Background: Elevated mammalian target of rapamycin (mTOR) signaling contributes to diabetic complications.
Results: mTOR inhibitor, rapamycin, improves metabolic status and cardiac function, attenuates oxidative stress, and alters antioxidant and contractile protein expression in type 2 diabetic mice.
Conclusion: Rapamycin may provide metabolic and cardiac benefits in diabetic mice.
Significance: mTOR inhibition may be an attractive novel therapeutic strategy for diabetes-related complications.
Keywords: Antioxidants, Cardiovascular Disease, Diabetes, mTOR, Oxidative Stress, Proteomics
Abstract
Elevated mammalian target of rapamycin (mTOR) signaling contributes to the pathogenesis of diabetes, with increased morbidity and mortality, mainly because of cardiovascular complications. Because mTOR inhibition with rapamycin protects against ischemia/reperfusion injury, we hypothesized that rapamycin would prevent cardiac dysfunction associated with type 2 diabetes (T2D). We also investigated the possible mechanisms and novel protein targets involved in rapamycin-induced preservation of cardiac function in T2D mice. Adult male leptin receptor null, homozygous db/db, or wild type mice were treated daily for 28 days with vehicle (5% DMSO) or rapamycin (0.25 mg/kg, intraperitoneally). Cardiac function was monitored by echocardiography, and protein targets were identified by proteomics analysis. Rapamycin treatment significantly reduced body weight, heart weight, plasma glucose, triglyceride, and insulin levels in db/db mice. Fractional shortening was improved by rapamycin treatment in db/db mice. Oxidative stress as measured by glutathione levels and lipid peroxidation was significantly reduced in rapamycin-treated db/db hearts. Rapamycin blocked the enhanced phosphorylation of mTOR and S6, but not AKT in db/db hearts. Proteomic (by two-dimensional gel and mass spectrometry) and Western blot analyses identified significant changes in several cytoskeletal/contractile proteins (myosin light chain MLY2, myosin heavy chain 6, myosin-binding protein C), glucose metabolism proteins (pyruvate dehydrogenase E1, PYGB, Pgm2), and antioxidant proteins (peroxiredoxin 5, ferritin heavy chain 1) following rapamycin treatment in db/db heart. These results show that chronic rapamycin treatment prevents cardiac dysfunction in T2D mice, possibly through attenuation of oxidative stress and alteration of antioxidants and contractile as well as glucose metabolic protein expression.
Introduction
Diabetes mellitus (DM)2 is a major metabolic disorder affecting a large population in the United States and across the world. DM is associated with increased morbidity and mortality, predominantly as a result of cardiovascular complications such as coronary artery disease, stroke, peripheral arterial disease, cardiomyopathy, and congestive heart failure (1). Endothelial dysfunction, proteomic/hormonal alterations, and metabolic distresses promote the development of diabetic cardiomyopathy (2). The diabetic heart undergoes structural remodeling to cope with the underlying changes; however, it ultimately fails because of deterioration of cardiac contractile force. The underlying pathological mechanisms of diabetic cardiomyopathy are still poorly understood, although there is accumulating evidence that the cardiac complications of DM are closely tied to metabolic disorders including hyperglycemia, hyperlipidemia, insulin resistance, impaired calcium homeostasis, increased oxidative stress, and renin-angiotensin-aldosterone system activation (3–5).
The serine-threonine kinase mammalian target of rapamycin (mTOR) serves as an intracellular sensor for energy metabolism, nutrient availability, and stresses and controls cellular growth and metabolism (6). Its role in regulating metabolic stress, aging, and cardiovascular diseases has received tremendous interest recently. mTOR interacts with several proteins to form two distinct complexes named mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (7). Both complexes share the catalytic mTOR subunit, mLST8 (mammalian lethal with sec-13 protein 8), DEPTOR (DEP domain containing mTOR-interacting protein), and Tti1-Tel2 complex. In contrast, the other components in mTORC1 are Raptor (regulatory-associated protein of mTOR) and PRAS40 (proline-rich AKT substrate). mTORC2 has Rictor (rapamycin-insensitive companion of mTOR) and mSin1 (mammalian stress-activated MAP kinase-interacting protein 1). The ribosomal protein p70S6K and 4FBP1 (eukaryotic initiation factor 4E-binding protein 1) are two downstream targets of mTORC1 (8). Rictor promotes the activity of mTORC2 by enabling it to phosphorylate AKT at Ser473 (9). We first reported that the mTOR inhibitor rapamycin (Sirolimus) induced protection against myocardial ischemia-reperfusion injury indicated by significantly reduced infarct size and reduction of necrosis and apoptosis following simulated ischemia-reoxygenation in cardiomyocytes (10). More recently, we showed that rapamycin triggers unique cardioprotective signaling including phosphorylation of ERK, STAT3, and endothelial NOS, in concert with increased Bcl-2 to Bax ratio and inactivation of GSK-3β (11). A previous study also showed that chronic treatment with the mTOR inhibitor Rapamune (4 weeks at 2 mg/kg/day, orally) attenuated chronically established left ventricular hypertrophy and cardiac fibrosis with preserved contractile function (12). Moreover, a recent study reported that selective activation of mTORC2 with concurrent inhibition of mTORC1 decreased cardiomyocyte apoptosis and tissue damage after myocardial infarction (13).
A persistent activation of mTORC1 signaling occurs in both genetic and diet-induced animal models of obesity and metabolic disorders in liver (14), skeletal muscle (14, 15), adipose tissue (16, 17), and heart (18, 19). Because mTOR dysregulation occurs in type 2 diabetes (T2D) and obesity, there are ongoing efforts to pharmacologically target mTOR signaling in protecting against diabetic-associated cardiovascular diseases. Systemic administration of rapamycin ameliorates diabetes-induced renal dysfunction and blocks the onset of type 1 and T2D (20, 21). In addition, several studies show that sirolimus-eluting stents are highly effective in reducing the risk for major cardiac events and are safe in diabetic patients with coronary artery disease (22, 23). Therefore, to investigate the cardioprotective mechanism of mTOR inhibition in T2D, the present study was designed to identify novel cardiac protein targets that may be altered by chronic treatment with rapamycin in a mouse model. By using an advanced proteomic approach with two-dimensional differential in-gel electrophoresis (2D-DIGE) and MALDI-TOF/TOF tandem mass spectrometry, we identified a number of proteins that are potentially responsible for cardioprotection and preservation of contractile function following rapamycin treatment in T2D mice.
EXPERIMENTAL PROCEDURES
Animals
Adult male leptin receptor null, homozygous db/db mice (strain: BKS.Cg-Dock7m+/+Leprdb/J) and respective background strain mice (C57BL/6J) were purchased from Jackson Laboratories (Bar Harbor, ME). The animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the Virginia Commonwealth University. The animal care and experiments were conducted under the Guide for the Care and Use of Laboratory Animals for Biomedical Research published by the National Institutes of Health (No. 85-23, revised 1996).
Experimental Groups and Treatment Protocol
Twenty adult (16–18 weeks) male db/db and corresponding background C57BL/6J mice were treated daily for 28 days with rapamycin (0.25 mg/kg in 5% DMSO intraperitoneally) or an equivalent volume of vehicle (5% DMSO intraperitoneally as control). The dose of rapamycin was chosen based on our previous studies on rapamycin-induced cardioprotection against myocardial ischemia-reperfusion injury (10, 11). During treatment period, fasting blood glucose levels and body weight were monitored weekly. Upon completion of treatment, the mice were anesthetized with pentobarbital sodium (70 mg/kg intraperitoneally), and the heart was quickly collected via thoracotomy, rinsed free of blood with saline, weighed, immediately frozen in liquid nitrogen, and stored at −80 °C until further proteomic experiments or Western blot analysis.
Measurement of Plasma Glucose, Triglycerides, and Insulin
At the time of sacrifice of the mice (after 4 h of fasting), blood was collected in a heparinized BD Vacutainer® tube and centrifuged at 1,000 × g for 15 min at 4 °C. The clear top yellow plasma layer was stored at −80 °C until analysis. The plasma glucose and triglycerides were assayed using commercially available colorimetric assay kits (Cayman Chemicals, Ann Arbor, MI). Plasma insulin concentration was measured using an ultrasensitive mouse insulin ELISA kit (Crystal Chem, Inc., Downers Grove, IL).
Proteomic Analysis
The 2D-DIGE and mass spectrometry protein identification were performed by Applied Biomics, Inc. (Hayward, CA) following the established protocols as described in our previous publications (24, 25). In brief, three heart tissues samples (100 mg) from C57BL control and db/db mice with DMSO (as control) or rapamycin treatment were sonicated with 2 ml of two-dimensional cell lysis buffer and centrifuged for collecting the supernatant. The protein sample (30 μg) was labeled with 1 μl of CyDye dilution. (Cy2, Cy3, and Cy5; Amersham Biosciences). The CyDyes stock (1 nmol/μl) were diluted 1:5 with dimethylformamide, incubated on ice for 30 min in dark, followed by addition of 1 μl of 10 mm lysine to stop the labeling reaction. The labeled samples were mixed with 2× two-dimensional sample buffer (8 m urea, 4% CHAPS, 20 mg/ml DTT, 2% pharmalytes, and a trace amount of bromphenol) followed by addition of 100 μl of destreak solution (GE Healthcare) and rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 20 mg/ml DTT, 1% pharmalytes, and a trace amount of bromphenol blue). The sample was loaded into a 13-cm immobilized pH gradient strip (pH 3–10, Linear; GE Healthcare/Amersham Biosciences), and isoelectric focusing was run under dark. Subsequently, the immobilized pH gradient strips were incubated in 10 ml of equilibration buffer 1 (50 mm Tris-HCl, pH 8.8, 6 m urea, 10 mg/ml DTT, 30% glycerol, 2% SDS, and a trace amount of bromphenol blue) for 15 min, followed by incubation in 10 ml of equilibration buffer 2 (50 mm Tris-HCl, pH 8.8, 6 m urea, 45 mg/ml iodacetamide, 30% glycerol, 2% SDS, and a trace amount of bromphenol blue) for 10 min. The immobilized pH gradient strips were transferred into 12% SDS gel and subjected to electrophoresis at 15 °C. Each gel was scanned immediately following SDS-PAGE using Typhoon Trio scanner (Amersham Biosciences). The scanned images were then analyzed by Image QuantTL software (GE Healthcare) and then subjected to in-gel analysis and cross-gel analysis using DeCyder software version 6.5 (GE Healthcare). The change in ratio of differential protein expression was obtained from in-gel DeCyder software analysis. Quantitative comparisons were then made between two individual samples for each of the three possible combinations. The pairwise volume ratios (i.e., C57 versus db/db, C57 versus db/db RAPA, and db/db versus db/db RAPA) were calculated for each protein spot and used to determine relative protein expression. The selected spots were picked up by Ettan Spot Picker (GE Healthcare) following the DeCyder software analysis and spot picking design. The selected protein spots were subjected to in-gel trypsin digestion, peptide extraction, and desalting, followed by MALDI-TOF/TOF to determine the protein identity (24, 25).
Western Blot Analysis
To confirm the results of 2D-DIGE, we performed Western blots to assess the expression levels of selected proteins, particularly the antioxidants and contractile and glucose metabolism proteins. Total soluble proteins were extracted from whole heart tissue with 1 ml of lysis buffer containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin, 0.2 mm PMSF, and halt protease and phosphatase inhibitor mixture (Thermo Fisher Scientific Inc., Rockford, IL). The homogenate was centrifuged at 14,000 × g for 15 min under 4 °C, and the supernatant was recovered. Protein (50 μg) from each sample was separated by SDS-PAGE and transferred onto nitrocellulose membrane. The membrane was incubated with primary antibody at dilution of 1:1,000 for each of the respective proteins, i.e., p-mTOR (Ser2448), mTOR, p-S6 (Ser235/236), S6, p-AKT (Ser473), AKT (Cell Signaling Technology, Danvers, MA), ferritin heavy chain, pyruvate dehydrogenase (PDH) E1α, PYGB, SOD2, myosin light chain 2 (MLY-2), myosin heavy chain 6α (MHC6α), Actin, GAPDH (Santa Cruz Biotechnology, Dallas, TX), and peroxiredoxin 5 (PRX-5; Proteintech Group, Inc., Chicago, IL). The membrane was washed and incubated with horseradish peroxidase-conjugated secondary antibody (1:2,000 dilutions) for 1 h at room temperature. The blots were developed using a chemiluminescent system (ECL Plus; Amersham Bioscience) and subsequently exposed to Kodak film. The optical density of the protein bands were quantified using ImageJ software.
Measurement of Cardiac Function by Echocardiography
Cardiac contractile function was measured before and after treatment with rapamycin using the Vevo 770TM imaging system (VisualSonics, Inc., Toronto, Canada). A 30-MHz probe was used to obtain two-dimensional, M-mode, and Doppler imaging from parasternal short axis view at the level of the papillary muscle and the apical four-chamber view (26). Left ventricular (LV) wall thickness, LV end systolic and end diastolic diameters, fractional shortening, and ejection fraction were calculated using Vevo analysis software (version 2.2.3).
Measurement of Oxidative Stress
Oxidative stress was measured by determining the glutathione levels in the myocardium using glutathione assay kit (BioVision, Mountain View, CA) according to the manufacturer's protocol. Briefly, cardiac tissue (60 mg) was homogenized in ice-cold glutathione assay buffer, and then 60 μl of the homogenate was added to the tube containing 20 μl of perchloric acid (6 n) and incubated in ice for 5 min. The sample was centrifuged at 13,000 × g at 4 °C for 2 min, and supernatant was collected for glutathione assay. The sample was neutralized with 3 n KOH and incubated with a fluorescence probe, o-phthalaldehyde for 40 min at room temperature. o-Phthalaldehyde reacts with GSH and generates fluorescence, which was detected at 340/420 nm (excitation/emission) using a fluorescence plate reader. The total glutathione was measured by adding a reducing agent that converts the oxidized glutathione (GSSG) to GSH. To measure GSSG specifically, a GSH quencher was added to remove GSH, and the reducing agent was added to destroy excess quencher and to convert GSSG to GSH. The GSH, GSSG, and total glutathione concentration were determined using standards curve of GSH.
Lipid Peroxidation Assay
Lipid peroxidation in heart was assayed by measuring malondialdehyde (MDA) and 4-hydroxynonenal using a lipid peroxidation assay kit (BioVision) according to the manufacturer's protocol. Briefly, the hearts were homogenized in MDA lysis buffer and then centrifuged at 13,000 × g for 10 min. Supernatant (200 μl) was incubated with thiobarbituric acid (600 μl) at 95 °C for 60 min in an Eppendorf tube. The solution was cooled down to room temperature, 200 μl of each reaction solution were transferred to a microplate, and absorbance was read at 532 nm.
Statistical Analysis
The differences between groups were analyzed with one-way analysis of variance followed by Student-Newman-Keul's post hoc test for pair-wise comparison. Probability value of p < 0.05 was considered statistically significant. All measurements are expressed as means ± S.E.
RESULTS
Effect of Rapamycin on Metabolic Status
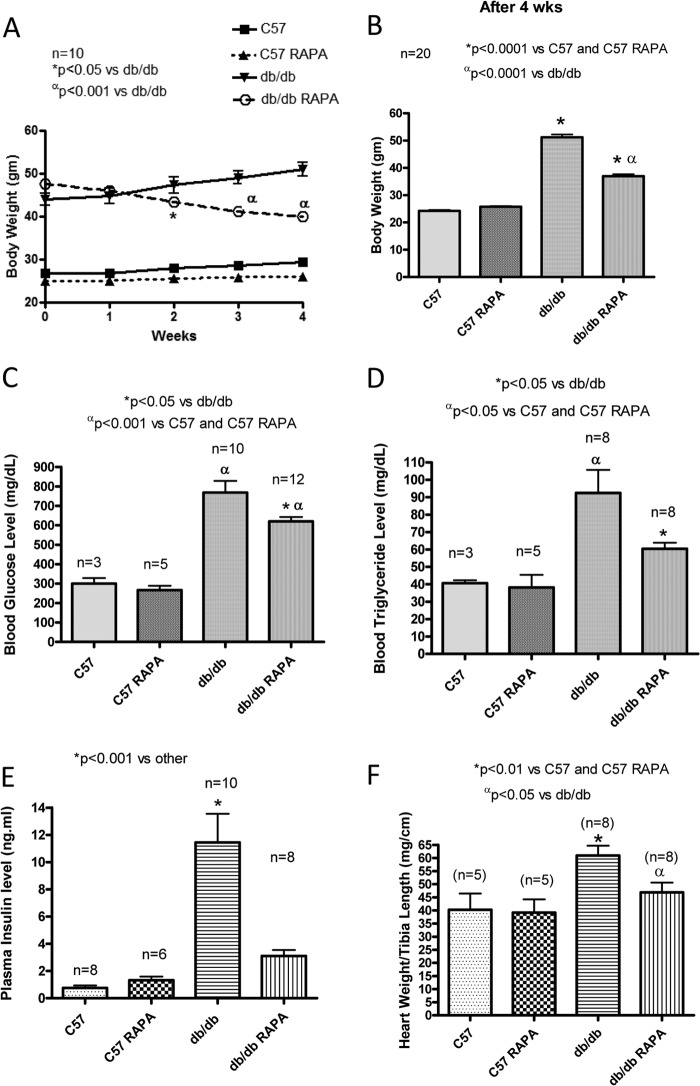
There were no significant changes in body weight, heart weight, plasma glucose, and triglyceride levels between control (DMSO-treated) and rapamycin-treated C57 mice during treatment periods (Fig. 1). The db/db mice had significantly higher body weight, ratio of heart weight to tibia length, plasma glucose, triglyceride, and insulin level as compared with the nondiabetic control C57 mice. Body weight was significantly reduced following the second week of rapamycin treatment in db/db mice as compared with db/db control (Fig. 1, A and B). The ratio of heart weight to tibia length was significantly higher in db/db mice compared with nondiabetic C57 mice. Although rapamycin treatment did not alter the ratio of heart to tibia length in nondiabetic C57 mice, it prevented the increase in heart weight/tibia length observed in db/db mice (Fig. 1F). Similarly, after 28 days of the treatment with rapamycin, the db/db mice showed a significant decrease in fasting plasma glucose (Fig. 1C), triglyceride (Fig. 1D), and insulin levels (Fig. 1E) as compared with the control nontreated db/db mice.
FIGURE 1.
Effect of chronic treatment of rapamycin on the metabolic parameters. A, body weight of C57 (wild type) and db/db mice during 4 weeks of treatment with RAPA (0.25 mg/kg/day, intraperitoneally). B–F, values were determined after 4 weeks of treatment with rapamycin. B, body weight. C, plasma glucose. D, triglyceride. E, insulin levels. F, ratio of heart weight to tibia length.
Effect of Rapamycin on Cardiac Function
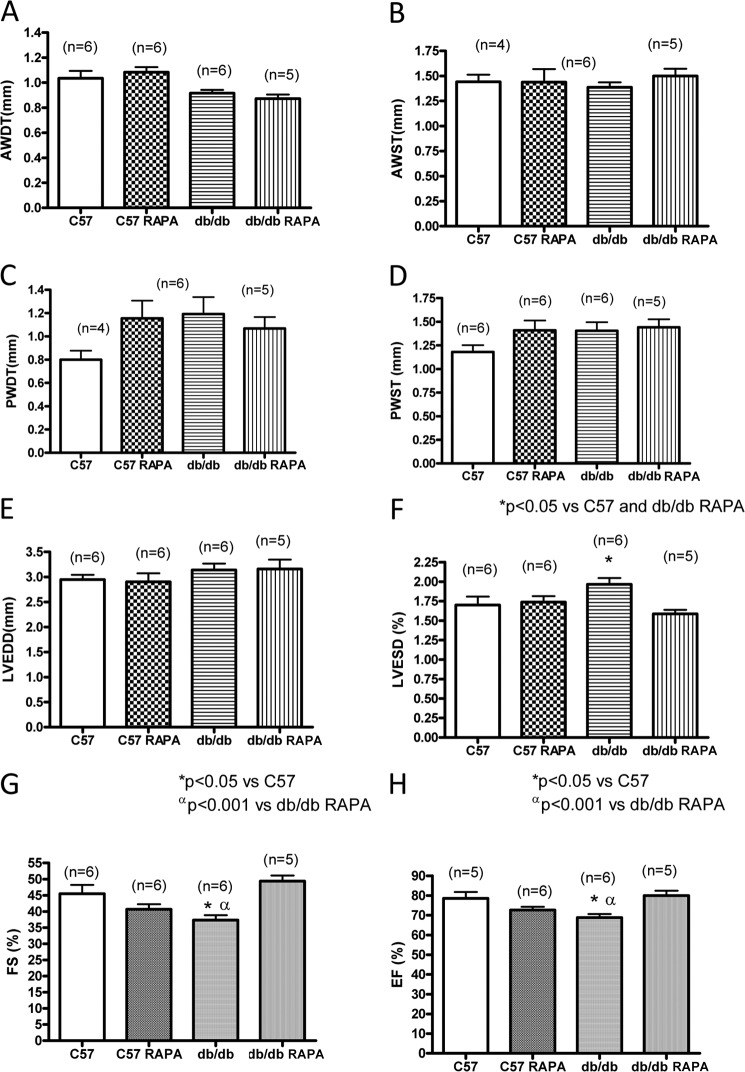
Rapamycin treatment did not alter cardiac function in C57 mice (Fig. 2). The hearts from db/db mice exhibited significant defect in cardiac function as noted by reduction of fractional shortening (37.3 ± 1.5%) and ejection fraction (68.8 ± 1.8%) as compared with the nondiabetic C57 control mice (fractional shortening, 48.3 ± 0.3%; ejection fraction, 78.7 ± 1.9%) (p < 0.05) (Fig. 2, G and H). However, rapamycin treatment significantly improved cardiac function (fractional shortening, 49.4 ± 1.7%; ejection fraction, 80.0 ± 2.5%) (Fig. 2, G and H). Although LV end diastolic diameter did not change between groups, LV end systolic diameters were significantly increased in db/db mice (2.0 ± 0.1%) as compared with C57 control (1.4 ± 0.1%) and db/db-rapamycin-treated mice (1.6 ± 0.2%) (Fig. 2, E and F). No differences in heart rate were observed between any of the groups.
FIGURE 2.
Cardiac function following 4 weeks of treatment with rapamycin. Cardiac function of C57 and db/db mice was assessed following 28 days of RAPA treatment by echocardiography. A, anterior wall diastolic thickness (AWDT). B, anterior wall systolic thickness (AWST). C, posterior wall diastolic thickness (PWDT). D, posterior wall systolic thickness (PWST). E, left ventricular end diastolic diameter (LVEDD). F, left ventricular end systolic diameter (LVESD). G, percentage of fractional shortening (FS). H, percentage of ejection fraction (EF).
Effect of Rapamycin on Phosphorylation of mTOR, S6, and AKT
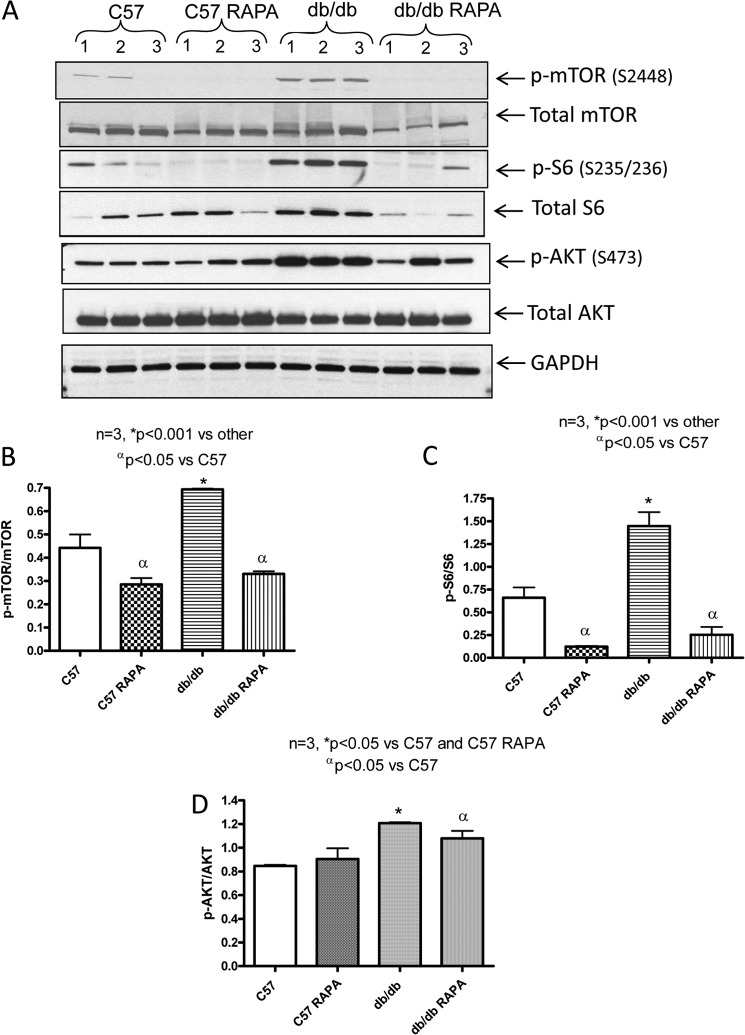
The phosphorylation of mTOR, ribosomal protein S6, and AKT were significantly enhanced in the hearts of db/db mice as compared with nondiabetic C57 mice (Fig. 3, A–D). Rapamycin treatment completely blocked the enhanced phosphorylation of mTOR and S6 in diabetic hearts. However, AKT phosphorylation in the nondiabetic C57 and diabetic db/db mice was not significantly altered after rapamycin treatment (Fig. 3, A and D).
FIGURE 3.
Phosphorylation level of mTOR, S6, and AKT in heart following 28 days of treatment with rapamycin. A, representative immunoblots of phospho-mTOR, mTOR, phospho-S6, S6, phosphor-AKT, and AKT in hearts of C57 and db/db mice following 28 days of RAPA treatment. B–D, densitometric analysis of the ratios of p-mTOR to mTOR (B), p-S6 to S6 (C), and p-AKT to AKT (D).
Role of Autophagy in Diabetic Heart
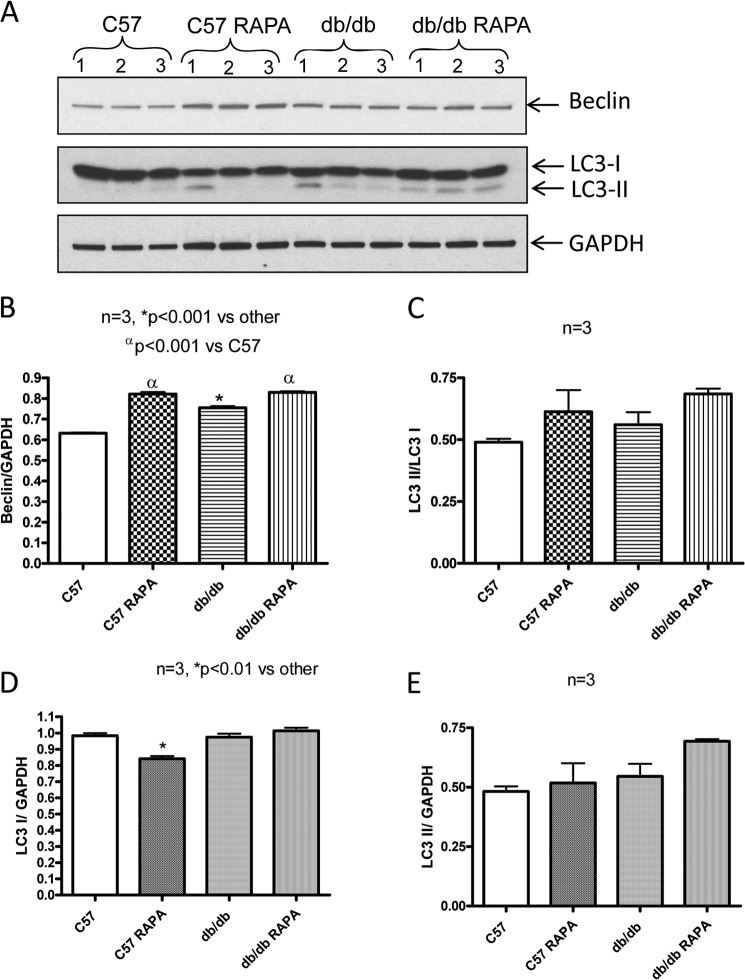
The cardiac expression of Beclin in C57 mice was increased following rapamycin treatment (Fig. 4, A and B). The expression of Beclin was also increased significantly in db/db mice as compared with C57 mice. However, rapamycin treatment further enhanced its expression in diabetic heart (Fig. 4, A and B). The ratio of LC3-II to LC3-I increased in the heart of C57 and db/db mice following rapamycin treatment, although this increase was not significant as compared with the nontreated control C57 or db/db (Fig. 4, A and C).
FIGURE 4.
Expression of Beclin and LC3 in heart following 28 days of treatment with rapamycin. A, representative immunoblots of Beclin and LC-3 in hearts of C57 and db/db mice following 28 days of RAPA treatment. B–E, densitometric analysis of the ratios of Beclin to GAPDH (B), ratios of LC3 II to LC3 I (C), LC3 I to GAPDH (D), and LC3 II to GAPDH (E).
Detection of Rapamycin-induced Alterations in Cardiac Protein Expression
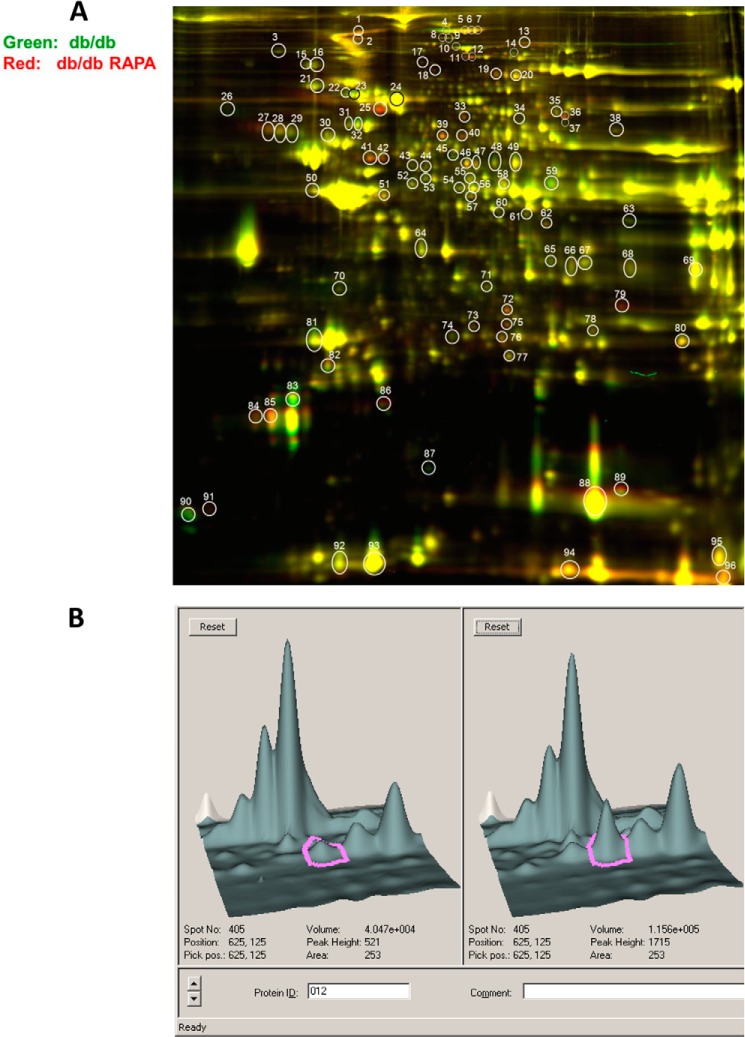
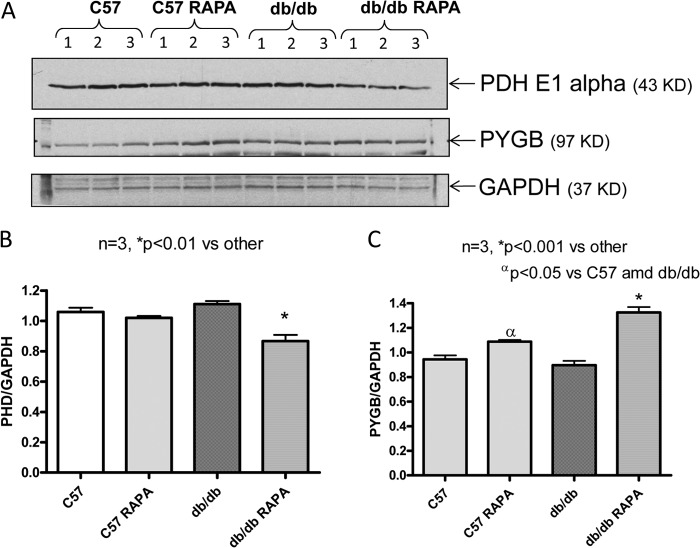
Following the 2D-DIGE and SDS-PAGE procedures, the scanned images from each gel were subjected to in-gel analysis to determine the protein differential expression. Quantitative pairwise comparisons of the protein volume ratios for each protein spot were made among the three treatment conditions (i.e., nondiabetic C57 versus db/db or db/db + rapamycin and db/db versus db/db + rapamycin). Fig. 5A shows a representative overlay image for in-gel comparison between the proteins in hearts from db/db (labeled in green dye) and db/db + rapamycin (labeled in red dye) mice. The numbered circles represent a total of 96 protein spots that are differentially expressed among the three groups. From these prominently identified spots, we further selected 30 protein spots that were significantly altered by rapamycin treatment (i.e., the ratio of fold, RAPA + db/db versus db/db heart greater than ±1.5) for protein identification using MALDI-TOF/TOF tandem mass spectrometry. Fig. 5B shows representative DeCyder three-dimensional images highlighting the substantial differences in protein abundance of PYGB (Spot# 12) between db/db and db/db + rapamycin mouse hearts.
FIGURE 5.
Scanned images of the two-dimensional gels. A, overlay image for gel comparison between heart from db/db (labeled in green dye) and heart from db/db mouse treated with RAPA for 28 days (labeled in red dye). The circled and numbered protein spots (30 spots in total 96 spots with a cutoff ratio of 1.5 for alteration in protein expression among the three treatment options) are subsequently isolated from the gels and identified by MALDI-TOF/TOF mass spectroscopy. B, representative three-dimensional DeCyder software integrated graph showing difference in PYGB (spot 12) between db/db and db/db + RAPA-treated mice.
As summarized in Table 1, among the 21 positively identified protein spots in the heart, 18 were significantly up-regulated (>1.5-fold), and three other spots were significantly down-regulated in rapamycin-treated db/db mice. To understand the functional significance of these proteins, we grouped them according to their relevant biological functions under four categories (Table 1). The first group comprises mitochondrial antioxidant enzymes: PRX-5 and ferritin heavy chain 1. PRX-5 was enhanced 6.48-fold in the hearts of db/db mice as compared with the WT C57. Moreover, it was further augmented by rapamycin treatment. Another antioxidant protein, ferritin heavy chain 1, was also increased by 1.59-fold in hearts of db/db mice as compared with the C57 mice. Rapamycin treatment further increased its levels in the db/db heart. The second group includes four cytoskeletal/contractile proteins, in which three myosin family members (i.e., myosin light polypeptide 2 (MLC-2), cardiac myosin-binding protein C (cMyBP-C), and myosin heavy polypeptide 6α (MHC6α)) were differentially affected by rapamycin treatment. The expression of nebulette was induced in rapamycin-treated diabetic heart. The third group has three proteins that play role in glucose metabolism. Two of these proteins (Pgm2 and PYGB) were induced, whereas the third one, PDH E1α, was reduced by rapamycin in the db/db heart (Table 1). Finally, the fourth group comprised 12 proteins with various cellular functions, of which hemoglobin β, carbonic anhydrase I and II, α-fetoprotein, fibrinogens, pantothenate kinase 4, and prolyl-4-hydroxylase β were significantly up-regulated, whereas heat shock protein β-7 was reduced by rapamycin.
TABLE 1.
The effects of chronic RAPA treatment on the cardiac proteins of type 2 diabetic mice
The following 21 positively protein spots were identified by proteomic analysis with 2D-DIGE and MALDI-TOF/TOF, which are differentially expressed among the three groups (wild-type control C57, db/db, and db/db + RAPA). The data are listed according to the high to low fold change ratio of protein abundance between db/db and db/db RAPA hearts. The cutoff ratio of change was ±1.5.
| Protein name | 2D-DIGE spot | Molecular mass | (db/db)/wild-type C57 ratio | (db/dbRAPA)/(db/db) ratio | RAPA effect |
|---|---|---|---|---|---|
| kDa | |||||
| Antioxidant proteins | |||||
| Peroxisomal membrane protein 20 (peroxiredoxin 5) | 89 | 17.0 | 6.48↑ | 1.71↑ | Induced |
| Ferritin heavy chain 1 | 86 | 21.1 | 1.59↑ | 1.57↑ | Induced |
| Cytoskeletal/contractile proteins | |||||
| Myosin, light polypeptide 2 | 83 | 18.9 | 2.06↑ | 1.98↓ | Reversed |
| Myosin, heavy polypeptide 6α | 1 | 223.4 | 1.36↓ | 1.81↑ | Reversed |
| Nebulette | 13 | 101.1 | 1.09↑ | 1.78↑ | Induced |
| Myosin-binding protein C | 7 | 140.5 | 1.62↓ | 1.50↑ | Reversed |
| Glucose metabolism proteins | |||||
| Pgm2 | 33 | 63.4 | 1.40↓ | 2.24↑ | Induced |
| Pyruvate dehydrogenase E1α | 59 | 43.2 | 1.02↑ | 1.71↓ | Reduced |
| PYGB | 11,12 | 67.0 | 1.01, 1.03↓ | 1.7, 2.7↑ | Induced |
| Proteins with other functions | |||||
| α-Fetoprotein | 25 | 47.2 | 1.32↓ | 3.09↑ | Induced |
| Fibrinogen-γ | 41,42 | 49.3 | 1.84↑, 1.11↓ | 2.22, 2.08↑ | Induced |
| Fibrinogen-α | 36 | 63 | 1.49↓ | 2.38↑ | Induced |
| Fibrinogen-β | 39,40 | 54.7 | 1.32↓, 1.3↑ | 1.86, 2.35↑ | Induced |
| Carbonic anhydrase I | 75 | 28.3 | 1.38↑ | 1.96↑ | Induced |
| Carbonic anhydrase II | 72 | 29 | 1.03↑ | 2.2↑ | Induced |
| Hemoglobin-β | 94 | 15.6 | 1.79↑ | 1.99↑ | Induced |
| Heat shock protein β-7 | 87 | 18.6 | 1.34↑ | 1.92↓ | Reduced |
| Serine (or cysteine) proteinase inhibitor | 26 | 46.6 | 1.81↓ | 1.9↓ | Reduced |
| Transferrin | 19 | 76.6 | 1.43↓ | 1.55↑ | Induced |
| Pantothenate kinase 4 | 18 | 86.3 | 1.02↓ | 1.52↑ | Induced |
| Prolyl 4-hydroxylase β | 27 | 58.9 | 1.85↓ | 1.5↑ | Induced |
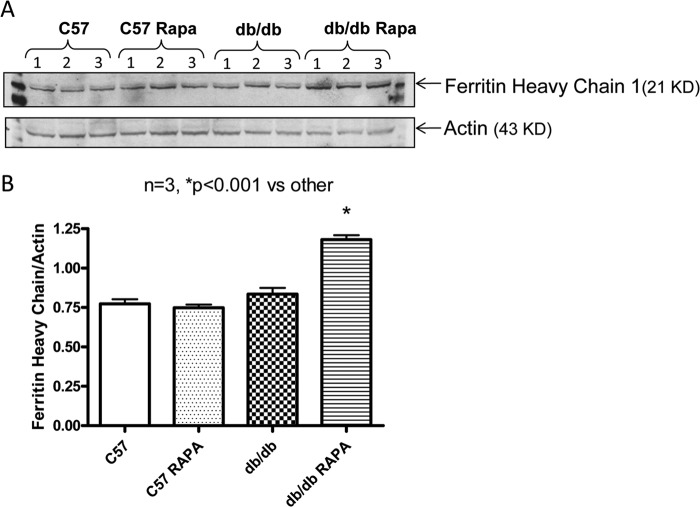
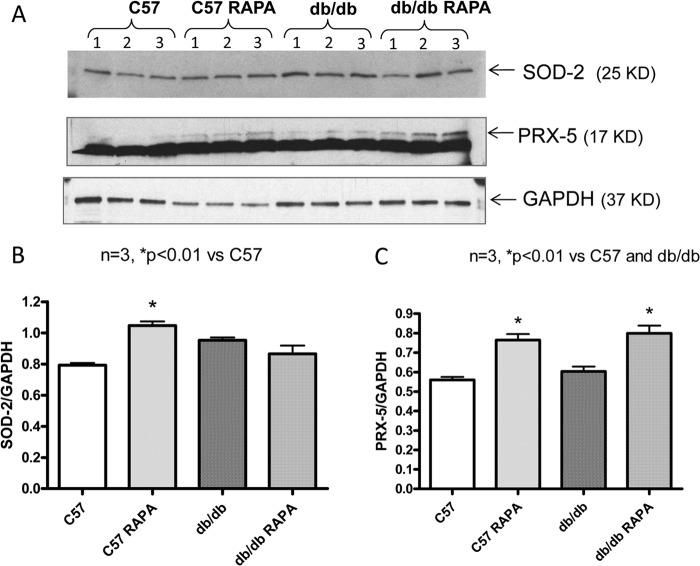
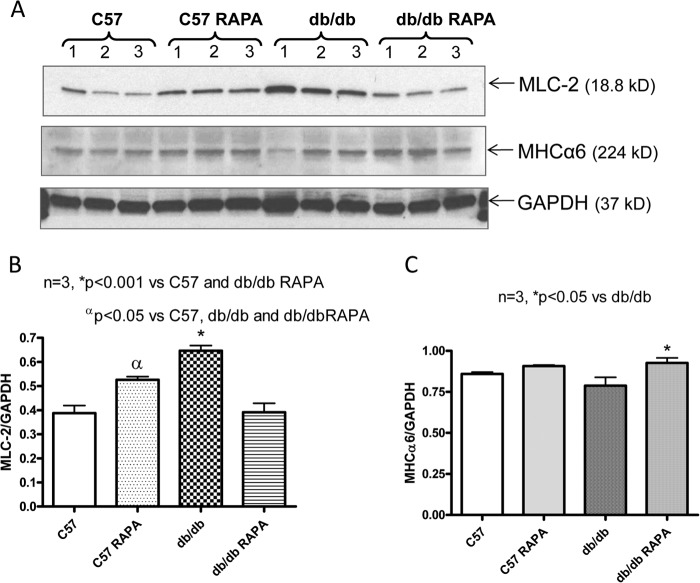
To confirm the 2D-DIGE results, the expression of six proteins with known functional significance were examined by Western blots. These proteins included antioxidant enzymes (ferritin heavy chain 1, PRX-5) (Figs. 6 and 7), contractile protein (MLC-2 and MHCα6) (Fig. 8), and metabolic protein (PDH E1α and PYGB) (Fig. 9). Western blots confirmed the induction of antioxidant proteins ferritin heavy chain 1 (Fig. 6) and PRX5 (Fig. 7) in the rapamycin-treated db/db hearts as compared with control db/db. The antioxidant protein, SOD2 level did not alter significantly in db/db hearts as compared with nondiabetic hearts with/without rapamycin treatment; however, rapamycin significantly induced it in hearts of C57 mice (Fig. 7, A and B). MLC-2 was significantly induced in the db/db hearts as compared with nondiabetic hearts (Fig. 8, A and B); even though the proteomic analysis reported conflicting effects of rapamycin on the expression of this protein, Western blot confirmed a significant reduction of MLC-2 expression and induction of MHCα6 in the rapamycin-treated db/db hearts as compared with control db/db hearts (Fig. 8, A–C). Moreover, the alteration of metabolic protein PDH E1α and PYGB, detected by 2D-DIGE proteomic analysis, was confirmed by Western blots (Fig. 9).
FIGURE 6.
Effect of rapamycin on the expression of ferritin heavy chain in the heart. A, representative immunoblots of ferritin heavy chain 1 and actin in hearts of C57 and db/db mice following 28 days of RAPA treatment. B, densitometric analysis of the ratio of ferritin heavy chain 1 to actin.
FIGURE 7.
Effect of rapamycin on the expression of SOD2 and PRX-5 proteins in the heart. A, representative immunoblots of SOD2, PRX-5, and GAPDH in hearts of C57 and db/db mice following 28 days of RAPA treatment. B and C, densitometric analysis of the ratios of SOD2 to GAPDH (B) and PRX-5 to GAPDH (C).
FIGURE 8.
Effect of rapamycin on the expression of myosin light chain-2 and myosin heavy chain α6 proteins in the heart. A, representative immunoblots of MLC-2, MHCα6, and GAPDH in hearts of C57 and db/db mice following 28 days of rapamycin treatment. B and C, densitometric analysis of the ratios of MLC-2 to GAPDH (B) and MHCα6 to GAPDH (C).
FIGURE 9.
Effect of rapamycin on the expression of pyruvate dehydrogenase E1α and PYGB proteins in the heart. A, representative immunoblots of pyruvate dehydrogenase E1α, PYGB, and GAPDH in hearts of C57 and db/db mice following 28 days of RAPA treatment. B and C, densitometric analysis of the ratios of pyruvate dehydrogenase E1α to GAPDH (B) and PYGB to GAPDH (C).
Effect of Rapamycin on Oxidative Stress
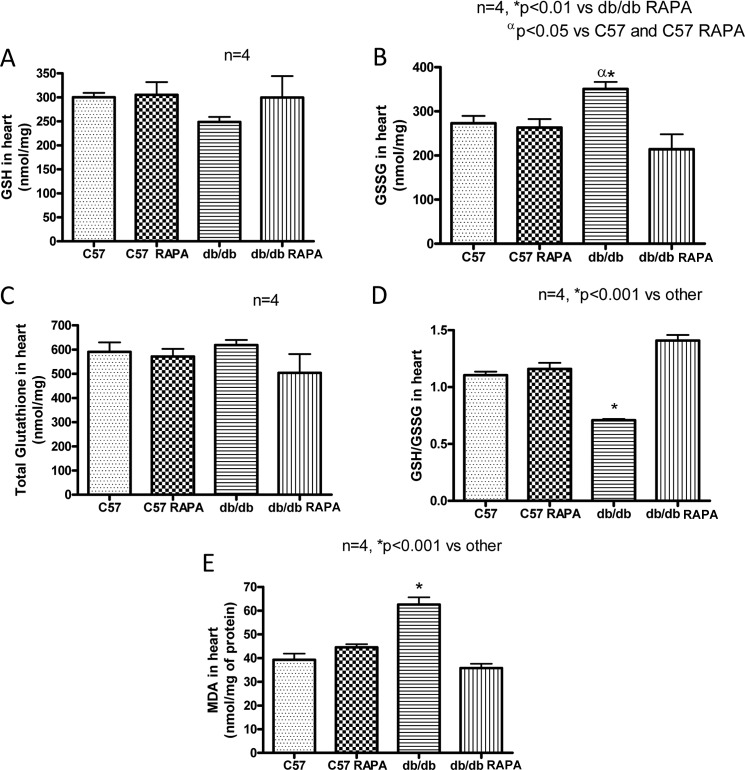
The cardiac levels of GSSG, reduced form (GSH), the total glutathione, and the GSH/GSSG ratio were not changed significantly in rapamycin-treated C57 mice as compared with the control C57 mice (n = 4; Fig. 10, A–C). GSSG level was significantly enhanced in the hearts of db/db as compared with the control C57 mice (p < 0.05, n = 4; Fig. 10B). Rapamycin treatment reduced GSSG level in the db/db hearts (Fig. 10B). The GSH level was slightly lower in db/db hearts, but total glutathione were not significantly altered between the groups (Fig. 10, A and C). However, the ratio of GSH/GSSG was significantly reduced in db/db hearts as compared with nondiabetic hearts. Treatment with rapamycin significantly restored the ratio of GSH to GSSG (p < 0.001 versus C57 and db/db, n = 4), indicating the attenuation of oxidative stress in T2D hearts (Fig. 10D). Similarly, the product of lipid peroxidation, MDA, also remained unaltered in the nondiabetic C57 hearts following rapamycin treatment (n = 4; Fig. 10E). However, MDA was significantly increased in db/db hearts as compared with nondiabetic C57 hearts (p < 0.001, n = 4), which was also attenuated by rapamycin treatment in T2D hearts (Fig. 10E).
FIGURE 10.
Effect of chronic rapamycin treatment on oxidative stress. A, GSH. B, GSSG. C, total glutathione. D, the ratio of GSH/GSSG level. E, MDA levels in hearts of C57 and db/db mice following RAPA treatment.
DISCUSSION
In the present study, we investigated the effect of mTOR inhibitor, rapamycin, on cardiac function in T2D mice and determined the associated changes in protein expression. For the first time, our results show that chronic treatment with low dose rapamycin (0.25 mg/kg/day, intraperitoneally) improves the metabolic status of diabetic mice with significant reduction in plasma glucose, insulin, and triglyceride levels (Fig. 1). Furthermore, our results demonstrate that rapamycin treatment prevents cardiac dysfunction (Fig. 2), attenuates oxidative stress (Fig. 10) and alters the expression of antioxidant and contractile proteins in T2D mice (Figs. 6–9).
Rapamycin preferentially inhibits mTORC1, although recent studies show that chronic treatment with the drug also inhibits mTORC2 (27, 28). Chronic treatment with the 8-fold higher dose of rapamycin (2 mg/kg/day) has been shown to impair whole body insulin sensitivity by disrupting mTORC2 and blocking the ability of mTORC2-AKT to inhibit hepatic gluconeogenesis (27). Therefore, a well controlled cardioprotective dose (0.25 mg/kg/day) (10, 11) and/or a more selective approach toward mTORC1 inhibition (without disrupting mTORC2) may be necessary for the treatment of diabetes-induced cardiovascular disease. Interestingly, our results show that rapamycin inhibits the T2D-enhanced phosphorylation of mTOR and S6 ribosomal protein (downstream target of mTORC1) in the heart, without interfering with phosphorylation of AKT (Ser473, target of mTORC2) (Fig. 3).
Previous studies have shown that myocardial autophagy is elevated in a murine model of T2D, which is correlated with plasma insulin and glucose levels as well as myocardial tissue insulin resistance (29, 30). However, other studies reported that diabetes did not increase the extent of autophagy in the heart following persistent ischemia (32, 33). By blocking the inhibitory effect of mTOR, rapamycin induces autophagy and protects cardiomyocytes from oxidative stress-induced toxicity (31). In the present study, the expression of Beclin was increased in diabetic heart, which was further enhanced following rapamycin treatment (Fig. 4). However, the autophagic marker, i.e., the ratio of LC3-II to LC3-I, did not change significantly following rapamycin treatment in both C57 and db/db hearts (Fig. 4). These results are consistent with previous studies showing mTORC1-regulated autophagy is resistant to inhibition by rapamycin in mammalian cells (34–36). Thus it appears that autophagy may not be significantly regulated by rapamycin treatment in the diabetic heart.
Our results also show that rapamycin treatment attenuates cardiac dysfunction in T2D mice, possibly by modifying contractile protein expression. Studies in humans and animal models of DM have demonstrated abnormal myofilament function (37) and impaired excitation-contraction coupling (38), which may depress myocardial function. Because diabetes is associated with a differential expression of myosin isozymes in the heart (39, 40), we utilized a proteomic approach with 2D-DIGE and MALDI-TOF/TOF-tandem mass spectrometry and identified four cytoskeletal/contractile proteins in db/db mouse hearts that were significantly altered after chronic treatment with rapamycin (Table 1). We found a coordinated down-regulation of a key contractile/cytoskeletal protein, MHC6α, in the db/db hearts, but up-regulation of MLC-2 (Table 1 and Fig. 8). Interestingly, chronic rapamycin treatment reversed the alteration of the cardiac expression of myosin isozymes. We also identified that the expression of cMyBP-C, a regulatory myofilament protein associated with the thick filament, is significantly reduced in T2D hearts (Table 1). Our data are consistent with reduced expression of MHC6α and MyBP-C in the hearts of diabetic mice (41) and rats (42). Previous studies reported that mutations in the MyBP-C gene frequently cause hypertrophic cardiomyopathy (43, 44), whereas its absence (cMyBP-C null mice) significantly attenuated cardiac function (45). Contractile abnormalities were also found in myosin-binding protein C (Mybpc3)-targeted knock-in mice with significant reduction in the cardiac expression of cMyBP-C (46). Interestingly, in the present study, rapamycin treatment enhanced the expression of cMyBP-C and also a thin filament-associated protein, nebulette (Table 1). Nebulette encodes a sarcomeric Z-disk protein that contributes to muscle force generation via its interaction with actin and cardiac tropomyosin-troponin complex, and participating in force transmission of the cardiac myofibril via the Z-disk assembly (47). Transgenic mice with cardiomyocyte-restricted expression of human mutant nebulette develop dilated cardiomyopathy at 6 months of age (48). Although rapamycin is a mTORC1 inhibitor, it potently inhibits only the phosphorylation of S6K, but not 4E-BP1 and cap-dependent translation, in mammalian cells (35, 36, 49). Therefore, rapamycin may not completely inhibit protein synthesis in the mammalian system. Alteration of contractile proteins after rapamycin treatment could be regulated by modification of other inhibitory proteins or other small molecules, such as microRNAs. Future investigations are needed to address this issue.
Increased production of reactive oxygen species in the diabetic heart is a contributing factor in the development and the progression of diabetic cardiomyopathy (50). Strategies that either reduce reactive oxygen species or augment myocardial antioxidant defense mechanisms have been shown to be efficacious in reducing diabetes-induced myocardial dysfunction (51–54). To study the effect of rapamycin in T2D-induced oxidative stress, we determined the levels of glutathione, a marker of oxidative stress. Our results show that GSSG was increased in the hearts of db/db mice, with a slight decline in GSH, as compared with the nondiabetic wild type mice (Fig. 10). Interestingly, rapamycin treatment reduced GSSG levels in the db/db mice and significantly reversed the decrease in GSH/oxidized glutathione ratio. The lipid peroxidation product MDA was also reduced in the T2D hearts following rapamycin treatment. However, rapamycin treatment did not alter cardiac level of GSH/oxidized glutathione or MDA in nondiabetic C57 mice, suggesting that the antioxidant effect of rapamycin is functional only under high oxidative stress in the diabetic heart. It is interesting that the previous studies also documented the antioxidant-like effect of rapamycin in different organs. For instance, this drug protects the mitochondria against oxidative stress and apoptosis in a rat model of Parkinson disease (55), improves endothelial function (56) and vascular contractility (57), and protects human corneal endothelial cells (58) by reducing oxidative stress. The potent antioxidant-like effect of rapamycin in T2D hearts suggests that this drug may have complimentary benefit in addition to mTORC1 inhibition for protection against myocardial dysfunction in T2D mice.
In the present study, the proteomic results identified the mitochondrial antioxidant enzyme PRX-5, which was enhanced in the diabetic heart, as compared with wild type heart. Interestingly, PRX-5 was further augmented following chronic rapamycin treatment (Table 1). We also confirmed its expression following rapamycin treatment in the diabetic heart by Western blot analysis (Fig. 7). PRXs are distributed in the cytosol, mitochondria, peroxisomes, and plasma and reduce hydrogen peroxide (59). Human mitochondrial PRX-5 protects from mitochondrial DNA damage induced by hydrogen peroxide (60). Our results also show the induction of SOD2 following rapamycin treatment in nondiabetic heart (Fig. 7). However, the expression of SOD2 was higher in T2D heart as compared with wild type mice and did not increase further after rapamycin treatment. The enhanced expression of SOD2 and PRX-5 in db/db hearts appears to be an adaptive response to oxidative stress associated with this T2D model. These results suggest that rapamycin treatment reduced oxidative stress in diabetic heart by augmenting the expression of PRX-5, but not SOD2.
Another powerful strategy to overcome reactive oxygen species toxicity is to limit the cellular availability of transition metals, most notably iron (61). Iron in the heme is necessary for the transport, binding, and release of oxygen and is essential to organism survival. However, it also donates electrons for the generation of superoxide radicals and participates in Fenton and Haber-Weiss reactions leading to formation of hydroxyl radical (61–63). An excess of free iron must be detoxified by sequestration in ferritin, the major intracellular iron storage protein (64). Ferritin has a high capacity to store free iron and serves to sequester and detoxify excess iron by oxidizing Fe(II) to Fe(III). It consists of two different subunits, ferritin heavy chain (FHC) and light chain. Only the FHC subunit has ferroxidase activity. A decrease in the abundance of FHC increases the levels of iron deposition and oxidative stress, which leads to cardiomyocytes cell death in failing hearts following ischemia or pressure overload (63). For the first time, we identified the induction of FHC in T2D mice as compared with nondiabetic mice, which was further intensified following rapamycin treatment (Table 1). Western blot confirmed rapamycin-mediated induction of FHC in diabetic heart (Fig. 6). Transferrins are iron-binding glycoproteins that also control the level of free iron in biological fluids. Transferrin imbalance, particularly its deficiency, can have serious health effects including the heart failure. In the present study, transferrin was also increased in the rapamycin-treated diabetic hearts. Taken together, these data suggest that rapamycin inhibits oxidative stress by increased expression of the antioxidant enzymes as well as iron-regulating proteins in the diabetic heart.
The proteomic study identified the alteration of three proteins involved in glucose metabolism following rapamycin treatment in the T2D hearts (Table 1). Pgm2 catalyzes the conversion of glucose 1-phosphate to the glycolytic intermediate glucose 6-phosphate. Although Pgm2 was reduced in the db/db hearts, we observed its complete recovery following rapamycin treatment. PDH E1α, which transforms pyruvate into acetyl-coA, did not change in the db/db hearts as compared with nondiabetic hearts but was significantly reduced following rapamycin treatment (Table 1 and Fig. 9). Chronic inhibition of PDH with the cardiac-specific overexpression of pyruvate dehydrogenase inhibitor, pyruvate dehydrogenase kinase 4, triggered an adaptive metabolic response in MHC-PDK4 transgenic mice (65). MHC-PDK4 transgenic mice were also resistant to high fat diet-induced cardiomyocyte lipid accumulation and also exhibited normal functional recovery after myocardial ischemia-reperfusion injury. This adaptive metabolic reprogramming is involved with the activation of AMP-activated protein kinase and the transcriptional co-activator, PGC-1α. The proteomic results also showed that rapamycin treatment significantly induced PYGB proteins (Table 1). The alterations of PDH E1α and PYGB were confirmed by Western blot analysis (Fig. 9).
We also identified the induction of carbonic anhydrase I and II (CA I and CA II) in T2D hearts following rapamycin treatment (Table 1). CA catalyzes CO2 hydration and the reverse reaction between H+ and HCO3− ions, which helps in the maintenance of intracellular pH (66, 67). CA II deficiency in humans is an autosomal recessive disease characterized by renal tubular acidosis, osteopetrosis, cerebral calcification, and growth retardation (68, 69). However, previous studies reported that CA II promotes cardiomyocyte hypertrophy and its inhibition reverts hypertrophy (70, 71). Further studies are required to understand the specific function of CA in rapamycin-treated diabetic hearts.
CoA is an essential cofactor for many metabolic pathways, including the major energy-producing pathways in the heart (72). Pantothenate kinase catalyzes pantothenic acid to CoA. Because insulin is a strong inhibitor of CoA synthesis, the CoA level was increased in diabetic hearts (73), which can also inhibit pantothenate kinase (72). Our proteomic approach revealed the induction of pantothenate kinase 4 in the rapamycin-treated T2D hearts (Table 1). However, the level of pantothenate kinase 4 was not altered in control db/db hearts as compared with nondiabetic heart. Similarly, we identified the induction of few other proteins in the rapamycin-treated T2D heart, i.e., β-prolyl 4-hydroxylase, β-hemoglobin, α-fetoprotein, and fibrinogens. Heat shock protein β-7 and serine proteinase inhibitor were reduced in the rapamycin-treated T2D hearts.
In the present study, we observed rapamycin-induced reduction in body weight, plasma glucose, triglyceride, and insulin levels in the db/db mice. However, rapamycin did not alter body weight and metabolic parameters or improve cardiac function in nondiabetic C57 mice. Even though these results would suggest that reduced body weight and improved metabolic status might have contributed to improved cardiac function with rapamycin treatment in diabetic mice, the proteomic data revealed that rapamycin altered contractile proteins, metabolic proteins, and antioxidant proteins only in the diabetic hearts. Therefore, body weight reduction or metabolic improvement may not be the sole reason for improvement of cardiac function with rapamycin treatment in diabetic mice. It has also been reported recently that long term administration of rapamycin to female db/db mice diminished obesity, without affecting food intake, and improved whole body insulin sensitivity (74). However, this phenotypic alteration following rapamycin treatment was not noticed in nondiabetic mice (db/+). It was suggested that the concurrent inhibition of leptin and mTOR signaling causes beneficial effects in rapamycin-fed db/db mice in terms of fat utilization and insulin signaling.
In summary, our results provide direct evidence that mTOR inhibition with rapamycin significantly improved the metabolic status as well as cardiac function in T2D mice. Rapamycin treatment also reduced oxidative stress and induced changes in several cytoskeletal/contractile proteins including MLY2, myosin heavy chain 6, myosin-binding protein C, glucose metabolism proteins (pyruvate dehydrogenase E1, PYGB, Pgm2), and antioxidant proteins (peroxiredoxin 5, ferritin heavy chain 1) that provide novel insights into the critical mechanisms by which rapamycin improves cardiac function in the T2D hearts. Based on these results, we propose that low dose rapamycin treatment may be a promising therapeutic modality in overcoming metabolic abnormalities and prevention of diabetic cardiomyopathy.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 HL51045, HL79424, HL93685, and HL118808 (to R. C. K.). This work was also supported by American Heart Association National Scientist Development Grant 10SDG3770011 (to F. N. S.) and A.D. Williams' Fund of Virginia Commonwealth University Grant UL1RR031990 (to A. D.).
- DM
- diabetes mellitus
- mTOR
- mammalian target of rapamycin
- mTORC
- mTOR complex
- T2D
- type 2 diabetes
- LV
- Left ventricular
- 2D-DIGE
- two-dimensional differential in-gel electrophoresis
- MDA
- malondialdehyde
- AKT
- protein kinase B
- MLC-2
- myosin light chain 2
- MHC6α
- myosin heavy polypeptide 6α
- cMyBP-C
- cardiac myosin-binding protein C
- PRX-5
- peroxiredoxin 5
- PDH
- pyruvate dehydrogenase
- PYGB
- glycogen phosphorylase
- Pgm2
- glucose phosphomutase
- CA
- carbonic anhydrase
- SOD2
- superoxide dismutase 2
- RAPA
- rapamycin
- GSSG
- oxidized glutathione
- FHC
- ferritin heavy chain.
REFERENCES
- 1. Plutzky J. (2011) Macrovascular effects and safety issues of therapies for type 2 diabetes 2. Am. J. Cardiol. 108, 25B–32B [DOI] [PubMed] [Google Scholar]
- 2. Adameova A., Dhalla N. S. (2014) Role of microangiopathy in diabetic cardiomyopathy. Heart Fail. Rev. 19, 25–33 [DOI] [PubMed] [Google Scholar]
- 3. Chong Z. Z., Maiese K. (2012) Mammalian target of rapamycin signaling in diabetic cardiovascular disease. Cardiovasc. Diabetol. 11, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ernande L., Derumeaux G. (2012) Diabetic cardiomyopathy. Myth or reality? Arch. Cardiovasc. Dis. 105, 218–225 [DOI] [PubMed] [Google Scholar]
- 5. Yang H., Jin X., Kei Lam C. W., Yan S. K. (2011) Oxidative stress and diabetes mellitus. Clin. Chem. Lab Med. 49, 1773–1782 [DOI] [PubMed] [Google Scholar]
- 6. Meric-Bernstam F., Gonzalez-Angulo A. M. (2009) Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 27, 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin D. E., Hall M. N. (2005) The expanding TOR signaling network. Curr. Opin. Cell Biol. 17, 158–166 [DOI] [PubMed] [Google Scholar]
- 8. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 9. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 10. Khan S., Salloum F., Das A., Xi L., Vetrovec G. W., Kukreja R. C. (2006) Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. J. Mol. Cell Cardiol. 41, 256–264 [DOI] [PubMed] [Google Scholar]
- 11. Das A., Salloum F. N., Durrant D., Ockaili R., Kukreja R. C. (2012) Rapamycin protects against myocardial ischemia-reperfusion injury through JAK2-STAT3 signaling pathway. J. Mol. Cell Cardiol. 53, 858–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao X. M., Wong G., Wang B., Kiriazis H., Moore X. L., Su Y. D., Dart A., Du X. J. (2006) Inhibition of mTOR reduces chronic pressure-overload cardiac hypertrophy and fibrosis. J. Hypertens. 24, 1663–1670 [DOI] [PubMed] [Google Scholar]
- 13. Völkers M., Konstandin M. H., Doroudgar S., Toko H., Quijada P., Din S., Joyo A., Ornelas L., Samse K., Thuerauf D. J., Gude N., Glembotski C. C., Sussman M. A. (2013) Mechanistic target of rapamycin complex 2 protects the heart from ischemic damage. Circulation 128, 2132–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khamzina L., Veilleux A., Bergeron S., Marette A. (2005) Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats. Possible involvement in obesity-linked insulin resistance. Endocrinology 146, 1473–1481 [DOI] [PubMed] [Google Scholar]
- 15. Drake J. C., Alway S. E., Hollander J. M., Williamson D. L. (2010) AICAR treatment for 14 days normalizes obesity-induced dysregulation of TORC1 signaling and translational capacity in fasted skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R1546–R1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ranieri S. C., Fusco S., Panieri E., Labate V., Mele M., Tesori V., Ferrara A. M., Maulucci G., De Spirito M., Martorana G. E., Galeotti T., Pani G. (2010) Mammalian life-span determinant p66shcA mediates obesity-induced insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 107, 13420–13425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 18. Sung M. M., Koonen D. P., Soltys C. L., Jacobs R. L., Febbraio M., Dyck J. R. (2011) Increased CD36 expression in middle-aged mice contributes to obesity-related cardiac hypertrophy in the absence of cardiac dysfunction. J. Mol. Med. 89, 459–469 [DOI] [PubMed] [Google Scholar]
- 19. Turdi S., Kandadi M. R., Zhao J., Huff A. F., Du M., Ren J. (2011) Deficiency in AMP-activated protein kinase exaggerates high fat diet-induced cardiac hypertrophy and contractile dysfunction. J. Mol. Cell Cardiol. 50, 712–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Inoki K., Mori H., Wang J., Suzuki T., Hong S., Yoshida S., Blattner S. M., Ikenoue T., Rüegg M. A., Hall M. N., Kwiatkowski D. J., Rastaldi M. P., Huber T. B., Kretzler M., Holzman L. B., Wiggins R. C., Guan K. L. (2011) mTORC1 activation in podocytes is a critical step in the development of diabetic nephropathy in mice. J. Clin. Invest. 121, 2181–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mori H., Inoki K., Masutani K., Wakabayashi Y., Komai K., Nakagawa R., Guan K. L., Yoshimura A. (2009) The mTOR pathway is highly activated in diabetic nephropathy and rapamycin has a strong therapeutic potential. Biochem. Biophys. Res. Commun. 384, 471–475 [DOI] [PubMed] [Google Scholar]
- 22. Baumgart D., Klauss V., Baer F., Hartmann F., Drexler H., Motz W., Klues H., Hofmann S., Völker W., Pfannebecker T., Stoll H. P., Nickenig G. (2007) One-year results of the SCORPIUS study. A German multicenter investigation on the effectiveness of sirolimus-eluting stents in diabetic patients. J. Am. Coll. Cardiol. 50, 1627–1634 [DOI] [PubMed] [Google Scholar]
- 23. de Waha A., Dibra A., Kufner S., Baumgart D., Sabate M., Maresta A., Schömig A., Kastrati A. (2011) Long-term outcome after sirolimus-eluting stents versus bare metal stents in patients with diabetes mellitus. A patient-level meta-analysis of randomized trials. Clin. Res. Cardiol. 100, 561–570 [DOI] [PubMed] [Google Scholar]
- 24. Koka S., Xi L., Kukreja R. C. (2012) Chronic treatment with long acting phosphodiesterase-5 inhibitor tadalafil alters proteomic changes associated with cytoskeletal rearrangement and redox regulation in Type 2 diabetic hearts. Basic Res. Cardiol. 107, 249. [DOI] [PubMed] [Google Scholar]
- 25. Xi L., Zhu S. G., Hobbs D. C., Kukreja R. C. (2011) Identification of protein targets underlying dietary nitrate-induced protection against doxorubicin cardiotoxicity. J. Cell Mol. Med. 15, 2512–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salloum F. N., Abbate A., Das A., Houser J. E., Mudrick C. A., Qureshi I. Z., Hoke N. N., Roy S. K., Brown W. R., Prabhakar S., Kukreja R. C. (2008) Sildenafil (Viagra) attenuates ischemic cardiomyopathy and improves left ventricular function in mice. Am. J. Physiol. Heart Circ. Physiol. 294, H1398–H1406 [DOI] [PubMed] [Google Scholar]
- 27. Lamming D. W., Ye L., Katajisto P., Goncalves M. D., Saitoh M., Stevens D. M., Davis J. G., Salmon A. B., Richardson A., Ahima R. S., Guertin D. A., Sabatini D. M., Baur J. A. (2012) Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 29. Mellor K. M., Bell J. R., Young M. J., Ritchie R. H., Delbridge L. M. (2011) Myocardial autophagy activation and suppressed survival signaling is associated with insulin resistance in fructose-fed mice. J. Mol. Cell Cardiol. 50, 1035–1043 [DOI] [PubMed] [Google Scholar]
- 30. Mellor K. M., Reichelt M. E., Delbridge L. M. (2011) Autophagy anomalies in the diabetic myocardium. Autophagy. 7, 1263–1267 [DOI] [PubMed] [Google Scholar]
- 31. Dutta D., Xu J., Kim J. S., Dunn W. A., Jr., Leeuwenburgh C. (2013) Upregulated autophagy protects cardiomyocytes from oxidative stress-induced toxicity. Autophagy 9, 328–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. French C. J., Tarikuz Zaman A., McElroy-Yaggy K. L., Neimane D. K., Sobel B. E. (2011) Absence of altered autophagy after myocardial ischemia in diabetic compared with nondiabetic mice. Coron. Artery Dis. 22, 479–483 [DOI] [PubMed] [Google Scholar]
- 33. Mellor K. M., Bell J. R., Ritchie R. H., Delbridge L. M. (2013) Myocardial insulin resistance, metabolic stress and autophagy in diabetes. Clin. Exp. Pharmacol. Physiol. 40, 56–61 [DOI] [PubMed] [Google Scholar]
- 34. Thoreen C. C., Sabatini D. M. (2009) Rapamycin inhibits mTORC1, but not completely. Autophagy 5, 725–726 [DOI] [PubMed] [Google Scholar]
- 35. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., Sabatini D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jweied E. E., McKinney R. D., Walker L. A., Brodsky I., Geha A. S., Massad M. G., Buttrick P. M., de Tombe P. P. (2005) Depressed cardiac myofilament function in human diabetes mellitus. Am. J. Physiol. Heart Circ. Physiol. 289, H2478–H2483 [DOI] [PubMed] [Google Scholar]
- 38. Pereira L., Matthes J., Schuster I., Valdivia H. H., Herzig S., Richard S., Gómez A. M. (2006) Mechanisms of [Ca2+]i transient decrease in cardiomyopathy of db/db type 2 diabetic mice. Diabetes 55, 608–615 [DOI] [PubMed] [Google Scholar]
- 39. Dillmann W. H. (1980) Diabetes mellitus induces changes in cardiac myosin of the rat. Diabetes 29, 579–582 [DOI] [PubMed] [Google Scholar]
- 40. Dillmann W. H. (1986) Diabetes mellitus and hypothyroidism induce changes in myosin isoenzyme distribution in the rat heart–do alterations in fuel flux mediate these changes? Adv. Exp. Med. Biol. 194, 469–479 [DOI] [PubMed] [Google Scholar]
- 41. Essop M. F., Chan W. A., Hattingh S. (2011) Proteomic analysis of mitochondrial proteins in a mouse model of type 2 diabetes. Cardiovasc. J. Afr. 22, 175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Howarth F. C., Qureshi M. A., Hassan Z., Al Kury L. T., Isaev D., Parekh K., Yammahi S. R., Oz M., Adrian T. E., Adeghate E. (2011) Changing pattern of gene expression is associated with ventricular myocyte dysfunction and altered mechanisms of Ca2+ signalling in young type 2 Zucker diabetic fatty rat heart. Exp. Physiol. 96, 325–337 [DOI] [PubMed] [Google Scholar]
- 43. Pohlmann L., Kröger I., Vignier N., Schlossarek S., Krämer E., Coirault C., Sultan K. R., El-Armouche A., Winegrad S., Eschenhagen T., Carrier L. (2007) Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ. Res. 101, 928–938 [DOI] [PubMed] [Google Scholar]
- 44. Vignier N., Schlossarek S., Fraysse B., Mearini G., Krämer E., Pointu H., Mougenot N., Guiard J., Reimer R., Hohenberg H., Schwartz K., Vernet M., Eschenhagen T., Carrier L. (2009) Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ. Res. 105, 239–248 [DOI] [PubMed] [Google Scholar]
- 45. Brickson S., Fitzsimons D. P., Pereira L., Hacker T., Valdivia H., Moss R. L. (2007) In vivo left ventricular functional capacity is compromised in cMyBP-C null mice. Am. J. Physiol. Heart Circ. Physiol. 292, H1747–H1754 [DOI] [PubMed] [Google Scholar]
- 46. Stöhr A., Friedrich F. W., Flenner F., Geertz B., Eder A., Schaaf S., Hirt M. N., Uebeler J., Schlossarek S., Carrier L., Hansen A., Eschenhagen T. (2013) Contractile abnormalities and altered drug response in engineered heart tissue from Mybpc3-targeted knock-in mice. J. Mol. Cell Cardiol. 63, 189–198 [DOI] [PubMed] [Google Scholar]
- 47. Maiellaro-Rafferty K., Wansapura J. P., Mendsaikhan U., Osinska H., James J. F., Taylor M. D., Robbins J., Kranias E. G., Towbin J. A., Purevjav E. (2013) Altered regional cardiac wall mechanics are associated with differential cardiomyocyte calcium handling due to nebulette mutations in preclinical inherited dilated cardiomyopathy. J. Mol. Cell Cardiol. 60, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Purevjav E., Varela J., Morgado M., Kearney D. L., Li H., Taylor M. D., Arimura T., Moncman C. L., McKenna W., Murphy R. T., Labeit S., Vatta M., Bowles N. E., Kimura A., Boriek A. M., Towbin J. A. (2010) Nebulette mutations are associated with dilated cardiomyopathy and endocardial fibroelastosis. J. Am. Coll. Cardiol. 56, 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang B. T., Ducker G. S., Barczak A. J., Barbeau R., Erle D. J., Shokat K. M. (2011) The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc. Natl. Acad. Sci. U.S.A. 108, 15201–15206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boudina S., Abel E. D. (2007) Diabetic cardiomyopathy revisited. Circulation 115, 3213–3223 [DOI] [PubMed] [Google Scholar]
- 51. Cai L., Wang Y., Zhou G., Chen T., Song Y., Li X., Kang Y. J. (2006) Attenuation by metallothionein of early cardiac cell death via suppression of mitochondrial oxidative stress results in a prevention of diabetic cardiomyopathy. J. Am. Coll. Cardiol. 48, 1688–1697 [DOI] [PubMed] [Google Scholar]
- 52. Matsushima S., Kinugawa S., Ide T., Matsusaka H., Inoue N., Ohta Y., Yokota T., Sunagawa K., Tsutsui H. (2006) Overexpression of glutathione peroxidase attenuates myocardial remodeling and preserves diastolic function in diabetic heart. Am. J. Physiol. Heart Circ. Physiol. 291, H2237–H2245 [DOI] [PubMed] [Google Scholar]
- 53. Matsushima S., Ide T., Yamato M., Matsusaka H., Hattori F., Ikeuchi M., Kubota T., Sunagawa K., Hasegawa Y., Kurihara T., Oikawa S., Kinugawa S., Tsutsui H. (2006) Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation 113, 1779–1786 [DOI] [PubMed] [Google Scholar]
- 54. Shen X., Zheng S., Metreveli N. S., Epstein P. N. (2006) Protection of cardiac mitochondria by overexpression of MnSOD reduces diabetic cardiomyopathy. Diabetes 55, 798–805 [DOI] [PubMed] [Google Scholar]
- 55. Jiang J., Jiang J., Zuo Y., Gu Z. (2013) Rapamycin protects the mitochondria against oxidative stress and apoptosis in a rat model of Parkinson's disease. Int. J. Mol. Med. 31, 825–832 [DOI] [PubMed] [Google Scholar]
- 56. Rajapakse A. G., Yepuri G., Carvas J. M., Stein S., Matter C. M., Scerri I., Ruffieux J., Montani J. P., Ming X. F., Yang Z. (2011) Hyperactive S6K1 mediates oxidative stress and endothelial dysfunction in aging. Inhibition by resveratrol. PLoS One 6, e19237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao G., Li J. J., Li Y., Li D., Wang Y., Wang L., Tang X. D., Walsh M. P., Gui Y., Zheng X. L. (2011) Rapamycin inhibits hydrogen peroxide-induced loss of vascular contractility. Am. J. Physiol. Heart Circ. Physiol. 300, H1583–H1594 [DOI] [PubMed] [Google Scholar]
- 58. Shin Y. J., Cho D. Y., Chung T. Y., Han S. B., Hyon J. Y., Wee W. R. (2011) Rapamycin reduces reactive oxygen species in cultured human corneal endothelial cells. Curr. Eye Res. 36, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 59. Fujii J., Ikeda Y. (2002) Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox. Rep. 7, 123–130 [DOI] [PubMed] [Google Scholar]
- 60. Banmeyer I., Marchand C., Clippe A., Knoops B. (2005) Human mitochondrial peroxiredoxin 5 protects from mitochondrial DNA damages induced by hydrogen peroxide. FEBS Lett. 579, 2327–2333 [DOI] [PubMed] [Google Scholar]
- 61. Pham C. G., Bubici C., Zazzeroni F., Papa S., Jones J., Alvarez K., Jayawardena S., De Smaele E., Cong R., Beaumont C., Torti F. M., Torti S. V., Franzoso G. (2004) Ferritin heavy chain upregulation by NF-B inhibits TNFα-induced apoptosis by suppressing reactive oxygen species. Cell 119, 529–542 [DOI] [PubMed] [Google Scholar]
- 62. Kukreja R. C., Hess M. L. (1992) The oxygen free radical system. From equations through membrane-protein interactions to cardiovascular injury and protection. Cardiovasc. Res. 26, 641–655 [DOI] [PubMed] [Google Scholar]
- 63. Omiya S., Hikoso S., Imanishi Y., Saito A., Yamaguchi O., Takeda T., Mizote I., Oka T., Taneike M., Nakano Y., Matsumura Y., Nishida K., Sawa Y., Hori M., Otsu K. (2009) Downregulation of ferritin heavy chain increases labile iron pool, oxidative stress and cell death in cardiomyocytes. J. Mol. Cell Cardiol. 46, 59–66 [DOI] [PubMed] [Google Scholar]
- 64. Torti F. M., Torti S. V. (2002) Regulation of ferritin genes and protein. Blood 99, 3505–3516 [DOI] [PubMed] [Google Scholar]
- 65. Chambers K. T., Leone T. C., Sambandam N., Kovacs A., Wagg C. S., Lopaschuk G. D., Finck B. N., Kelly D. P. (2011) Chronic inhibition of pyruvate dehydrogenase in heart triggers an adaptive metabolic response. J. Biol. Chem. 286, 11155–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schroeder M. A., Ali M. A., Hulikova A., Supuran C. T., Clarke K., Vaughan-Jones R. D., Tyler D. J., Swietach P. (2013) Extramitochondrial domain rich in carbonic anhydrase activity improves myocardial energetics. Proc. Natl. Acad. Sci. U.S.A. 110, E958–E967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sterling D., Reithmeier R. A., Casey J. R. (2001) A transport metabolon. Functional interaction of carbonic anhydrase II and chloride/bicarbonate exchangers. J. Biol. Chem. 276, 47886–47894 [DOI] [PubMed] [Google Scholar]
- 68. Lai L. W., Chan D. M., Erickson R. P., Hsu S. J., Lien Y. H. (1998) Correction of renal tubular acidosis in carbonic anhydrase II-deficient mice with gene therapy. J. Clin. Invest. 101, 1320–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sly W. S., Hewett-Emmett D., Whyte M. P., Yu Y. S., Tashian R. E. (1983) Carbonic anhydrase II deficiency identified as the primary defect in the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. Proc. Natl. Acad. Sci. U.S.A. 80, 2752–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alvarez B. V., Johnson D. E., Sowah D., Soliman D., Light P. E., Xia Y., Karmazyn M., Casey J. R. (2007) Carbonic anhydrase inhibition prevents and reverts cardiomyocyte hypertrophy. J. Physiol. 579, 127–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brown B. F., Quon A., Dyck J. R., Casey J. R. (2012) Carbonic anhydrase II promotes cardiomyocyte hypertrophy. Can. J. Physiol. Pharmacol. 90, 1599–1610 [DOI] [PubMed] [Google Scholar]
- 72. Robishaw J. D., Berkich D., Neely J. R. (1982) Rate-limiting step and control of coenzyme A synthesis in cardiac muscle. J. Biol. Chem. 257, 10967–10972 [PubMed] [Google Scholar]
- 73. Reibel D. K., Wyse B. W., Berkich D. A., Neely J. R. (1981) Regulation of coenzyme A synthesis in heart muscle. Effects of diabetes and fasting. Am. J. Physiol. 240, H606–H611 [DOI] [PubMed] [Google Scholar]
- 74. Deepa S. S., Walsh M. E., Hamilton R. T., Pulliam D., Shi Y., Hill S., Li Y., Remmen H. V. (2013) Rapamycin modulates markers of mitochondrial biogenesis and fatty acid oxidation in the adipose tissue of db/db mice. J. Biochem. Pharmacol. Res. 1, 114–123 [PMC free article] [PubMed] [Google Scholar]