Abstract
MicroRNAs, or miRNAs, are endogenously encoded small RNAs that play a key role in diverse plant biological processes. Jatropha curcas L. has received significant attention as a potential oilseed crop for the production of renewable oil. Here, a sRNA library of mature seeds and three mRNA libraries from three different seed development stages were generated by deep sequencing to identify and characterize the miRNAs and pre-miRNAs of J. curcas. Computational analysis was used for the identification of 180 conserved miRNAs and 41 precursors (pre-miRNAs) as well as 16 novel pre-miRNAs. The predicted miRNA target genes are involved in a broad range of physiological functions, including cellular structure, nuclear function, translation, transport, hormone synthesis, defense, and lipid metabolism. Some pre-miRNA and miRNA targets vary in abundance between the three stages of seed development. A search for sequences that produce siRNA was performed, and the results indicated that J. curcas siRNAs play a role in nuclear functions, transport, catalytic processes and disease resistance. This study presents the first large scale identification of J. curcas miRNAs and their targets in mature seeds based on deep sequencing, and it contributes to a functional understanding of these miRNAs.
Introduction
Increased energy consumption is a key concern of contemporary society as a result of rapidly diminishing fossil fuel deposits and increasing levels of carbon dioxide released into the atmosphere. Thus, environmentally friendly sources of fuels, such as bioethanol and biodiesel, are promising alternatives for fossil fuels. In this context, great interest has been generated regarding the potential of Jatropha curcas L. for biodiesel production. This species belongs to the Euphorbiaceae family and is found in almost all tropical areas; it occurs on a large scale in tropical and temperate regions [1], [2]. J. curcas has potential for biodiesel production because it is perennial, drought-resistant and has a high oil content (∼40%). Additionally, this crop can be grown in degraded soils (non-agricultural lands), which controls erosion without competing for food production habitats [3], [4].
In spite of having the potential for high fuel production, improving the quality of J. curcas seed oil remains challenging. The desired traits include increased oil content, decreased unsaturated fatty acid content (to increase the oxidative stability), decreased free fatty acid content (to prevent soap formation and increase biodiesel productivity) and decreased 18 carbon fatty acid content (to reduce viscosity) [5]. Furthermore, reducing seed toxicity and increasing pest tolerance are also desirable [1], [5].
MicroRNAs (miRNAs) are small non-coding RNAs that act as post-transcriptional regulators of gene expression [6]. They are typically transcribed by RNA Polymerase II as long polyadenylated transcripts, with an imperfect stem-loop structure known as pri-miRNA, which is recognized and processed by DICER-Like1 (DCL1) into an miRNA precursor (pre-miRNA). This sequence is further processed to generate mature miRNAs, which are composed of single-stranded RNA molecules of approximately 21 nucleotides (nt) in length [7]–[9]. To regulate protein-coding genes, the mature miRNA binds to sites in the 3′ and 5′ untranslated regions (UTR) or the coding sequence (CDS), leading to mRNA degradation or translational inhibition, depending on the degree of complementarity between the miRNA and its target transcript [8], [10]. This regulation plays critical roles in plant development and growth as well as a range of physiological processes, including abiotic and biotic stress responses [11], [12], and in lipid metabolism [13], [14].
At present, there have only been two recent studies regarding the identification of miRNAs from J. curcas. One study used a small RNA cloning methodology and did not provide the precursor sequence from Jatropha [15]. The other study used known plant miRNAs from Viridiplantae to search for J. curcas miRNAs in publicly available EST and GSS databases; therefore, only conserved miRNAs were identified by this approach [16]. In the present study, conserved and novel miRNAs from J. curcas were identified through the deep sequencing of small RNAs (sRNA) from mature seeds. The targets of these miRNAs were predicted in silico, and the results indicate that miRNAs are involved in a wide range of physiological processes in seeds, including growth, hormone signaling, stress resistance and lipid metabolism, among others. In addition, polyadenylated transcript sequencing (mRNA-seq) libraries from three seed development stages (immature, intermediate and mature) were used to identify and characterize the abundance of pre-miRNAs and miRNA targets, providing important information related to regulatory timing.
Material and Methods
J. curcas Seed Collection and RNA Isolation
For the RNA isolation, fruits from J. curcas plants grown in an open environment at Embrapa Clima Temperado (Pelotas, RS, Brazil) were collected at 10–20 (immature seeds), 20–40 (intermediate seeds) and 40–60 (mature seeds) days after flower opening (DAF). The seeds were dissected from their fruits and immediately frozen in liquid nitrogen and then stored at −80°C. Total RNA was isolated from a pool of seeds from each stage with Trizol (Invitrogen, CA, USA), according to the manufacturer’s protocol. RNA quality was evaluated by electrophoresis on a 1% agarose gel, and the RNA concentration was determined by Nanodrop (Nanodrop Technologies, Wilmington, DE, USA).
Small RNA and mRNA-seq Library Construction using Deep Sequencing
Total RNA (>10 µg) was isolated from immature, intermediate and mature seeds and sent to Fasteris SA (Plan-les-Ouates, Switzerland) for processing and sequencing with an Illumina HiSeq2000. To generate the mRNA-seq libraries, polyadenylated transcript sequencing was performed as follows: Poly(A) mRNAs were purified, cDNA was synthesized using a Poly(T) primer shotgun to generate 500 nt inserts, 3p and 5p adapters were bound, and a cDNA colony template library was generated by PCR amplification, and 100 single-end bases were sequenced by Illumina sequencing. Illumina sequencing output data were sequence tags of 100 bases. Three mRNA-seq libraries were constructed from immature seeds (L1), from intermediate seeds (L2) and from mature seeds (L3).
Only RNA from mature seeds was used to produce the small RNA library. In brief, RNA bands corresponding to a size range of 20–30 nt were separated and purified from the acrylamide gel and subsequently bound to 3p and 5p adapters in two separate subsequent steps, each followed by acrylamide gel purification. Then, the cDNAs were synthesized and a cDNA colony template library was generated by PCR amplification for Illumina sequencing.
Sequence Data Analysis
Figure S1 summarizes the overall data analyses performed with the sRNA library and mRNA-seq libraries. First, all low quality reads (FASTq value <13) were removed, and 5p and 3p adapter sequences were trimmed using the Genome Analyzer Pipeline (Fasteris). The remaining low quality reads with ‘n’ were removed using the PrinSeq script [17]. Sequences shorter than 18 nt and longer than 25 nt were excluded from further analysis. sRNAs derived from Viridiplantae rRNAs, tRNAs, snRNAs and snoRNAs (from the tRNAdb [18]; SILVA rRNA [19] and NONCODE v3.0 [20] databases); cpRNA from J. curcas; and mtRNA from Ricinus communis [from the NCBI GenBank database (http://ftp.ncbi.nlm.nih.gov)] were identified by mapping with Bowtie v 0.12.7 [21]. rRNAs, tRNAs, snRNAs and snoRNAs were excluded from the sRNA library that was used in further miRNA predictions and analyses.
The mRNA-seq libraries were first filtered for low quality “n” reads, and J. curcas coding sequence reads were obtained from the Jatropha genome database (http://www.kazusa.or.jp/jatropha). L1, L2 and L3 were pooled to produce mRNA contigs using the CLC Genome Workbench version 4.0.2 (CLCbio, Aarhus, Denmark) algorithm for de novo sequence assembly, with the default parameters (similarity = 0.8, length fraction = 0.5, insertion/deletion cost = 3, mismatch cost = 3), originating from the L1–L2–L3 library.
Sequence data from this article can be found in the GenBank data libraries under accession number(s) GSM1226039 (L1), GSM1226040 (L2), GSM1226038 (L3) and GSM1226041 (sRNA dataset).
Prediction of Conserved and Novel miRNAs
To identify phylogenetically conserved miRNAs, reads from the sRNA library derived from mature seeds were mapped to a set of all mature Viridiplantae unique miRNAs obtained from the miRBase database (Release 19, August 2012) using Bowtie v 0.12.7 [21]. Only perfectly matched sequences were considered to be known miRNAs. To search for novel miRNAs, reads from the sRNA library derived from mature seeds were matched against contigs assembled from the L1–L2–L3 library using SOAP2 [22]. The SOAP2 output was filtered with an in house filter tool to separate pre-miRNA candidate sequences by using an anchoring pattern of one or two blocks of aligned sRNAs with a perfect match. MFOLD (http://mfold.bioinfo.rpi.edu/cgi-bin/rnaform1.cgi) was employed to predict hairpin structures with default parameters. Sequences were considered to be pre-miRNA if the RNA sequence could form an appropriate stem-loop structure with a mature miRNA sitting in one arm of the hairpin structure; the secondary structure had a high negative minimum folding free energy (MFE, 17–110 kcal/mol), using RNAstructure 5.3 [23], and a high negative minimum folding free energy index (MFEI, higher than 0.5). All putative pre-miRNAs were verified by a BLASTn algorithm from NCBI databases and the miRBase database (Release 19, August 2012). The frequency of identified miRNAs was obtained by aligning the conserved and novel precursors identified in this study and the sRNA library using Bowtie v 0.12.7, with the default parameters. The SAM files from Bowtie were then processed using in house Python scripts to count the frequencies of each read and map them into the three libraries. The most frequent miRNA for each precursor was designated as miRNA, while the others were designated as isomiRNAs.
miRNA Targets Prediction
mRNA contigs from the L1–L2–L3 library were clustered by using Gene Indices clustering tools (http://compbio.dfci.harvard.edu/tgi/software/) [24] to reduce sequence redundancy. The clustering output was passed on to a CAP3 assembler [25] for multiple alignment and consensus building. Contigs that could not reach the threshold set and fell into a random assembly and remained as a list of singletons.
The predicted target genes of conserved and novel miRNAs was performed with a psRNAtarget [26] by aligning mature sequences against assembled J. curcas unigenes from the L1–L2–L3 library. Default parameters were used, and a maximum expectation of 4.0 was applied for the search with the most abundant miRNA. A maximum expectation of 5.0 was used to search the isomiRNA target genes, which were related to lipid metabolism pathways. An annotation of predicted targets was performed by using BLASTX from Blast2GO v2.3.5 software [27] based on their sequence similarity with previously identified and annotated genes from the NR and Swiss-Prot/Uniprot protein databases. The annotation was improved by analyzing conserved domains/families using the InterProScan tool, and Gene Ontology (GO) terms for the cellular component, molecular function and biological processes were determined by using the GOslim tool in the blast2GO software. Transcript orientations were obtained from the BLAST outputs.
In silico Expression Analysis of Pre-miRNAs and miRNA Predicted Targets
To calculate the frequency of pre-miRNAs, mRNA reads from each individual library (L1, L2 and L3) were aligned in Bowtie v 0.12.7 by using the default parameters and allowing zero mismatches. For reference, all identified pre-miRNAs in this study were used. The SAM files from Bowtie were then processed, as discussed above. The scaling normalization method proposed by Robinson and Oshlack [28] was used for data normalization. Both R packages, EdgeR [29] and A–C test [30], were independently used to assess whether the pre-miRNA was differentially represented. In brief, EdgeR uses a negative binomial model to estimate the over dispersion from the pre-miRNA count. The dispersion parameter of each pre-miRNA was estimated by tagwise dispersion. The differential expression is then assessed for each pre-miRNA by using an adapted exact test for over dispersed data. The A-C test computes the probability that two independent counts of the same pre-miRNA came from similar samples. Pre-miRNAs were considered to be differentially represented if they had a p-value ≤0.001 in both statistical tests. The same method was adopted to evaluate the expression profile of predicted miRNAs targets, allowing two mismatches (one in the seed and another in the rest of the sequence).
siRNA Prediction
siRNAs were identified by aligning J. curcas 24-nt sRNAs against the contigs from the L1–L2–L3 library. Putative contigs with a typical sRNA distribution pattern along the matching sequences [31] were further subjected to annotation using Blast2GO software, as described above.
Results and Discussion
Deep Sequencing of sRNAs and mRNA Libraries
To identify the conserved and novel miRNAs in J. curcas seeds, an sRNA library from mature seeds was constructed and sequenced by Illumina technology, resulting in a total of 16,771,931 reads. After removing the 3p and 5p adapter sequences and filtering out low quality “n” sequences, sRNAs within a 1–44 nt range were obtained, in which the majority were 18–26 nt in length (Table 1). Sequences shorter than 18 nt and longer than 25 nt were removed, resulting in 13,953,403 reads (Table 2), from which 5,400,278 reads were unique tags (38.7%). Approximately 80% (4,328,139) of the unique tags corresponded to singletons (Table S1).
Table 1. Raw data from sequencing Jatropha curcas sRNAs.
| Read length (nt) | Number of reads | Percentage (%) of reads |
| 0 | 12,163 | 0.073 |
| 1–17 | 1,347,333 | 8.330 |
| 18–26 | 14,175,504 | 84.190 |
| 27–44 | 516,355 | 3.790 |
| Remaining | 720,576 | 4.960 |
Table 2. Data from reads of the sRNA database of mature Jatropha curcas seeds filtered with non-coding RNAs.
| Size | Total reads* | % | rRNA | tRNA | snRNA | snoRNA | cpRNA | mtRNA | all filters | |||||||
| totalreads | % | totalreads | % | totalreads | % | totalreads | % | totalreads | % | totalreads | % | totalreads | % | |||
| 18 | 369,493 | 2.65 | 132,827 | 35.95 | 9,763 | 2.64 | 283 | 0.08 | 167 | 0.05 | 6,069 | 1.64 | 2,269 | 0.61 | 151,378 | 40.97 |
| 19 | 445,999 | 3.20 | 131,575 | 29.50 | 22,091 | 4.95 | 241 | 0.05 | 130 | 0.03 | 10,314 | 2.31 | 2,043 | 0.46 | 166,394 | 37.31 |
| 20 | 503,781 | 3.61 | 107,113 | 21.26 | 14,525 | 2.88 | 240 | 0.05 | 102 | 0.02 | 27,221 | 5.40 | 2,534 | 0.50 | 151,735 | 30.12 |
| 21 | 2,869,361 | 20.56 | 164,287 | 5.73 | 5,755 | 0.20 | 959 | 0.03 | 78 | 0.00 | 51,618 | 1.80 | 14,261 | 0.50 | 236,958 | 8.26 |
| 22 | 2,271,978 | 16.28 | 117,565 | 5.17 | 7,860 | 0.35 | 374 | 0.02 | 61 | 0.00 | 106,135 | 4.67 | 3,714 | 0.16 | 235,709 | 10.37 |
| 23 | 1,335,058 | 9.57 | 125,303 | 9.39 | 7,860 | 0.59 | 257 | 0.02 | 33 | 0.00 | 23,003 | 1.72 | 2,279 | 0.02 | 158,735 | 11.89 |
| 24 | 5,760,030 | 41.28 | 102,477 | 1.78 | 9,347 | 0.16 | 356 | 0.01 | 27 | 0.00 | 31,370 | 0.54 | 3,447 | 0.06 | 147,024 | 2.55 |
| 25 | 397,703 | 2.85 | 87,646 | 22.04 | 6,541 | 1.64 | 178 | 0.04 | 17 | 0.00 | 8,911 | 2.24 | 4,192 | 1.05 | 107,485 | 27.03 |
| Total | 13,953,403 | 100.00 | 968,793 | 6.94 | 83,742 | 0.60 | 2,888 | 0.02 | 615 | 0.00 | 264,641 | 1.90 | 34,739 | 0.25 | 1,355,418 | 9.71 |
*Total reads before filtering with non-coding and organellar small RNAs. The small RNAs were clustered according to their origin as follows: ribosome (rRNA), transporter (tRNA), small nuclear (snRNA), small nucleolar (snoRNA), mitochondrial (mtRNA) and chloroplastic (cpRNA).
Non-coding RNAs were also removed from the sRNA library for further analysis (Table 2). The sequence analysis showed that rRNA had the highest read frequency of all filtered sRNA classes, with 6.94% of the total reads. The majority of these rRNA sequences were found in the dataset with 21 nt-long sequences. Interestingly, 35.95% of the 18 nt sequences represented rRNAs. cpRNA sequences were the second most abundant filtered sequences after those from rRNA, corresponding to 1.9% of the total reads. tRNAs, mtRNAs, snRNAs and snoRNA were less frequent in the sRNA library. Taken together, 9.71% of the sRNA library was filtered with these RNA types, leaving 12,597,985 reads.
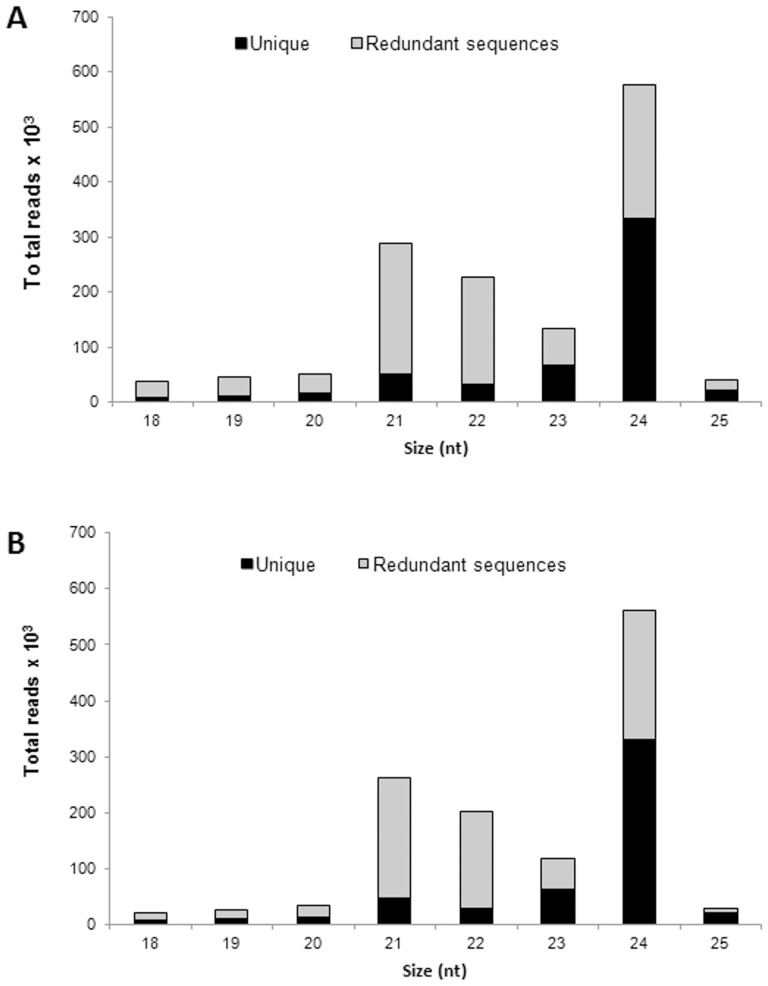
The length distribution of redundant and non-redundant sRNAs reads indicated that the most abundant and diverse sequences are within 21 (20.89%) and 24 nt (44.55%), a typical size range for Dicer-like (DCL)-derived products [9]. This distribution pattern for the small RNA size is similar to that of seeds from other species, such as Arabidopsis [32], peanut [13], barley [33], soybean [34] and canola [14], which implies that J. curcas possesses similar small RNA biogenesis processing components to other plant species. The same length distribution pattern was observed before and after filtering the sRNA library (Figure 1), indicating that the small RNA library was of high quality.
Figure 1. Total number of redundant and unique reads in the sRNA library of J. curcas mature seeds.
(A) Data before filtering with non-coding RNAs and organelle RNAs. (B) Data after filtering with non-coding RNAs and organelle RNAs.
In this study, a specific mRNA transcriptome of J. curcas seeds was produced and used as a sequence reference for further analyses. Three mRNA seqs were obtained from seeds as follows: the L1 (immature seed) library yielded 43,328,830 reads, the L2 (intermediate mature) library yielded 35,062,185 reads, and the L3 (mature seed) library yielded 16,653,188 reads. All three libraries were pooled for de novo assembly (L1–L2–L3). The resulting 61,863 contigs had an average length of 755 bp, and the size ranged between 100 bp and 15,706 bp, with 27,520 contigs constituting more than 500 bp in length.
Identification of Conserved miRNAs and Pre-miRNAs
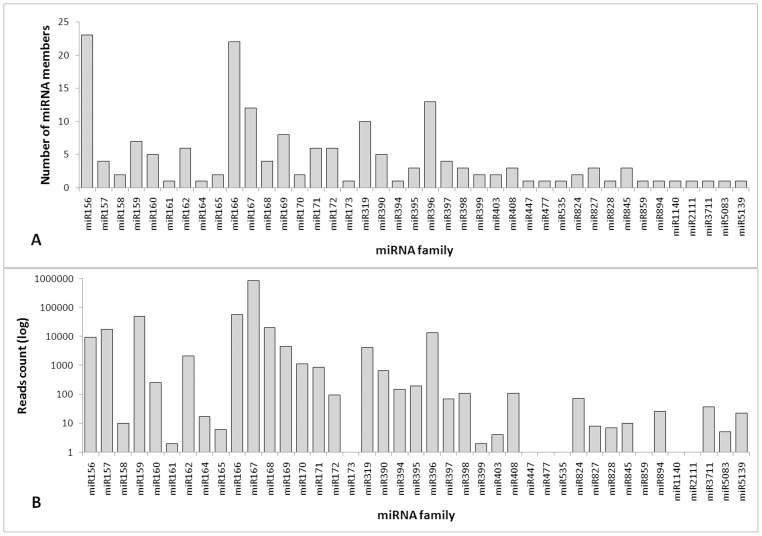
Several studies have reported miRNA conservation across different plant taxa [35], [36]. To identify homologous miRNAs in the J. curcas sRNA library, a set of 2,585 unique mature plant miRNA sequences were extracted from the miRBase database and used for alignment against the sRNA library. Only sequences with exactly the same size and nucleotide composition were considered. A strict criterion of sharply defined distribution patterns for one or two block-like anchored sRNAs and at least 10 reads of a single miRNA sequence were used to predict novel miRNAs (see methods). The read depth distribution along putative pre-miRNAs was previously shown to be a reliable guide for differentiating possible miRNAs from contaminant sequences, such as the degradation products of mRNAs or transcripts that are simultaneously expressed in both sense and antisense orientations [14], [33], [37]. In total, 1,021,895 reads perfectly matched 177 conserved miRNAs belonging to 41 families, with an average of approximately 4 miRNA members per family (Table S2). Overall, the Jcu_MIR167 family was the most abundant conserved miRNA family present in J. curcas seeds, accounting for 842,066 reads, and the largest families were Jcu_MIR156 and Jcu_MIR166, with 23 and 21 members, respectively. Of the remaining miRNA families, 19 contained between 2 to 8 members, and 16 were represented by a single member (Figure 2 and Table S2). Furthermore, the results indicate that different members of the same miRNA family have clearly different expression levels (Table S2). For example, Jcu_MIR166 presents members ranging from 1 to 35,439 reads. Interestingly, the conserved miRNAs represented the most abundant J. curcas miRNAs and were distributed throughout seven families (MIR156, MIR157, MIR159, MIR166, MIR167, MIR168 and MIR396). These abundant miRNA families are largely found in Viridiplantae, indicating a fundamental role in plant life maintenance (Table S2).
Figure 2. Known miRNA families identified in mature J. curcas seeds.
(A) The total number of miRNA members (isomiRNAs) from each miRNA family. (B) The number of total read counts of each miRNA family.
In a study performed by Wang et al. [15], six conserved miRNAs were identified by the cloning approach from RNA libraries derived from theleaves and developing seeds of J. curcas. Two (Jcu_MIR166_3p and Jcu_MIR167_5p) of them were also identified in the present study, and three conserved miRNAs were identified only in the library from leaves. Moreover, the miRNA JcumiR004, which was identified as a novel plant miRNA, according to Wang et al. [15], was annotated in the present study as Jcu_MIR171_5p because the mature sequence showed a perfect match with the MIR171 from several species (Table S2, sequence UGAUUGAGCCGUGCCAAUAUC). Therefore, the discrepancies in identifying conserved miRNAs from the present study and the one performed by Wang et al. [15] correspond mostly to a difference in selected tissues and methods. During the cloning approach, there was a chance miRNAs with low expression level would not be detected. A more recent work by Vishwakarma and Jadeja [16] also focused on the identification of conserved miRNAs from J. curcas after transcript and partial genome sequence analysis. These authors were able to identify 24 predicted miRNAs belonging to five miRNA families (Jcu_MIR166, Jcu_MIR167, Jcu_MIR1096, Jcu_MIR5368 and Jcu_MIR5021). A lower number of miRNA families were identified by these authors relative to the present study, most likely because they used known plant miRNAs from Viridiplantae to search the conserved J. curcas miRNAs homologs in publicly available (and relatively small) EST and GSS databases compared to the database from the RNAseq generated in the present study.
To identify putative conserved pre-miRNA sequences, the sRNA library was matched against a set of de novo assembled contigs from three developmental stages of J. curcas seeds (L1–L2–L3 library). The candidate pre-miRNAs were predicted by exploring the secondary structure, the minimum folding free energy (MFE) and the minimum folding free energy index (MFEI). Candidate mRNA sequences with a stem-loop hairpin structure showing MFE values of 40–100 kcal/mol, MFEI values higher than 0.85 and more than 10 miRNA reads anchored in the same orientation in the 5p and/or 3p arm in a two block-like pattern were considered putative pre-miRNAs. The precursor identity was determined by BLAST searches against mature miRNAs in miRBase and the NCBI database. As a result, 41 known full-length plant pre-miRNA sequences were identified along with 18 miRNAs anchored in the 3p-arm and 19 miRNAs in the 5p-arm (Figure S2). The precursors had an average length of 154 bp, a CG content of 43.45%, an MFE of −53.13 and an MFEI of −0.86 (Table S3), which were similar to the pre-miRNA characteristics in other plant species [14], [35], [36]. Twenty conserved pre-miRNAs did not generate a hairpin structure according to MFOLD (http://mfold.bioinfo.rpi.edu/cgi-bin/rnaform1.cgi).
At the present, there is no miRNA sequence for J. curcas available in miRNA databases, and there is only one recently published report regarding the identification of miRNAs through the cloning of sRNA sequences, which did not provide the precursor sequence [15]. In this context, the use of deep sequencing technologies represents a powerful large scale approach for the reliable identification of miRNAs in J. curcas [14], [32], [33], [38], [39]. In the present study, the deep sequencing of an sRNA library from J. curcas mature seeds and three mRNAseq libraries from three stages of seed development allowed for the identification of conserved and species-specific miRNAs and pre-miRNAs.
Identification of Novel miRNAs and Pre-miRNAs
In addition to conserved miRNAs, 16 sequences with characteristic hairpin-like structures were BLASTed against miRBase and NCBI databases, and no homologies with previously known plant miRNAs were found; these sequences characterize novel pre-miRNAs in plants. The identified pre-miRNAs had an average length of 162 bp and average MFE, MFEI and % CG content of −58.98, −0.96 and 41.50, respectively (Figure S3 and Table S4). Ten miRNAs were anchored in the 3p-arm and 15 miRNAs in the 5p-arm of these pre-miRNAs. The most abundant novel miRNA yielded 11,899 reads (Jcu_nMIR001), and it is the sixth most abundant miRNA in J. curcas, suggesting an important role in this tissue. The majority of novel miRNAs are 21 nt longer (Table S5), as was observed for conserved miRNAs. Interestingly, only one (JcumiR006) of the 46 novel miRNAs identified by Wang et al. [15] through cloning was identified in the present study (corresponding to JcuMIR0015_5p in the present study, sequence GGCAUGGGCGAUAUGGGCAAGA). This difference in the identified miRNAs is most likely a result of the chosen method, as explained earlier. Similarly, members of the Jcu_MIR166 and Jcu_MIR167 families demonstrated by Vishwakarma and Jadeja [16] were also identified in the present study. However, we were unable to identify six other members from the other families in our sRNA library, suggesting that these specific microRNAs may not be expressed or may not be detectable in mature seeds.
Identification of IsomiRNAs
It was previously shown that miRNA variants, which are known as isomiRNAs, are detectable by high-throughput sequencing [40]–[43]. They show additional nucleotides in the 5′or 3′ terminus compared to the canonical or most abundant mature miRNAs. IsomiRNAs are considered to be a consequence of inaccuracies in Dicer pre-miRNA processing. Length heterogeneity can also arise by the exonuclease ‘nibbling’ of the ends, which produces a shorter template product, or by the post-transcriptional addition of one or more bases [44]. In the present study, the alignment of the sRNA library with identified J. curcas pre-miRNAs allowed for the estimation of the number and abundance of each isomiRNA corresponding to conserved (Table S2 and Figure S2) and novel J. curcas miRNA families (Figure S3 and Table S5).
IsomiRNAs were produced from both known and novel predicted pre-miRNAs of J. curcas. It was possible to observe that miRNA families differ significantly from each other in the number and abundance of isomiRNAs, as was observed in other studies [14], [34], [36]. The known pre-miRNA Jcu_MIR168 and the novel pre-miRNA Jcu_nMIR001 produced more isomiRNAs than the other pre-miRNAs. Unexpectedly, a variant of the novel Jcu_nMIR001 showed more than 10,000 reads because species-specific miRNAs usually present low levels of expression compared to the conserved miRNAs. The predicted targets of Jcu_nMIR001 miRNA are ribosomal proteins, which could explain the high abundance of this miRNA. It is also interesting to note that in some conserved miRNA families, the most abundant miRNA was not the canonical miRNA described for other species. This wide variation suggests that the same miRNA family is involved in divergent functions and may be necessary at different levels, according to the species, timing, tissue and/or other situations, such as environmental conditions and stresses. It has been shown that isomiRNAs can be expressed in a cell-specific manner, and numerous recent studies suggest that at least some isomiRNAs may affect target selection, miRNA stability, or loading into the RNA-induced silencing complex (RISC) [44].
Abundance of J. curcas Pre-miRNAs during Seed Development
Because pre-miRNAs are processed as mRNAs species, in silico approaches can be used as a reliable tool for estimating the abundance of pre-miRNAs. Although this analysis does not directly predict the abundance of mature miRNA or the isomiRNAs, the number of precursor sequences present in seed developmental stages can provide information about the overall variation of the miRNA set generated from each precursor. The abundance analysis revealed that the majority of pre-miRNAs were present at similar levels in all libraries, suggesting that they are necessary during all seed development processes (Tables S3 and S4). Nevertheless, the abundance of some pre-miRNAs varied from one library to another, suggesting that the miRNAs from these precursors may be associated with the mRNA silencing involved in specific seed stage process, such as growth or maturation. Among the pre-miRNAs that were differentially represented, Jcu_MIR390 was only detected in L3 seeds; Jcu_MIR169a was only detected in L2 seeds; Jcu_MIR167a, Jcu_MIR172 and Jcu_MIR845 were only detected in L1 seeds (Table S3); Jcu_MIR156d, Jcu_MIR167b and Jcu_MIR319 were better represented in the L2 and L3 libraries; and Jcu_MIR390 was better represented in L3. Among the novel J. curcas miRNAs, Jcu_nMIR001 was better represented at the beginning of seed development, and Jcu_nMIR003, Jcu_nMIR007 and Jcu_nMIR016 were more represented in the intermediate seed stage (Table S4). It is also important to note that some novel pre-miRNAs were highly abundant, such as Jcu_nMIR001, which was the most expressed precursor during immature and mature seed stages, and Jcu_nMIR003, which was the most expressed in the intermediate stage, indicating that they play an important role in this plant. In summary, the prediction of pre-miRNA abundance among libraries increased the understanding and implications of post-transcriptional regulation in J. curcas seeds.
siRNA in J. curcas Seeds
In contrast to the biogenesis and action of miRNA, siRNAs are 24-nt long sequences produced by double-stranded RNAs that can act at the transcriptional level through DNA methylation and histone modification and at the post-transcriptional level through the regulation of gene expression [45]. Several siRNAs have been recognized to play important roles in plant stress tolerance [46], [47]. Because of the large abundance of 24-nt sequences in the J. curcas sRNA library, we investigated the presence of siRNAs. The sRNAs that were 24 nt were matched against contigs assembled from L1–L2–L3 libraries. Putative contigs with typical sRNA distribution patterns along the matching sequences were further subjected to annotation (see methods). As a result, 42 siRNA precursors were identified and annotated (Table S6). This analysis indicates that siRNAs play a role in nuclear functions and in transport, catalytic processes and disease resistance. As expected, transposons and retroelements were relatively abundant among siRNA precursors, supporting their mechanism of action in guide chromatin-based events and resulting in transcriptional silencing [48]. It was reported that siRNA precursors can also be formed by cellular RNA-dependent RNA polymerase activity (RdRp) [49]. In fact, most J. curcas siRNA precursors were annotated as RdRps. Out of these precursors, the majority are associated with nuclear functions and play roles in transport, catalytic processes and disease resistance.
Prediction of J. curcas miRNA Targets
Because the roles of miRNAs during plant development and in species-specific adaptation processes are executed through the cleavage or translation repression of target genes [6], miRNA target prediction is critical for gaining insight into the regulatory functions of miRNAs. In this study, the putative target genes of J. curcas miRNAs were predicted by using the web-based computer server psRNATarget, based on perfect or near perfect complementarity between miRNAs and their targets. The most abundant mature miRNAs from each family (isomiRNAs were not considered) were aligned with a set of unigenes generated from assembled J. curcas contigs from all seed development mRNA-seq libraries (L1–L2–L3). A total of 57 sequences were predicted as potential targets of 28 known plant miRNAs and 12 novel miRNAs, with an average of 1.8 targets per miRNA (Table S7).
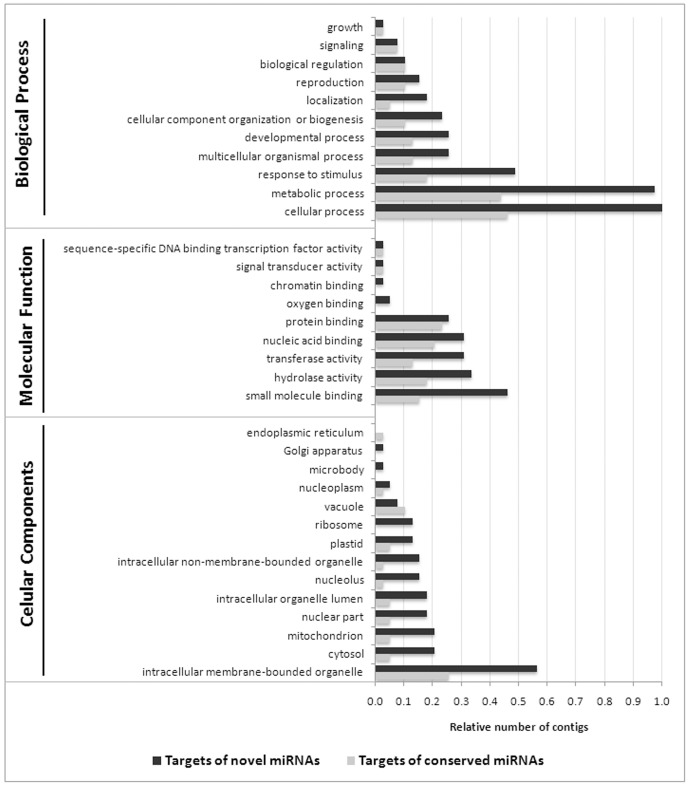
All of the identified targets were analyzed by using BLASTX against protein databases, followed by a GO analysis to evaluate their putative functions. According to the categorized GO annotation, 109 genes are involved in cellular components, with the majority of conserved and novel miRNA targets localized in intracellular membrane-bounded organelles. In the molecular functions category, 107 genes participate in catalytic or signaling transduction activities and binding activities with proteins and nucleic acids (Figure 3). With respect to biological processes, 216 genes primarily participate in stimulus responses and different cellular and metabolic processes, suggesting that the novel and conserved J. curcas miRNAs are involved in a broad range of physiological functions. These functions include participation in plant growth and development (pentatricopeptide repeat-containing protein, auxin response factor 10, seed maturation protein, etc.), lipid metabolism (phosphatidylserine decarboxylase and glycerophosphoryl diester), nutrient/cellular transport (amino acid transporter, high affinity nitrate transporter, metal transporter nramp6-like, etc.), and, primarily, in defense (tir-nbs-lrr resistance protein, cc-nbs-lrr resistance protein, cytosolic class I small heat shock protein partial, proline synthetase associated, dehydration responsive element binding proteins, etc.) (Table S7). The miRNAs targets were differentially represented during seed development, suggesting that they are regulated according to seed needs. For example, caffeoyl-o-methyltransferase and seed maturation protein are the most represented targets at the end of seed development, and subtilisin-like protease-like and auxin response factor 10 are better represented at the beginning of seed development than the other targets.
Figure 3. Targets of the miRNAs identified in mature seeds of J. curcas.
The percentage (%) of contigs for each Gene Ontology (GO) term is relative to the total number of contigs from each gene category.
The auxin response factor (ARF) is a plant-specific family of DNA binding proteins involved in hormone signal transduction [50], [51]. The ARF gene and the F-box family proteins, also previously described in relation to auxin signaling [52], were predicted targets of conserved J. curcas miRNAs and were highly abundant in immature seeds. These genes, as well as some other predicted targets, such as proteins associated with nucleotide synthesis, ribosomal proteins, metal transporters and membrane proteins, may play a role in seed growth and formation. Another important gene targeted by Jcu_MIR156, Jcu_MIR168, Jcu_MIR403, Jcu_MIR472 and the novel Jcu_nMIR005 and Jcu_nMIR009 is the pentatricopeptide repeat gene (PPR), which participates in the regulation of gene expression and was present at considerable levels in all seed developmental stages, especially immature J. curcas seeds (Table S7). PPR belongs to a large gene family implicated in post-transcriptional processes, such as splicing, editing, processing and translation, in specific organelles, such as mitochondria and chloroplasts [53].
Several predicted targets of J. curcas miRNAs are involved in abiotic stresses, including genes associated with proline and phenylpropanoid synthesis, hormones and responses to dehydration and high temperatures. The dehydration-responsive element/C-repeat (DRE/CRT) was predicted as the target of Jcu_MIR156 and Jcu_MIR168. DRE/CRT has been identified as a cis-acting element involved in one of the ABA independent regulatory systems of abiotic stress response. In Arabidopsis, MIR156 and MIR168 were described as dehydration stress-responsive miRNAs [54]. An analysis of J. curcas DREB gene expression was performed by Tang et al. [55], and the expression was induced by cold, salt and drought stresses. Although only a few reads corresponding to this gene were detected in the present study, it could be interesting to investigate the role of miRNAs in the regulation of DREB expression in other tissues and during abiotic stresses, especially in J. curcas plants that are well-known for their tolerance to drought stress [56].
In addition to abiotic stress, miRNAs from J. curcas seeds may also participate in biotic stresses. The recognition of an invading microbial pathogen is often followed by the synthesis of specific plant disease resistance proteins (R) [57] that possess activities that can lead to plant cell death through the familiar hypersensitive response (HR), a characteristic feature of many plant defense mechanisms [58]. Genes encoding R proteins (cc-nbs-lrr resistance protein, disease resistance rpp13-like protein 1-like and tir-nbs-lrr resistance protein) and an HR protein were the predicted targets of Jcu_MIR159 and Jcu_MIR472 and were highly abundant at the end of seed development (Table S7), most likely as a mechanism to protect the mature seed against pathogens, thereby preventing unsuccessfully germination.
Prediction of J. curcas miRNA Targets Involved in Lipid Metabolism
Jatropha has emerged as a promising biodiesel crop, but increasing the oil content and improving the oil quality are still challenging [2]–[4]. To investigate the involvement of isomiRNAs in regulating the lipid metabolism of J. curcas, the putative target genes of isomiRNAs with more than 10 reads were searched. This approach allowed for the identification of 12 miRNA targets related to lipid metabolic pathways (Table S8). This is the first report of miRNA associations with genes from the lipid metabolism in J. curcas. When estimating the sequence abundance of the target contigs, only hydroxysteroid 11-beta-dehydrogenase 1-like protein was better represented at the end of seed development, and phosphatidylserine decarboxylase, diphosphomevalonate decarboxylase, 1-phosphatidylinositol-bisphosphate, lipid binding, phosphatidylserine synthase 2 and cyclopropane-fatty-acyl-phospholipid synthase were more represented at the beginning of seed formation.
LPAT, or 1-acyl-sn-glycerol-3-phosphate acyltransferase, was the predicted target of Jcu_MIR001 and Jcu_MIR007 isomiRNA. This enzyme provides the key intermediate in membrane phospholipid and storage lipid biosynthesis in developing seeds [59]. Through the analysis of miRNA target abundance, it was possible to observe that LPAT was more represented in intermediate seeds. It was proposed that increasing the LPAT expression in seeds leads to a greater flux of intermediates through the Kennedy pathway and enhanced triacylglycerol accumulation [43]. Glycerophosphoryl diester (predicted target of Jcu_MIR001) and glycerol-3-phosphate dehydrogenase (predicted target of Jcu_MIR403) provide glycerol-3-phosphate for TAG assembly in the Kennedy pathway, and they were also more represented in intermediate seeds. These results confirm the general observation that storage lipid biosynthesis usually occurred at the middle-late stage during seed development [60] and suggest that the seed already reached maturity by the intermediate stage. Interestingly, a seed maturation protein target of Jcu_MIR472 was better represented in the intermediate stage, corroborating this finding. The same LPAT and GPDH expression pattern during seed development was observed by Xu et al. [61] and Guo et al. [43] by using real time PCR, which confirmed these results. In the study by Xu et al. [61], the authors verified that the gene encoding fatty acid desaturase exhibited low expression in all seed developmental stages, as was observed in the present study, suggesting that the transcript accumulation of this gene is not necessarily linked to triacylglycerol biosynthesis in developing Jatropha seeds.
Conclusions
There is some uncertainty about the genetic contribution to divergent phenotypes and the response to growth conditions and stress in Jatropha genotypes because there are several reports indicating low genetic diversity among these plants. Epigenetic polymorphisms were suggested to be involved [62] but may not explain all of the observed the variation. The present study identified the J. curcas miRNAs that are involved in a wide range of physiological functions and therefore may also contribute to this phenotypic variation. The identification of a large set of miRNAs and their targets as well as siRNAs in J. curcas seeds contributes to the elucidation of complex miRNA-mediated regulatory systems, which control seed development and other physiological processes. Several Jatropha miRNAs were predicted to regulate genes associated with lipid metabolism; these miRNAs are promising candidates for improving the yield and quality of J. curcas seed oil. However, further studies are necessary to search for more novel miRNAs and to validate their target by expression analysis during seed development and also under specific environmental and physiological conditions.
Supporting Information
Flow chart of the methodology adopted to identify J. curcas miRNAs.
(PDF)
Predicted secondary structures of known miRNA precursors in J. curcas . Locations and expressions of small RNAs mapped onto these precursors are presented. Read sequences corresponding to miRNA candidates, which are located in the 5p and 3p arms and labeled in red and purple, respectively. Values on the left side of the miRNA sequences represent the miRNA length (Jn) and read counts (x n) in the mature seed library.
(PDF)
Predicted secondary structures of novel miRNA precursors in J. curcas . The locations and the expression of small RNAs mapped onto these precursors are shown here. The sequences of miRNA candidates located in the 5p and 3p arms are labeled in red and purple, respectively. Values on the left side of the miRNA sequence represent miRNA length (Jn) and read counts (x n) in the mature seed library.
(PDF)
Abundance of small RNAs from each size according to the number of reads.
(XLSX)
Known miRNAs identified in the J. curcas sRNA library. This identification was performed by selecting perfect homologies to plant-conserved miRNAs, as deposited in the miRBase database (release 19, August 2012).
(XLSX)
Putative known miRNA precursors in J. curcas and their abundance during seed development.
(XLSX)
Putative new J. curcas miRNA precursors and their abundance during seed development.
(XLSX)
Novel miRNAs identified in the sRNA library of J. curcas.
(XLSX)
Putative precursors of siRNA in J. curcas.
(XLSX)
Predicted targets of known and new miRNAs in mature J. curcas seeds. The abundance of these targets during seed development is shown here.
(XLSX)
Predicted targets of known and new isomiRNAs related to the lipid metabolism of J. curcas. The abundance of targets during seed development is shown here.
(XLSX)
Funding Statement
This research was supported by Conselho Nacional de Desenvolvimento Científico e Tencológico - CNPq grants numbers (559636/2009-1 and 307868/2011-7), Ministerio de Ciência e Tecnologia - MCT and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior- CAPES. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carels N (2009) Jatropha curcas: A Review. Adv Bot Res 50: 39–86. [Google Scholar]
- 2. Divakara BN, Upadhyaya HD, Wani SP, Laxmipathi CL (2010) Biology and genetic improvement of Jatropha curcas L. Appl Energ. 87: 732–742. [Google Scholar]
- 3. Berchmans HJ, Hirata S (2008) Biodiesel production from crude Jatropha curcas L. seed oil with a high content of free fatty acids. Bioresour Technol 99: 1716–1721. [DOI] [PubMed] [Google Scholar]
- 4. Becker K, Makkar HPS (2008) Jatropha curcas: A potential source for tomorrow’s oil and biodiesel. Lipid Tech 20: 104–107. [Google Scholar]
- 5. King AJ, He W, Cuevas A, Freudenberger M, Ramiaramanana D, et al. (2009) Potential of Jatropha curcas as a source of renewable oil and animal feed. J. Exp. Bot. 60: 2897–2905. [DOI] [PubMed] [Google Scholar]
- 6. Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 7. Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 60–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartel DP (2009) MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voinnet O (2009) Origin, biogenesis and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- 10. Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- 11. Chen H, Li Z, Xiong L (2012) A plant microRNA regulates the adaptation of roots to drought stress. FEBS Letters 586: 1742–1747. [DOI] [PubMed] [Google Scholar]
- 12. Schommer C, Bresso EG, Spinelli SV, Palatnik JF (2012) MicroRNAs in plant development and stress responses. Signaling and Communication in Plants 15: 29–47. [Google Scholar]
- 13. Chi X, Yang Q, Chen X, Wang J, Pan L, et al. (2011) Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS ONE 6: e27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Körbes AP, Machado RD, Guzman F, Almerão MP, de Oliveira LFV, et al. (2012) Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS ONE 7: e50663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang CM, Liu P, Sun F, Li L, Liu P, et al. (2012) Isolation and identification of miRNAs in Jatropha curcas . Int J Biol Sci 8: 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vishwakarma NP, Jadeja VJ (2013) Identification of miRNA encoded by Jatropha curcas from EST and GSS. Plant Signal Behav 8: 2, e23152. [DOI] [PMC free article] [PubMed]
- 17. Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jühling F, Mör M, Hartmann RK, Sprinz M, Stadler PF, et al. (2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: 159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pruesse E, Quast C, Knitte K, Fuchs BM, Ludwig W, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He G, Elling AA, Deng XW (2011) The Epigenome and Plant Development. Annu Rev Plant Biol 62: 411–435. [DOI] [PubMed] [Google Scholar]
- 21. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- 23. Mathews DH (2004) Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10: 1178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pertea G, Huang X, Liang F, Antonescu V, Sultana R, et al. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652. [DOI] [PubMed] [Google Scholar]
- 25. Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: 9–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conesa A, Götz S (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson MD, McCarthy DJ, Smyth GK (2010) EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995. [DOI] [PubMed] [Google Scholar]
- 31. Molina LG, Cordenonsi FG, Morais GL, de Oliveira LFV, Carvalho JB, et al. (2012) Metatranscriptomic analysis of small RNAs present in soybean deep sequencing libraries. Genet Mol Biol 35: 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, et al. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schreiber AW, Shi BJ, Huang CY, Langridge P, Baumann U (2011) Discovery of barley miRNAs through deep sequencing of short reads. BMC Genomics 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song QX, Liu YF, Hu XY, Zhang WK, Ma B, et al. (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biology 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. The Plant Journal 46: 243–259. [DOI] [PubMed] [Google Scholar]
- 36. Sunkar R, Agadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guzman F, Almerão MP, Körbes AP, Loss-Morais G, Margis R (2012) Identification of microRNAs from Eugenia uniflora by high-throughput sequencing and bioinformatics analysis PLoS ONE. 7: e49811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yian Z, Li C, Han X, Shen F (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414: 60–66. [DOI] [PubMed] [Google Scholar]
- 39. Moxon S, Schwach F, Maclean D, Dalmay T, Studholme DJ, et al. (2008) A tool kit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253. [DOI] [PubMed] [Google Scholar]
- 40. Kulcheski FR, de Oliveira LF, Molina LG, Almerão MP, Rodrigues FA, et al. (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, et al. (2009) Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ebhardt A, Fedynak A, Fahlman RP (2010) Naturally occurring variations in sequence length creates microRNA isoforms that differ in argonaute effector complex specificity. Silence 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gu K, Yi C, Tian D, Sangha JS, Hong Y, et al. (2012) Expression of fatty acid and lipid biosynthetic genes in developing endosperm of Jatropha curcas . Biotechnology for Biofuels 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, et al. (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot 63: 1025–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, et al. (2010) An endogenous, systemic RNAi pathway in plants. The EMBO Journal 29: 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimi Biophys Acta - Gene Regulatory Mechanisms 1819: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He S, Liu C, Skogerbo G, Zhao H, Wang J, et al. (2008) NONCODE v2.0: decoding the non-coding. Nucleic Acids Res 36: 170–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahlquist P (2002) RNA-Dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273. [DOI] [PubMed] [Google Scholar]
- 50. Yang JH, Han SJ, Yoon EK, Lee WS (2006) Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res 34: 1892–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218. [DOI] [PubMed] [Google Scholar]
- 52. Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biol 1: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmitz-Linneweber C, Small I (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670. [DOI] [PubMed] [Google Scholar]
- 54. Liu H, Tian X, Li Y, Wu C, Zheng C (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana . RNA 14: 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tang M, Liu X, Deng H, Shen S (2011) Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. Plant Sci 181: 623–631. [DOI] [PubMed] [Google Scholar]
- 56. Wang WG, Li R, Liu B, Li L, Wang SH, et al. (2011) Effects of low nitrogen and drought stresses on proline synthesis of Jatropha curcas seedling. Acta Physiol Plant 33: 1591–1595. [Google Scholar]
- 57. Belkhadir Y, Subramaniam R, Dangl JL (2004) Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr Opin Plant Biol 7: 391–399. [DOI] [PubMed] [Google Scholar]
- 58. Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cellular Microbiol 6: 201–211. [DOI] [PubMed] [Google Scholar]
- 59. Maisonneuve S, Bessoule JJ, Lessire R, Delseny M, Roscoe TJ (2010) Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol 152: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hills MJ (2004) Control of storage-product synthesis in seeds. Curr Opin Plant Biol 7: 302–8. [DOI] [PubMed] [Google Scholar]
- 61. Xu R, Wanga R, Liu A (2011) Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in developing seeds of Jatropha (Jatropha curcas L.). Biomass and bioenergy 35: 1683–1692. [Google Scholar]
- 62. Yi C, Zhang S, Liu X, Bui HT, Hong Y (2010) Does epigenetic polymorphism contribute to phenotypic variances in Jatropha curcas L.? BMC Plant Biology 10: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart of the methodology adopted to identify J. curcas miRNAs.
(PDF)
Predicted secondary structures of known miRNA precursors in J. curcas . Locations and expressions of small RNAs mapped onto these precursors are presented. Read sequences corresponding to miRNA candidates, which are located in the 5p and 3p arms and labeled in red and purple, respectively. Values on the left side of the miRNA sequences represent the miRNA length (Jn) and read counts (x n) in the mature seed library.
(PDF)
Predicted secondary structures of novel miRNA precursors in J. curcas . The locations and the expression of small RNAs mapped onto these precursors are shown here. The sequences of miRNA candidates located in the 5p and 3p arms are labeled in red and purple, respectively. Values on the left side of the miRNA sequence represent miRNA length (Jn) and read counts (x n) in the mature seed library.
(PDF)
Abundance of small RNAs from each size according to the number of reads.
(XLSX)
Known miRNAs identified in the J. curcas sRNA library. This identification was performed by selecting perfect homologies to plant-conserved miRNAs, as deposited in the miRBase database (release 19, August 2012).
(XLSX)
Putative known miRNA precursors in J. curcas and their abundance during seed development.
(XLSX)
Putative new J. curcas miRNA precursors and their abundance during seed development.
(XLSX)
Novel miRNAs identified in the sRNA library of J. curcas.
(XLSX)
Putative precursors of siRNA in J. curcas.
(XLSX)
Predicted targets of known and new miRNAs in mature J. curcas seeds. The abundance of these targets during seed development is shown here.
(XLSX)
Predicted targets of known and new isomiRNAs related to the lipid metabolism of J. curcas. The abundance of targets during seed development is shown here.
(XLSX)