APE1. Bottom, ChIP assay for APE1-nCaRE sequence association. Percentage of immunoprecipitated nCaRE DNA relative to that present in total input chromatin. Data were further normalized to the amount of immunoprecipitated protein. Western blot analysis was performed on total cell extracts (input) and immunoprecipitated material (IP) with specific antibody for FLAG and APE1. IB, immunoblot. (B) ChIP analysis on mutated human SIRT1 promoter. Top, base composition of the nCaRE SIRT1-B mutated sequence used for site-directed mutagenesis of SIRT1 promoter. Divergent sequences in the mutant nCaRE are bold. Bottom, HeLa cells were cotransfected with vector expressing APE1WT and, alternatively, wild-type or mutated hSIRT1 promoter. The histogram represents the amount of hSIRT1 promoter sequence that was immunoprecipitated. Data are percentage of input and are normalized to the amount of APE1 immunoprecipitated, as evaluated by Western blot analysis. (C) hSIRT1 promoter is activated in presence of APE1, as shown in the reporter assay. Western blot analysis showing the normalization of protein levels. (D) Analysis of SIRT1 mRNA level with qPCR in clones expressing APE1WT or APE1 silenced (CL.3) cells. Western blot analysis on protein extract of clones showing the suppression of endogenous APE1 expression upon 10 d of treatment with doxycycline.
We then evaluated whether APE1 binding to SIRT1 promoter plays a role in SIRT1 transcriptional regulation by performing promoter-reporter assays. HeLa cells were cotransfected with a luciferase reporter vector bearing the SIRT1 promoter and APE1WT FLAG-tagged vector (Figure 4C). SIRT1 promoter-reporter assays showed that there was a significant increase in the luciferase signal detected in the presence of APE1 compared with that of the promoter alone. We evaluated the effect of APE1 silencing on endogenous SIRT1 mRNA expression levels through an inducible shRNA knockdown strategy (Vascotto et al., 2009a, b). Endogenous APE1 knockdown (CL.3) caused a significant reduction in the SIRT1 endogenous expression levels (Figure 4D), which was rescued in cells reconstituted with a siRNA-resistant APE1 cDNA expression plasmid (WT). These data demonstrated a positive effect of APE1 on SIRT1 transcriptional activation. Although unexpected, since previous data reported a transcriptional repressive role for APE1 through nCaRE sequence binding (Okazaki et al., 1991; Fuchs et al., 2003), these data agree with our previous observations from gene expression profiling analysis (Vascotto et al., 2009a), in which reduced expression of SIRT1 in APE1-knockdown HeLa cells was apparent.
Oxidative stress induces SIRT1 transcription via recruitment of BER enzymes
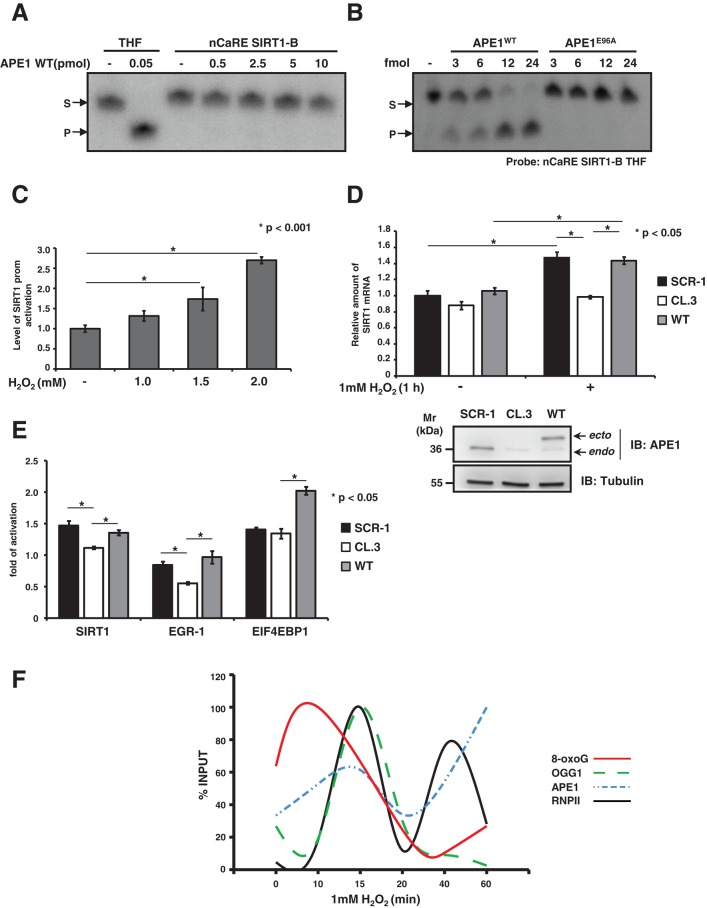
To better understand, at the molecular level, the transcriptional function exerted by APE1 through binding to nCaRE-B sequences, we investigated whether APE1’s positive role in SIRT1 transcription relied on its enzymatic activity on DNA. First, we observed that under basal conditions APE1 has no endonuclease activity on the SIRT1 nCaRE-B sequence, as assessed through an oligonucleotide cleavage assay (Figure 5A). APE1 cleavage, occurring at any site on the cruciform nCaRE-B sequence, should lead to a site-specific, single-stranded break that can be detected by the appearance of an extra fragment in the cleavage assay. Even using an increasing amount of purified recombinant APE1 protein, we did not detect any nuclease activity that, on the contrary, was readily visible when using a radiolabeled 26-mer ds oligonucleotide containing a tetrahydrofuran mimicking an AP site (here referred to as THF; Berquist et al., 2008). Substitution of the guanine residue at position 12, within the predicted loop of the nCaRE-B sequence, with a THF residue resulted instead in efficient APE1 cleavage activity on the nCaRE-B sequence. This activity depends on APE1 catalytic function, since the catalytically inactive APE1E96A mutant (Izumi et al., 1999) was unable to efficiently cleave the same substrate (Figure 5B).
FIGURE 5:
Recruitment of BER enzymes on the SIRT1 promoter. (A) APE1 endonuclease activity on ds, nCaRE SIRT1-B radiolabeled oligonucleotide. A radiolabeled ds, THF-containing deoxyoligonucleotide (THF) was used as control. Reactions were performed with increasing amounts (picomoles) of recombinant APE1WT protein. (B) APE1 AP endonuclease activity on nCaRE SIRT1-B THF-containing probe incubated with increasing amounts of recombinant APE1WT protein or a catalytic inactive APE1 mutant (APE1E96A). (C) Reporter assay with HeLa cells transfected with hSIRT1 firefly reporter vector and challenged with increasing doses of H2O2 for 1 h as indicated. (D) qPCR analysis of SIRT1 mRNA levels in clones expressing APE1WT or APE1-silenced cells APE1CL.3 after 1 mM H2O2 treatment for 1 h. Western blot analysis on the protein extracts. (E) qPCR analysis of SIRT1, EGR-1, and EIF4EBP mRNA levels in APE1CL.3 or APE1WT clones after 1 mM H2O2 treatment for 1 h. Data shown are reported as fold of activation after H2O2 treatment. (F) Results of four independent ChIP analyses relative to accumulation of 8-oxodeoxyguanine, OGG1, APE1, and RNA polymerase II protein on the SIRT1 promoter after 1 mM H2O2 treatment for different times (as reported). Data are percentage of input and normalized to quantity of DNA immunoprecipitated by α-tubulin (α-tub). See Supplemental Figure S4 for detailed information.
Prompted by these findings, we hypothesized that the APE1-positive function that we observed on the SIRT1 promoter may be ascribed to the APE1 catalytic activity on nCaRE-B sequences. This can occur after specific stimuli, such as oxidative stress, which can lead to abasic site formation on nCaRE-B sequences (Amente et al., 2010; Francia et al., 2012). It is well known that SIRT1 expression and function are regulated by external stressors, including exposure to genotoxic agents (Cohen et al., 2004; Kim and Um, 2008; Yamamori et al., 2010). We therefore measured SIRT1 transcription after oxidative stress, such as that generated by H2O2 exposure. First, we demonstrated that SIRT1 promoter activation was increased by H2O2 treatment in a concentration-dependent manner (Figure 5C). Next we examined the effect of APE1 silencing or re-expression on SIRT1 transcriptional activation in HeLa cell clones upon H2O2 treatment (Figure 5D and Supplemental Figure S6). We treated control (SCR-1), APE1-silenced (CL.3), and APE1WT cell clones with 1 mM H2O2 for 1 h. SIRT1 mRNA levels were then evaluated by quantitative PCR (qPCR) and compared with those for untreated clones. On oxidative treatment, SIRT1 mRNA significantly increased, and, of note, this response was higher in the presence of APE1WT protein, whereas it was lower in the case of APE1-knockdown–expressing cells. The residual activation of SIRT1 mRNA expression, which was apparent also in APE1-knockdown cells, might be ascribable to the presence of residual endogenous APE1 protein and/or of further limiting factors, as already speculated (Chung et al., 1996; Kuninger et al., 2002; Bhattacharyya et al., 2009). The observed induction of SIRT1 mRNA transcription in HeLa cells upon oxidative stress correlates with a parallel increase in its protein levels. Confirmation of the biological relevance of our findings comes from the observation of concomitant increased deacetylation activity of SIRT1 protein toward its substrate, that is, K382 of p53 (Vaziri et al., 2001; Supplemental Figure S6).
We also confirmed the general relevance of our model by testing other hypothetical APE1 target genes containing an nCaRE-B element in their promoters and resulting dysregulation in the APE1-kd cell model (Figure 1 and Supplemental Table S2; Vascotto et al., 2009a). To this aim, we evaluated by qPCR the expression levels, upon H2O2 treatment, of EGR-1 and eukaryotic translation initiation factor 4E–binding protein 1 (EIF4EBP1) in the HeLa cell inducible-kd (CL.3) clone and the reconstituted cell clone (WT) used here (Figure 5E). Similar results were obtained using another genotoxic agent, methyl methanesulfonate (Supplemental Figure S9). These data, showing inducible expression of these genes dependent on APE1 expression to a similar extent of that observed in the case of SIRT1, were suggestive of a general mechanism of gene activation upon DNA damage that involves APE1 binding to nCaRE-B elements.
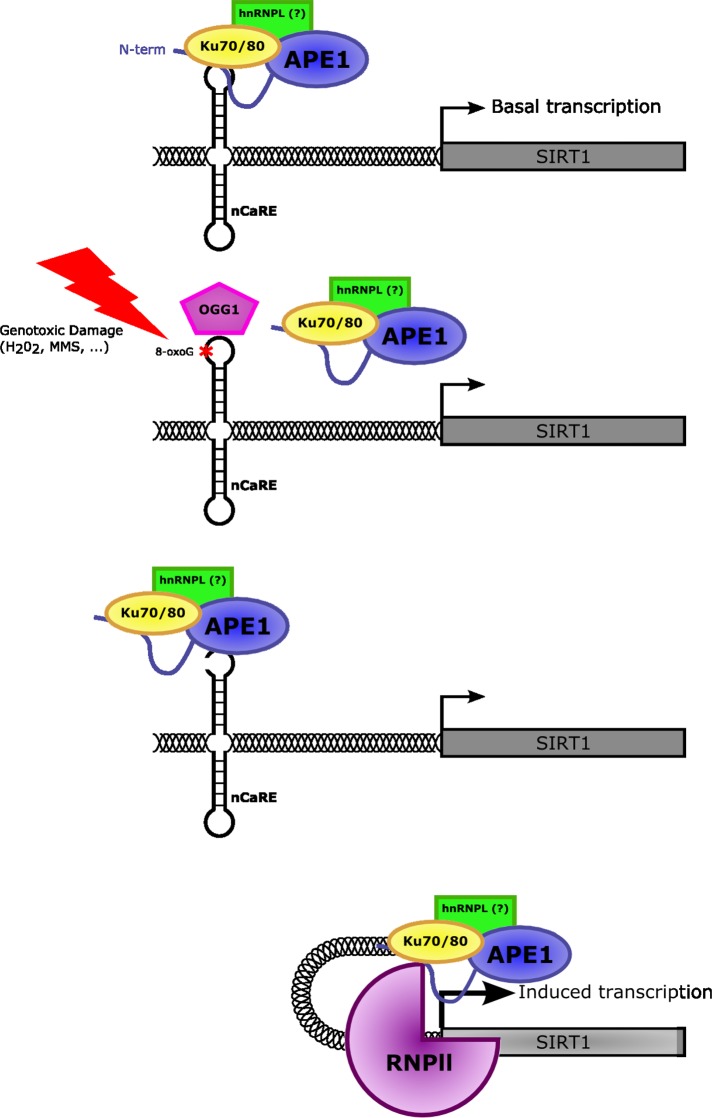
To find a relationship between SIRT1 transcription induced by oxidative stress and the APE1 regulatory activity on the nCaRE-B sequences located within the promoter, we studied the dynamics of oxidative repair enzyme recruitment on the SIRT1 nCaRE-B sequence at early time upon H2O2 treatment. DNA base oxidation determines the formation of 8-oxodeoxyguanine (8-oxodG), which is recognized by the DNA glycosylase OGG1. This enzyme initiates the BER pathway by removing the 8-oxodG lesion, which is further processed by APE1, which cleaves the apurinic site. To assess the dynamics of occupation of the nCaRE-B sequence on the SIRT1 promoter by these enzymes upon oxidative stress, we performed a time-course ChIP analysis on the SIRT1 nCaRE-B sequence after 1 mM H2O2 treatment (Figure 5F and Supplemental Figure S7). We immunoprecipitated SIRT1 nCaRE-B sequence with antibodies against 8-oxodG, OGG1, and APE1. We observed that the signal of 8-oxodG promptly reached its plateau 10 min after H2O2 treatment and subsequently decreased, concomitant with accumulation of OGG1, which is recruited immediately after (15 min), in accordance with the BER processes. Occupancy by APE1 follows OGG1 recruitment. To demonstrate a direct link between DNA repair and SIRT1 transcriptional initiation, we examined the assembly of RNA polymerase II (RNPII) on the SIRT1 nCaRE-B sequence. Under basal condition, RNPII was found with relatively low abundance on the SIRT1 nCaRE-B sequence; conversely, the polymerase was progressively recruited as soon as the oxidative stress began (15 min after H2O2 treatment). Afterward (40 min after H2O2 addition), RNAPII was again recruited on the SIRT1 promoter, showing a wave-like behavior. In concert with this observation, we also noticed augmented interaction between APE1 and RNPII in accordance with the kinetics observed during ChIP analysis (Supplemental Figure S8). This oxidatively induced recruitment of RNPII to the SIRT1 promoter may suggest that oxidative stress can trigger SIRT1 transcriptional activation. One could envision a mechanism in which H2O2, causing oxidation of the guanine at the SIRT1 nCaRE-B sequence, promotes recruitment of components of the base excision repair system—OGG1 and APE1, together with proteins involved in nCaRE-B binding such as Ku70. When recruited to the SIRT1 promoter, APE1 (through its endonuclease activity) produces nicks on the SIRT1 nCaRE-B sequence, possibly favoring the DNA relaxation necessary for the formation of chromatin loops that bring RNAPII at the transcription start site (Figure 6); this last enzyme in turn can initiate transcription.
FIGURE 6:
Mechanistic model for the role of APE1 in oxidatively mediated SIRT1 transcription. Under normal conditions, APE1, together with other protein factors, is bound to the nCaRE element present within the SIRT1 promoter involved in the basal activation of SIRT1 transcription. Conversely, upon oxidative stress conditions, DNA oxidation determines the formation of 8-oxodeoxyguanine (8-oxoG) lesions at the nCaRE sequence present in the SIRT1 promoter, which are recognized and processed by BER enzymes, including APE1. The nicks introduced at the chromatin level by APE1 during 8-oxoG removal might promote the formation of chromatin loops moving the active form of RNA polymerase II closer to the TSS of the gene, turning on the transcription.
DISCUSSION
After its cloning by independent groups, first as a DNA-repair enzyme (Demple et al., 1991; Robson and Hickson, 1991) and then as a redox coactivator protein (Xanthoudakis and Curran, 1992), a number of articles described different APE1 functions, elucidating its involvement in several biological contexts. As the main apurinic/apyrimidinic endonuclease in mammalian cells, APE1 is classically known for its essential function as a DNA-repair enzyme in the BER pathway. Besides this crucial role in the maintenance of genome stability, APE1 was demonstrated to be involved in redox signaling and the regulation of gene expression (Tell et al., 2010a; Wilson and Simeonov, 2010), supporting the notion that it is a multifunctional protein, with features that go beyond the classic activities of a DNA-repair enzyme. Of note, its multifunctional nature suggests APE1 as an ideal candidate protein linking DNA-damage sensing/repair and transcriptional regulation of genes during cell response to genotoxic damage. Among these non-canonical activities, another interesting APE1 function is its ability to bind the nCaRE sequence of some gene promoters, thus acting as a transcriptional regulator. Okazaki's group was the first to identify two nCaRE sequences within the PTH gene promoter (nCaRE-A and nCaRE-B; Okazaki et al., 1991). The presence of these elements was also described in the regulatory region of a few other genes, such as human APE1 (Izumi et al., 1996), rat atrial natriuretic polypeptide (Okazaki et al., 1992), human renin (Fuchs et al., 2003), and Bax (Bhattacharyya et al., 2009). Besides these few genes, no further evidence has been provided. However, since nCaRE-B elements are present within ALU repeats, which are widely distributed throughout the expressed genome, it is expected that APE1 could regulate the expression of a large number of genes. Here we performed an unbiased investigation of the whole human genome, searching for putative genes whose transcription may be mediated through APE1’s ability to bind nCaRE-B elements. In particular, although nCaRE elements seem to be active also at downstream regions (Izumi et al., 1996), here we specifically focused on nCaRE-B elements present only on the upstream sequence of human genes. In the near future, we plan to extend this approach to downstream and intron regions. Bioinformatic analyses revealed a number of genes potentially regulated by APE1, which are involved in several pathways related to gene expression (Figure 1). Among the 57 candidate genes retrieved from our bioinformatic analysis, we chose to study the human deacetylase SIRT1, which bears two nCaRE-B elements in its promoter. Increased interest in SIRT1 was due to recent articles showing that this deacetylase controls the acetylation status of APE1 lysine residues K6-7 (Yamamori et al., 2010) and K27-35, thus modulating the subnuclear distribution of this protein and coordinating its enzymatic functions in the BER pathway (Lirussi et al., 2012). Therefore we hypothesized the existence of a possible autoregulatory loop between the two proteins: APE1 modulates SIRT1 expression, which, in turn, regulates APE1 function through deacetylation.
To investigate APE1’s transcriptional regulatory function on SIRT1 expression, we first examined APE1’s ability to bind the nCaRE-B sequences found in the SIRT1 promoter. Through different in vitro approaches, we showed that APE1 is able to bind SIRT1 nCaRE-B sequences (Figures 2 and 3). In particular, by a combination of EMSA and SPR analyses with limited proteolysis experiments, we demonstrated that the APE1 N-terminal domain is essential for the proper binding of these elements. The essential role of this protein domain in DNA binding is remarkable, particularly if one considers the phylogenesis of the nCaRE-B elements and that of the APE1 N-terminal region. nCaRE-B sequences are present within ALU repeats, which belong to the short interspersed nucleotide element family of repetitive sequences that originally derived from the reverse transposition of 7SL RNA. This event took place in the genome of an ancestor of Supraprimates (Kriegs et al., 2007): these repetitive elements have been found exclusively in primates (Deininger et al., 1981), scandentians (Nishihara et al., 2002), and rodents (Krayev et al., 1980), all members of the placental mammalian clade Supraprimates (Euarchontoglires; Murphy et al., 2001). Similarly, information on the sequence homology of the APE1 N-terminal domain across species points to the recent phylogenetic acquisition of this region. Sequence conservation of this domain is very high in mammals but almost absent in other organisms, with the exception of Danio rerio, Dictyostelium, and Drosophila. Accordingly, it could be envisioned that once ALU elements appeared in primates and were stabilized in their genomes, progressively losing their transcriptional potential, these organisms needed to evolve novel mechanisms to cope with the acquired RNA Pol II regulatory sites present within the ALU region. The concomitant acquisition of the APE1 N-terminal domain in mammals could explain new modulatory functions toward these DNA elements. The observation that specific K residues (K24-27) within this reduced APE1 portion seem to be required for the correct binding of nCaRE-B (unpublished data) nicely fits with this hypothesis. The zebrafish homologue of APE1, which shares <40% of the N-terminal amino acid sequence with the human protein and lacks two of five K residues in this region, is indeed no longer able to stably bind these nCaRE-B sequences (Figure 2B; Poletto et al., 2013). Similar results were obtained using a human recombinant APE1 mutant protein bearing specific K-to-A multiple substitution at K27/31/32/35, in which the positive charges at the amino acid side chain were removed to mimic a condition similar to that exerted by K acetylation (unpublished data).
The APE1 N-terminal domain seems required for stable binding to the SIRT1 nCaRE-B elements, even though it is not sufficient (Poletto et al., 2013). EMSA analysis, performed with nuclear extracts of HeLa cells expressing the deletion form of APE1, demonstrated that APE1 is part of a multiprotein complex, not the limiting factor in the binding reaction. As speculated by Okazaki and also in later work, the binding affinity observed for the multiprotein complex is higher than that detected when using purified APE1 protein alone (Figure 2). This suggested that other factors are necessary and cooperate with APE1 to fully exert this function, confirming previous observations (Chung et al., 1996; Kuninger et al., 2002). The two subunits of Ku antigen (Ku70 and Ku80) were among the protein factors already described by Chung et al. (1996) to be involved in the specific binding of nCaRE-A sequences. Here we demonstrated that Ku70 binding is not exclusively limited to the nCaRE-A elements, since we identified this protein in the complex that binds to the nCaRE-B sequence of SIRT1. The Ku heterodimer is a main component of the nonhomologous end-joining pathway that repairs DNA double-strand breaks (DSBs), which are generally produced upon extensive oxidative and infrared damage to DNA (Lieber, 2010). The peculiar structure of Ku allows recognition and tight binding to DSBs, together with the recruitment of DNA-PKcs and other factors to form the active protein kinase complex DNA-PK that facilitates processing and ligation of broken ends (Walker et al., 2001; Postow, 2011). Its involvement in nCaRE binding is not clear, but emerging evidence suggests a biological role in its noncanonical functions (Adelmant et al., 2012). We envisage that Ku70 association with nCaRE elements could further facilitate APE1 binding, especially after DNA damage, since we observed increased interaction between the two proteins upon oxidative stress (Supplemental Figure S8).
We also better characterized the topology of the APE1-nCaRE complex. The palindromic nature of nCaRE-B sequences was described by Okazaki et al. (1991), who suggested the possible involvement of a dimeric nuclear protein in this process. Here we suggest that the SIRT1 nCaRE-B, due to its palindromic sequence, can potentially fold into a cruciform-like structure, and APE1 binding activity toward these elements strongly relies on the secondary conformation adopted by the oligonucleotide, as established for other DNA and RNA substrates (Figure 3B and Supplemental Figure S4; Poletto et al., 2013). Formation of similar cruciform-like structures by palindromic sequences has been described in eukaryotic cells, and their biological consequences have been related to different processes, including regulation of transcriptional events when present in close proximity to gene promoters (Pearson et al., 1994; Shlyakhtenko et al., 2000; Alvarez et al., 2002; Cunningham et al., 2003; Kurahashi et al., 2004). We therefore hypothesize a similar mechanism for SIRT1 transcriptional regulation. However, the requirement of a specific recognition motif cannot be excluded since mutations in the SIRT1 nCaRE-B sequence, determining only a partial disruption of the oligonucleotide secondary structure, did not completely affect APE1-binding activity in vivo (Figure 4B).
We further studied APE1’s transcriptional function on the SIRT1 promoter. We found that APE1 is able to bind the nCaRE-B sequence of SIRT1 promoter in vivo and confirmed this through ChIP-Seq analysis (unpublished data). In particular we observed that APE1 positively affects SIRT1 gene transcription. Although APE1 was implicated in the repression of PTH gene transcription in a Ca2+-dependent manner, APE1 overexpression activated the SIRT1 promoter (Figure 4C) apparently through a Ca2+-independent mechanism. This unexpected positive function on the transcription of an nCaRE-containing promoter was also reported in other work, in which the authors suggested that the role of the nCaRE sequences in the context of different promoters and cells conditions could affect nCaRE activity (Bhakat et al., 2003; Fuchs et al., 2003). Of interest, we noticed that the positive effect of APE1 was particularly pronounced during oxidative stress, through its binding to SIRT1 nCaRE-B sequences (Figure 5D). Treatment with H2O2 leads to activation of the SIRT1 promoter, which determines an increase of the corresponding transcriptional expression in an APE1-dependent manner. This positive transcriptional effect was also observed when we looked at the expression of other genes that have nCaRE-B elements in their promoters (Figure 5E), thus corroborating the hypothesis of a general mechanism of SIRT1 gene activation upon DNA damage that involves functional activation of APE1. These findings are in line with data from Yamamori et al. (2010) showing that genotoxic insult stimulates SIRT1 expression and therefore its deacetylase activity on APE1 K6/7, favoring APE1 binding to XRCC1. Of interest, these authors sowed that a decrease of APE1 acetylation at later times after oxidative treatment is usually accompanied by SIRT1 up-regulation. Together these findings are in accordance with a model of a positive autoregulatory loop between the two proteins. Thus SIRT1 seems to be involved in a feedback mechanism that shuts off the cellular response mediated by APE1 acetylation (Yamamori et al., 2010). Further proof of APE1 and SIRT1 collaboration comes from the recent demonstration that APE1 stimulates SIRT1 activity in endothelial cells by reducing thiol moieties of cysteine residues in SIRT1, thus protecting endothelium from oxidative stress (Jung et al., 2013).
It has been suggested that DNA oxidation could trigger positive transcription in the context of Myc-mediated transcription through the involvement of BER enzymes, including APE1 (Perillo et al., 2008; Gillespie et al., 2010). Similarly, we speculated that APE1’s positive effect on SIRT1 transcription might depend on APE1 endonuclease activity over the nCaRE-B elements present within the SIRT1 promoter. We propose a model in which oxidative-mediated DNA repair and gene transcription are linked (Figure 6). During oxidative stress, DNA oxidation determines the formation of 8-oxodG lesions, which are recognized and processed by enzymes of the BER pathway, including APE1. In our model, the oxidative burst is an early event, essential for formation of a productive transcription initiation complex, which relies on initial recruitment of BER enzymes. The nicks introduced at the chromatin level by APE1 during 8-oxodG removal might promote the local relaxation required for formation of chromatin loops moving the active form of RNA polymerase II closer to the TSS of the gene, as previously recruited by APE1 on the nCaRE-B sequence, turning on the transcription. Our data therefore can be generalized to a regulatory model for all genes that contain nCaRE-B elements. Accordingly, a new hypothesis can be proposed for the molecular activation of specific genes during early response to DNA damage. This model links DNA-repair enzymes and transcriptional regulation effectors and may constitute a general model for explaining the adaptive cell response to oxidative stress involving gene regulation and DNA damage in which APE1 plays a central role.
MATERIALS AND METHODS
Bioinformatic analysis
All human and mouse DNA sequences were retrieved from the Ensembl database, release 56 (www.ensembl.org/), by using a dedicated program written in Perl that collects entries from this archive. A sequence window was selected that contains 5′ genomic DNA of every gene coding for a protein. This region extends from 6000 base pairs upstream and 1000 base pairs downstream of each transcriptional start site. Gene Ontology (GO) annotations were obtained from the Ensembl database by using the data-mining tool BioMart (www.ensembl.org/; Spudich et al., 2007). Human and mouse promoter regions were scanned for significant similarities to nCaRE-B by using Glsearch as program for local alignment (available in the Fasta3 program package; Pearson, 2000). Glsearch was chosen because it calculates an alignment that is global in the query and local in the library. The following nCaRE-B sequences were used as query (Izumi et al., 1996):
| Name | Sequence (5′ to 3′) |
|---|---|
| nCaRE-B (PTH) | TTTTTGAGACAGGGTCTCACTCTG |
| nCaRE-B1 (APE1) | TTTTGAGACAGTCTCAGCTCTG |
| nCaRE-B2 (APE1) | TTTTGAGACAGAGTTTCACTCTTG |
Alignments were computed with Altschul and Gish's statistical estimates, which are more suitable for searching short query sequences (–z 3 option; Altschul, 1991). We selected only those promoter genes that showed one or more matches for nCaRE-B sequences, allowing up to two mismatches in the case of the human genome and up to three mismatches in the case of the mouse genome; in fact, in the latter case, most alignments were found with three mismatches. From mouse promoter genes that contained nCaRE-B elements, we retrieved only orthologous human/mouse genes, as obtained from the BioMart Ensembl database. For the microarray filter, we cross-checked human genes selected from alignment search with microarray data obtained from the gene expression profile of HeLa cells silenced for APE1 by RNA interference (Vascotto et al., 2009a). The GO filter identified coregulated human genes, as determined by microarray analysis, studying the prevalence of their GO annotation terms. This analysis was obtained by using a Perl program kindly provided by Corà et al. (2004), which performs an exact Fisher's test based on hypergeometric distribution to determine whether the term appears in the set significantly more often than what is expected by chance. This program uses four different entries: 1) a file containing the whole GO database structure (OBO version 1.2; www.geneontology.org/); 2) the list of genes from the whole human genome, 4) a list of all genes with all the GO terms associated with them (as obtained from Ensembl), and 4) the set of genes to be tested. In general, a GO annotation term was considered to be significantly overexpressed when the corresponding p value (not corrected for multiple testing) was <1E-4. Phylogenetic footprinting analysis consisted of the last selection from significant data obtained from the GO filter of that gene also present in the mouse orthologous data set.
Gene annotation co-occurrence analysis
Gene identifiers corresponding to the list of 57 putative genes regulated by APE1 were submitted to GeneCodis (http://genecodis.cnb.csic.es/), a Web-based tool for ontological analysis (Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009; Tabas-Madrid et al., 2012), selecting Homo sapiens as the source for annotations and Biological Process as the GO category to perform the gene annotation cooccurrence analysis.
Cell culture and transient transfection experiments
HeLa cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Euroclone, Milan, Italy), 100 U/ml penicillin, and 10 μg/ml streptomycin sulfate. One day before transfection, cells were seeded in 10-cm plates at a density of 3 × 106 cells/plate. Cells were then transiently transfected with plasmids of interest by using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's instructions. Cells were harvested 48 h after transfection.
Inducible APE1 knockdown and generation of APE1 knock-in cell lines
Inducible silencing of endogenous APE1 and reconstitution with mutant proteins in HeLa cell clones was performed as described (Vascotto et al., 2009a, b). For inducible shRNA experiments, doxycycline (1 μg/ml) (Sigma-Aldrich, St. Louis, MO) was added to the cell culture medium, and cells were grown for 10 d.
Plasmids and expression of recombinant proteins
Plasmid containing the human SIRT1 promoter was kindly provided by K. Irani, University of Pittsburgh (Pittsburgh, PA). This plasmid consists of a fragment of the human SIRT1 promoter (–1266 to +137 relative to transcription start site) cloned into the pGL4.1 firefly luciferase reporter vector (Promega, Madison, WI; Yamamori et al., 2010). The human SIRT1 promoter carrying the mutation at nCaRE-B sequences was generated with a Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA), using the primers SIRT1-B mut, forward, 5′-TCATCTAGGTTTTATTTATATATTTTTTTGCTAAGGAGCGTCGCTCTTGCTGCCCAGGCTGGTGTG-3′, and SIRT1-B mut, reverse, 5′-CACACCAGCCTGGGCAGCAAGAGCGACGCTCCTTAGCAAAAAAATATATAAATAAAACCTAGATGA-3′.
Expression and purification of recombinant proteins from E. coli were performed as previously described (Vascotto et al., 2009b; Fantini et al., 2010).
Antibodies and Western blot analysis
For Western blot analyses, the indicated amounts of cell extracts were resolved in 10% SDS–PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell, BioScience, Dassel, Germany). Membranes were blocked with 5% (wt/vol) nonfat dry milk in phosphate-buffered saline containing 0.1% (vol/vol) Tween 20 and probed with monoclonal anti-FLAG antibody (Sigma), monoclonal anti-APE1 antibody (Vascotto et al., 2009a), monoclonal anti-Ku70 (sc-12729; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti–RNA polymerase II (Abcam, Cambridge, MA), monoclonal anti-SIRT1 (Abcam), monoclonal anti-p32 (Santa Cruz Biotechnology), and polyclonal anti-p53(acetyl K382) (Abcam). Blots were developed by using the enhanced chemiluminescence procedure (GE Healthcare, Piscataway, NJ) or Western Lightning Ultra (Perkin Elmer, Waltham, MA). Data normalization was performed by using a monoclonal anti-tubulin antibody (Sigma). Blots were quantified by using a Chemidoc XRS video densitometer (Bio-Rad, Hercules, CA).
Alignments and secondary structure predictions
Multiple alignments were performed using CLUSTALW2 (www.ebi.ac.uk/Tools/msa/clustalw2/).
Potential secondary structures for SIRT1-B nCaRE-B oligonucleotide were determined by using the mfold Web Server program (http://mfold.rna.albany.edu/?q=mfold). Structure predictions were run by setting the program parameters as close as possible to the conditions used in binding assays (37°C and 50 mM monovalent cation).
Chromatin immunoprecipitation analysis
ChIP assay was performed by using a protocol described previously (Lirussi et al., 2012) on HeLa cells cotransfected with hSIRT1 promoter and APE1-FLAG–tagged expressing vectors.
Preparation of nuclear cell extracts
Nuclear protein extracts were prepared as described earlier (Ziel et al., 2005).
Electrophoretic mobility shift assay analysis
APE1 binding to nucleic acids was assessed as already described (Fantini et al., 2010), with some modifications. Briefly, the indicated amount of recombinant proteins or 5 μg of the reported nuclear extract was incubated at 37°C for 15 min with 250 pmol of unlabeled poly(dT) or 250 ng of sonicated salmon sperm DNA (Sigma). A 2.5-pmol amount of 32P-labeled, ds oligonucleotides was then added, incubated for additional 15 min, and further separated onto a native 6% wt/vol polyacrylamide gel at 150 V for 4 h. When performing supershift assays, 5 μl of monoclonal anti-APE1 (Vascotto et al., 2009a), anti-Ku-70 (sc-12729; Santa Cruz Biotechnology) or anti-P2Y6 (Alomone Labs, Jerusalem, Israel) was preincubated with HeLa nuclear extract from APE1SCR-1 clone at 4°C for 3 h.
Oligonucleotides used for EMSA were the following:
| Name | Sequence (5′ to 3′) |
|---|---|
| nCaRE-B SIRT1-A | Forward TTTTTGAGACAGAGTTTCACTCTTG |
| Reverse CAAGAGTGAAACTCTGTCTCAAAAA | |
| nCaRE-B SIRT1-B | Forward TTTTTGAGACGGAGTTTCGCTCTTG |
| Reverse CAAGAGCGAAACTCCGTCTCAAAAA | |
| Poly(dT) | Forward TTTTTTTTTTTTTTTTTTTTTTTT |
T7 endonuclease I footprinting
Footprinting analysis on the SIRT1 nCaRE-B sequence was conducted using T7 endonuclease I. Briefly, 5′-32P-end-labeled nCaRE-B SIRT1-B was digested with 5 U of T7 endonuclease I at 37°C for 1 h and then incubated with 35 pmol of APE1 recombinant protein for 15 min at 37°C. The reaction mixtures were then loaded and separated for 2 h onto a denaturing 8 M urea sequencing gel. After separation, the gel was incubated for 30 min in a 10% methanol and 10% acetic acid solution and then wrapped in Saran wrap and exposed to film for autoradiography.
Determination of AP endonuclease activity
Determination of APE1 AP endonuclease activity was performed using an oligonucleotide cleavage assay, as described previously (Vascotto et al., 2009b). The indicated amount of recombinant APE1 protein was incubated with a 5′-32P-end-labeled 26-mer ds oligonucleotide containing a single THF artificial AP site at position 14, which is cleaved to a 14-mer in the presence of AP endonuclease activity. Alternatively, a 5′-32P-end-labeled ds SIRT1-B nCaRE-B oligonucleotide or a 5′-32P-end-labeled ds nCaRE SIRT1-B oligonucleotide, bearing a single THF residue at position 12 (bold) 5′-TTTTTGAGACGGAGTTTCGCTCTTG-3′ (Integrated DNA Technologies, Munich, Germany), was used. The conversion of the radiolabeled THF-containing oligonucleotide substrate (S) to the shorter product (P) was evaluated on a denaturing 20% polyacrylamide gel.
Reporter assays
For reporter assay experiments, we used a human SIRT1 promoter plasmid allowing for promoter activity measurements upon luciferase assay, as already described (Yamamori et al., 2010). To this purpose, 2.5 × 104 HeLa cells were seeded in a 96-well plate and cotransfected with 15 ng of human SIRT1 promoter, 0.3 ng of a constitutive Renilla reporter plasmid, and 75 ng of a vector expressing FLAG-tagged APE1 protein. When performing luciferase assays upon H2O2, cells were challenged with increasing amounts of H2O2 in serum-free medium for 1 h at 37°C, and then firefly and Renilla luciferase activities were measured 24 h after the treatment by using the Dual-Glo Luciferase assay system (Promega), according to manufacturer's recommendations. Firefly activity was normalized to Renilla activity to correct for differences in transfection efficiency. Results are from triplicate experiments.
Quantitative PCR
Total RNA from cell lines was extracted with the SV Total RNA isolation System kit (Promega). One microgram of total RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer's instructions. Quantitative reverse-transcription PCR was performed with a CFX96 Real-Time System (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). Primer sequences for human SIRT1 were those reported in Yamamori et al. (2010). Human glyceraldehyde-3-phosphate dehydrogenase was used as internal control; sense, 5′-CCCTTCATTGACCTCAACTACATG-3′; antisense, 5′-TGGGATTTCCATTGATGACAAGC-3′.
Surface plasmon resonance analysis
Real-time binding assays were performed on a Biacore 3000 SPR instrument (GE Healthcare). Biotinylated ds oligonucleotides were immobilized on an SA-chip at the desired level, as a result of their injection at a concentration of 500 nM in HBS (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 150 mM NaCl, 3.4 mM EDTA, and 0.005% [vol/vol] P20 surfactant, 0.1 mM Tris(2-carboxyethyl)phosphine) at 10 μl/min as flow rate. Flow cell 2 contained 100 RU of poly(dT); flow cells 3 and 4 contained 77 and 60 RU of SIRT1-B nCaRE-B and SIRT1-B nCaRE-B mutated, respectively; and flow cell 1 (with streptavidin) was left blank to be used as a reference surface. APE1 and its deletion mutant APE1NΔ33 were serially diluted in running buffer to the indicated concentrations and injected at a flow rate of 20 μl/min for 4.5 min at 20°C. Disruption of any complex that remained bound after a 3-min dissociation was achieved by using an injection of 1 M NaCl at 20 μl/min for 1 min. The BIAevaluation analysis package, version 4.1 (GE Healthcare), was used to subtract blank signal and evaluate kinetic and dissociation constants. Kinetic parameters were estimated assuming a 1:1 binding model and using Evaluation, version 7 4.1 (GE Healthcare). An affinity steady-state model was applied to fit the RUmax data versus protein concentrations, and fitting was performed with Prism, version 4.00 (GraphPad, San Diego, CA; Fantini et al., 2010).
Limited proteolysis
Suitable experimental conditions were chosen by testing proteolysis with different enzyme/substrate values; no preventive removal of DNA was performed. Thus limited proteolysis experiments on recombinant APE1 were conducted in 50 mM NH4HCO3, pH 7.5 (reaction buffer), at 37°C by using an enzyme-to-substrate ratio ranging from 1:500 to 1:5000 (wt/wt). Three identical aliquots of APE1 (500 pmol) were combined with reaction buffer or DNA nCaRE-B ds oligonucleotides (PTH or nCaRE SIRT1-B; 5:1 mol DNA/protein) dissolved in reaction buffer to generate samples (100-μl final volume each), which were incubated for 15 min at 37°C before protease addition. After digestion started, the extent of proteolysis was monitored on a time-course basis by sampling 10 μl of the mixture at time intervals ranging from 5 to 120 min. Reaction samples were immediately quenched with 5% formic acid and then frozen in dry ice before liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) analysis.
LC-ESI-MS analysis
APE1 digests were analyzed with a Q-TOF Premier mass spectrometer (Waters, Milford, MA) equipped with a nanospray source. Peptide mixtures were separated on an Atlantis C18 column (100 μm × 100 mm, 3 μm), using a linear gradient ranging from 30 to 60% acetonitrile in 1% formic acid, over a period of 50 min at a flow rate of 800 nl/min. Spectra were acquired in the m/z = 650–2500 range. Data were processed by using MassLynx software (Waters). Mass calibration was performed using multiply charged ions from horse heart myoglobin (Sigma). Depending on polypeptide size, mass values are reported as monoisotopic or average values. Observed mass values are assigned to specific polypeptides by using Paws software (Proteometrics, New York, NY), based on APE1 sequence and selectivity of the protease used for protein digestion.
Statistical analysis
Statistical analyses were performed by using the Excel (Microsoft, Redmond, WA) data analysis program for Student's t test analysis. p < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
We thank Mattia Poletto for helpful suggestions during recombinant protein purification and critical comments on the manuscript. This work was supported by grants from the Italian Association for Cancer Research (IG10269 and IG14038) and the Ministry of Education, Universities and Research, Italy (FIRB_RBRN07BMCT and PRIN2008_CCPKRP_003 to G.T.; FIRB_ RBNE08YFN3_003 and PRIN2008_CCPKRP_002 to A.S.). This project was funded under the Cross-Border Cooperation Program Italy-Slovenia 2007–2013 by the European Regional Development Fund and national funds.
Abbreviations used:
- 8-oxodG
8-oxodeoxyguanine
- APE1
apurinic apyrimidinic endonuclease 1
- BER
base excision repair
- GO
Gene Ontology
- LC-ESI-MS
liquid chromatography–electrospray ionization–mass spectrometry
- nCaRE
negative calcium responsive element
- PTH
parathyroid hormone
- SIRT1
sirtuin-1
- SPR
surface plasmon resonance
- THF
tetrahydrofuran
- zAPE1
zebrafish APE1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0286) on December 19, 2013.
REFERENCES
- Adelmant G, et al. DNA ends alter the molecular composition and localization of Ku multicomponent complexes. Mol Cell Proteomics. 2012;11:411–421. doi: 10.1074/mcp.M111.013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF. Amino acid substitution matrices from an information theoretic perspective. J Mol Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D, Novac O, Callejo M, Ruiz MT, Price GB, Zannis-Hadjopoulos M. 14-3-3sigma is a cruciform DNA binding protein and associates in vivo with origins of DNA replication. J Cell Biochem. 2002;87:194–207. doi: 10.1002/jcb.10294. [DOI] [PubMed] [Google Scholar]
- Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- Barnes T, Kim W, Mantha AK, Kim S, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beernink PT, Segelke BW, Hadi MZ, Erzberger JP, Wilson DM, 3rd, Rupp B. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J Mol Biol. 2001;307:1023–1034. doi: 10.1006/jmbi.2001.4529. [DOI] [PubMed] [Google Scholar]
- Berquist BR, McNeill DR, Wilson DM. Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J Mol Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Izumi T, Yang S, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Chattopadhyay R, Burnette BR, Cross JV, Mitra S, Ernst PB, Bhakat KK, Crowe SE. Acetylation of apurinic/apyrimidinic endonuclease-1 regulates Helicobacter pylori-mediated gastric epithelial cell apoptosis. Gastroenterology. 2009;136:2258–2269. doi: 10.1053/j.gastro.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a Web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Stubbe J. Bleomycins: towards better therapeutics. Nat Rev Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- Chung U, et al. The interaction between Ku antigen and REF1 protein mediates negative gene regulation by extracellular calcium. J Biol Chem. 1996;271:8593–8598. doi: 10.1074/jbc.271.15.8593. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Corà D, Cunto FD, Provero P, Silengo L, Caselle M. Computational identification of transcription factor binding sites by functional analysis of sets of genes sharing overrepresented upstream motifs. BMC Bioinformatics. 2004;5:57. doi: 10.1186/1471-2105-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham LA, Coté AG, Cam-Ozdemir C, Lewis SM. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol Cell Biol. 2003;23:8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déclais A, Fogg JM, Freeman ADJ, Coste F, Hadden JM, Phillips SEV, Lilley DMJ. The complex between a four-way DNA junction and T7 endonuclease I. EMBO J. 2003;22:1398–1409. doi: 10.1093/emboj/cdg132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Jolly DJ, Rubin CM, Friedmann T, Schmid CW. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981;151:17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Matsumoto Y, Wilson DM., 3rd Nucleotide sequence and DNA secondary structure, as well as replication protein A, modulate the single-stranded abasic endonuclease activity of APE1. J Biol Chem. 2006;281:3889–3898. doi: 10.1074/jbc.M511004200. [DOI] [PubMed] [Google Scholar]
- Fantini D, et al. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res. 2010;38:8239–8256. doi: 10.1093/nar/gkq691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie MN, Pastukh VM, Ruchko MV. Controlled DNA “damage” and repair in hypoxic signaling. Respir Physiol Neurobiol. 2010;174:244–251. doi: 10.1016/j.resp.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MA, Morera S, Rothwell DG, Fortelle EDL, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, de Cabo R. AsSIRTing the DNA damage response. Trends Cell Biol. 2008;18:77–83. doi: 10.1016/j.tcb.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RP, Adamson ED. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- Izumi T, Henner WD, Mitra S. Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry. 1996;35:14679–14683. doi: 10.1021/bi961995u. [DOI] [PubMed] [Google Scholar]
- Izumi T, Malecki J, Chaudhry MA, Weinfeld M, Hill JH, Lee JC, Mitra S. Intragenic suppression of an active site mutation in the human apurinic/apyrimidinic endonuclease. J Mol Biol. 1999;287:47–57. doi: 10.1006/jmbi.1999.2573. [DOI] [PubMed] [Google Scholar]
- Jung S, Kim C, Kim Y, Naqvi A, Yamamori T, Kumar S, Kumar A, Irani K. Redox factor-1 activates endothelial SIRTUIN1 through reduction of conserved cysteine sulfhydryls in its deacetylase domain. PLoS One. 2013;8:e65415. doi: 10.1371/journal.pone.0065415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Um S. SIRT1: roles in aging and cancer. BMB Rep. 2008;41:751–756. doi: 10.5483/bmbrep.2008.41.11.751. [DOI] [PubMed] [Google Scholar]
- Krayev AS, Kramerov DA, Skryabin KG, Ryskov AP, Bayev AA, Georgiev GP. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980;8:1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Kuninger DT, Izumi T, Papaconstantinou J, Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Inagaki H, Yamada K, Ohye T, Taniguchi M, Emanuel BS, Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lirussi L, et al. Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Mol Biol Cell. 2012;23:4079–4096. doi: 10.1091/mbc.E12-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHaffie GS, Ralston SH. Origin of a negative calcium response element in an ALU-repeat: implications for regulation of gene expression by extracellular calcium. Bone. 1995;17:11–14. doi: 10.1016/8756-3282(95)00131-v. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Terai Y, Okada N. Characterization of novel Alu- and tRNA-related SINEs from the tree shrew and evolutionary implications of their origins. Mol Biol Evol. 2002;19:1964–1972. doi: 10.1093/oxfordjournals.molbev.a004020. [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Ando K, Igarashi T, Ogata E, Fujita T. Conserved mechanism of negative gene regulation by extracellular calcium. Parathyroid hormone gene versus atrial natriuretic polypeptide gene. J Clin Invest. 1992;89:1268–1273. doi: 10.1172/JCI115711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Zajac JD, Igarashi T, Ogata E, Kronenberg HM. Negative regulatory elements in the human parathyroid hormone gene. J Biol Chem. 1991;266:21903–21910. [PubMed] [Google Scholar]
- Parkinson MJ, Lilley DM. The junction-resolving enzyme T7 endonuclease I: quaternary structure and interaction with DNA. J Mol Biol. 1997;270:169–178. doi: 10.1006/jmbi.1997.1128. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Ruiz MT, Price GB, Zannis-Hadjopoulos M. Cruciform DNA binding protein in HeLa cell extracts. Biochemistry. 1994;33:14185–14196. doi: 10.1021/bi00251a030. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Poletto M, Vascotto C, Scognamiglio PL, Lirussi L, Marasco D, Tell G. Role of the unstructured N-terminal domain of the human apurinic/apyrimidinic endonuclease 1 (hAPE1) in the modulation of its interaction with nucleic acids and nucleophosmin (NPM1) Biochem J. 2013;452:545–557. doi: 10.1042/BJ20121277. [DOI] [PubMed] [Google Scholar]
- Postow L. Destroying the ring: freeing DNA from Ku with ubiquitin. FEBS Lett. 2011;585:2876–2882. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzone G, Vitale RM, Scaloni A, Rossi M, Amodeo P, Guagliardi A. Structural characterization of the functional regions in the archaeal protein Sso7d. Proteins. 2007;67:189–197. doi: 10.1002/prot.21220. [DOI] [PubMed] [Google Scholar]
- Robson CN, Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaloni A, Miraglia N, Orrù S, Amodeo P, Motta A, Marino G, Pucci P. Topology of the calmodulin-melittin complex. J Mol Biol. 1998;277:945–958. doi: 10.1006/jmbi.1998.1629. [DOI] [PubMed] [Google Scholar]
- Scaloni A, Monti M, Acquaviva R, Tell G, Damante G, Formisano S, Pucci P. Topology of the thyroid transcription factor 1 homeodomain-DNA complex. Biochemistry. 1999;38:64–72. doi: 10.1021/bi981300k. [DOI] [PubMed] [Google Scholar]
- Shankar R, Grover D, Brahmachari SK, Mukerji M. Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol Biol. 2004;4:4–37. doi: 10.1186/1471-2148-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Hsieh P, Grigoriev M, Potaman VN, Sinden RR, Lyubchenko YL. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J Mol Biol. 2000;296:1169–1173. doi: 10.1006/jmbi.2000.3542. [DOI] [PubMed] [Google Scholar]
- Spudich G, Fernández-Suárez XM, Birney E. Genome browsing with Ensembl: a practical overview. Brief Funct Genomic Proteomic. 2007;6:202–219. doi: 10.1093/bfgp/elm025. [DOI] [PubMed] [Google Scholar]
- Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010a;67:3589–3608. doi: 10.1007/s00018-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Pellizzari L, Cimarosti D, Pucillo C, Damante G. Ref-1 controls pax-8 DNA-binding activity. Biochem Biophys Res Commun. 1998;252:178–183. doi: 10.1006/bbrc.1998.9548. [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Wilson DM, 3rd, Lee CH. Intrusion of a DNA repair protein in the RNome world: is this the beginning of a new era. Mol Cell Biol. 2010b;30:366–371. doi: 10.1128/MCB.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vascotto C, et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics. 2009a;9:1058–1074. doi: 10.1002/pmic.200800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vascotto C, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009b;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol Life Sci. 2010;67:3621–3631. doi: 10.1007/s00018-010-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung S, Kim C, Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Igarashi T, Muramatsu M, Fukagawa M, Motokura T, Ogata E. Hypocalcemia increases and hypercalcemia decreases the steady-state level of parathyroid hormone messenger RNA in the rat. J Clin Invest. 1989;83:1053–1056. doi: 10.1172/JCI113946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: impact on DNA integrity and gene expression. FASEB J. 2005;19:387–394. doi: 10.1096/fj.04-2805com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.