Background: Sar1 mediates the onward transport of ER cargo.
Results: Sar1B promotes VLDL secretion, whereas Sar1A antagonizes this activity, and a deficit of both reduces cholesterol biosynthesis.
Conclusion: Sar1B independently of and through its lipoprotein secretion function promotes the expression of genes regulating cholesterol biosynthesis.
Significance: Sar1B-mediated transport activities contribute to both the functional integrity of the ER membrane and blood cholesterol levels.
Keywords: Apolipoproteins, Cholesterol Regulation, Endoplasmic Reticulum (ER), Lipoprotein Secretion, Transcriptomics
Abstract
Triglycerides and cholesterol are essential for life in most organisms. Triglycerides serve as the principal energy storage depot and, where vascular systems exist, as a means of energy transport. Cholesterol is essential for the functional integrity of all cellular membrane systems. The endoplasmic reticulum is the site of secretory lipoprotein production and de novo cholesterol synthesis, yet little is known about how these activities are coordinated with each other or with the activity of the COPII machinery, which transports endoplasmic reticulum cargo to the Golgi. The Sar1B component of this machinery is mutated in chylomicron retention disorder, indicating that this Sar1 isoform secures delivery of dietary lipids into the circulation. However, it is not known why some patients with chylomicron retention disorder develop hepatic steatosis, despite impaired intestinal fat malabsorption, and why very severe hypocholesterolemia develops in this condition. Here, we show that Sar1B also promotes hepatic apolipoprotein (apo) B lipoprotein secretion and that this promoting activity is coordinated with the processes regulating apoB expression and the transfer of triglycerides/cholesterol moieties onto this large lipid transport protein. We also show that although Sar1A antagonizes the lipoprotein secretion-promoting activity of Sar1B, both isoforms modulate the expression of genes encoding cholesterol biosynthetic enzymes and the synthesis of cholesterol de novo. These results not only establish that Sar1B promotes the secretion of hepatic lipids but also adds regulation of cholesterol synthesis to Sar1B's repertoire of transport functions.
Introduction
Whole-body triglyceride and cholesterol homeostasis are of fundamental importance to human health. Their failure manifests in multiple diseases, ranging from life-threatening conditions in infancy (1–4), to severe coronary heart disease in young adults (5, 6), and to indolent disorders of middle and old age (7–11). In higher organisms, the ability of tissues to accommodate large fluctuations in dietary triglyceride and cholesterol intake is well developed, involving cross-talk between the cellular processes that govern the delivery of these lipids into the circulation via triglyceride-rich, apoB-containing lipoproteins, their uptake, de novo lipogenesis, and cholesterol biosynthesis (12). From the study of familial hypobetalipoproteinemia (OMIM +107730) and abetalipoproteinemia (OMIM 200100), it is evident that both apolipoprotein (apo)B and the microsomal triglyceride transfer protein (MTTP)4 are obligatory for the production of chylomicrons and very low density lipoproteins (VLDL) within the endoplasmic reticulum (ER) of enterocytes and hepatocytes, respectively (1, 13–15). Moreover, human liver, in contrast to the intestine, uses apoB100 rather than the shorter apoB48 for exporting lipid into the circulation (12). Sar1B, a coat protein II (COPII) component, has also been shown to be central to the lipid economy by the discovery that its mutations cause the rare recessive disorder chylomicron retention disease (CMRD) (OMIM 246700) (2). However, it is not clear why this GTPase is obligatory for delivery of chylomicrons into the circulation and whether this relates to the usage of apoB48 rather than apoB100 for their production or insufficient Sar1A in the intestine of CMRD patients to compensate for loss of Sar1B function.
It is also not known why some CMRD children develop hepatic steatosis (16–18), despite severe intestinal fat malabsorption; and it is very odd that affected individuals develop severe hypocholesterolemia (2, 16–18), because therapeutic reduction of intestinal cholesterol absorption only modestly affects circulating cholesterol levels, due to compensatory rises in endogenous de novo cholesterol biosynthesis (12, 19, 20). Indeed, Western-type diets provide ∼400 mg of cholesterol per day, and our bodies synthesize ∼1 g de novo (21, 22). Hence, blood cholesterol levels reflect both dietary and endogenously synthesized cholesterol.
Cholesterol synthesis is a multistep reaction that is thought to occur in virtually all nucleated cells (23). In this context, it may be relevant that SAR1B expression has been detected in multiple tissues (2); most of the enzymes synthesizing cholesterol de novo reside in the ER membrane (24–28), including HMG-CoA reductase (29), the target of statins, a highly successful class of cholesterol-lowering drugs (30, 31). Hepatic biosynthesis may be especially sensitive to intestinal cholesterol absorption because of the liver's central position in directing cholesterol into VLDL or bile (32). After uptake by enterocytes, cholesterol is packed with triglycerides into chylomicrons and secreted into the lymph. In the circulation, the triglycerides are rapidly hydrolyzed, and the released glycerol and fatty acids taken up by peripheral tissues, whereas the cholesterol-enriched, highly atherogenic remnant particles are captured by the liver (33, 34). From internal cellular endosomal compartments, the cholesterol may be returned to the plasma membrane (35) or be transported to the ER (36, 37).
In ER membranes, cholesterol may suppress the activities of one or more cholesterol biosynthetic enzymes or activate acyl-CoA:cholesterol acyltransferase (ACAT) 2 (38–41). It may also impede binding between the COPII protein Sec24 and the sterol regulatory element-binding protein (SREBP) escort protein Scap (42, 43), thereby blocking export of the ER membrane-bound Scap-SREBP2 complex to the Golgi apparatus (44). Here, SREBP2 undergoes proteolytic cleavage, culminating in the release of its amino-terminal domain (45), known to transcriptionally activate genes regulating cholesterol homeostasis (46–50). However, it is not known whether the Sar1B component of the COPII transport machinery contributes to the ER export of SREBP2, de novo cholesterol synthesis, or the secretion of cholesteryl esters via VLDL.
Studies with Sar1 bound to nonhydrolysable analogues of GTP and a GTP-restricted form of Sar1 (i.e. Sar1:H79G) have defined several aspects of the COPII vesicle assembly process (51–54). In outline, Sar1-GDP is recruited to the cytosolic face of the ER membrane. Here, the guanine-nucleotide exchange protein Sec12 swaps the GDP on Sar1 for GTP. Next, the Sar1-GTP recruits Sec23/24 to the ER membrane to form a pre-budding complex that captures both ER cargo and Sec13/31 (55, 56), culminating in polymerization of the COPII coat and fission of the COPII vesicle from the ER, along with its captured cargo. This process, including the sorting of ER cargo into COPII pre-budding complexes, is coupled to the hydrolysis of Sar1's GTP; Sec12 continually reactivates Sar1, promoting continued pre-budding complex assembly and cargo capture, whereas unchecked GTP hydrolysis triggers coat disassembly (52–55).
In CMRD, most SAR1B mutations alter an amino acid in Sar1B's GDP/GTP binding pocket (2, 17, 18), suggesting that the intestinal fat malabsorption defect is attributable to breakdown/absence of the Sar1 GTPase cycle that normally secures capture of nascent chylomicrons into COPII pre-budding complexes and their subsequent onward transport. However, it is not known whether this breakdown relates to a specific aspect(s) of the apoB lipoprotein/COPII vesicle working relationship, and it is unclear to what extent Sar1B's homologue Sar1A can compensate for loss of Sar1B in this partnership. Additionally, there are no data regarding the contributions of the Sar1A- and Sar1B-GTPase cycles in securing VLDL secretion and cholesterol homeostasis.
Here, we demonstrate that ubiquitously expressed Sar1B (2) acts at two control points crucial to cholesterol metabolism. First, Sar1B promotes hepatic apoB100-lipoprotein secretion (as well as apoB48-lipoprotein secretion), and this is paired with expression changes in APOB and MTTP, but not of genes involved in triglyceride synthesis. The apoB lipoprotein secreting activity of Sar1B is antagonized by Sar1A. This is more pronounced for the more triglyceride-rich apoB lipoproteins, indicating that only Sar1B has the attributes to mediate the assembly of COPII pre-budding complexes of the right structures and composition to secure export of such nascent apoB lipoproteins and hence efficient delivery of triglyceride and cholesterol moieties into the circulation. Second, deficiency of Sar1B, but not Sar1A, in apoB lipoprotein-producing cells reduces ER export of Srebp2, whereas aggregate levels of Sar1 determine the rate of de novo cholesterol synthesis. The study indicates that Sar1B, both independently of and through its promotion of apoB-mediated lipid secretion activities, contributes to the regulation of cellular, as well as plasma, triglyceride and cholesterol levels.
EXPERIMENTAL PROCEDURES
Production of Stable Cell Lines, Knockdown, and Tissue Culture
Human SAR1A and SAR1B sequences were PCR-amplified from Marathon-ready human liver cDNA (Clontech) using gene-specific primers. FLAG epitope tags (Asp-Tyr-Lys-Asp4-Lys) were fused in-frame to the carboxyl termini of cDNAs and juxtaposed to a terminator codon. PCR and appropriate restriction sites were used to manipulate the SAR1 sequences. Site-directed mutagenesis was performed by a standard two-step PCR-based strategy (57). All constructs were sequenced before use. Transfection of pcDNA4/TO vectors into McArdle-RH7777 (58) and CHO (Invitrogen) cell lines stably transformed with pcDNA6/TR was performed with either FuGENE (Roche Applied Science) or Lipofectamine Plus Reagent (Invitrogen). Individual clones were picked, expanded, and screened for recombinant protein expression using serial dilutions of cell lysates, anti-FLAG M2 antibody (Sigma-Aldrich), a goat anti-mouse IgG horseradish peroxidase-conjugated secondary antibody (Bio-Rad), and Femto-enhanced chemiluminescence (Pierce). Blots were scanned on a flatbed scanner and immunoreactive products quantified using Adobe Photoshop. Cell lines (two per construct) were selected to have comparable levels of recombinant protein expression.
Knockdowns were performed with pre-designed siRNA (Qiagen) using HiPerFect transfection reagent (Qiagen) according to the manufacturer's fast forward protocol. Volumes of transfection reagents and concentrations of siRNA were optimized for each pre-designed FlexiTube siRNA (Qiagen). The best results were obtained using a final concentration of 5 nm siRNA. For the double-knockdown experiments, 5 nm of each isoform was used. The scrambled siRNA sequence was 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (Qiagen).
McArdle-RH7777 cells were cultured at 37 °C, 5% CO2 in DMEM containing 20% heat-inactivated FCS (Labtech International), 4500 mg/liter d-glucose, 2 mm l-glutamine (Invitrogen), and 1% penicillin/streptomycin (Invitrogen). CHO cell lines were cultured in Ham's F-12 media (Sigma) containing 10% heat-inactivated FCS, 2 mm l-glutamine, and 1% penicillin/streptomycin. Blasticidin (10 μg/ml; Sigma) and Zeocin (250 μg/ml; Invitrogen) were added to the media of cells stably transfected with the tetracycline repressor (pcDNA6/TR) and operon (pcDNA4/TO) vectors (Invitrogen).
Metabolic Labeling, Immunoprecipitations, and Sucrose Gradient Ultracentrifugation of ApoB Lipoproteins
Prior to labeling for 1 h with 125 Ci/ml l-[35S]methionine (ICN Flow) at 37 °C, cells were incubated with complete DMEM containing 1 μg/ml tetracycline between 24 and 48 h and then in serum-free media supplemented with 0.8 mm oleic acid (Sigma) complexed to 3% fatty acid-free BSA overnight and cysteine/methionine-free media (Invitrogen) containing oleate for 30 min. After removal of label, incubations were continued in serum-free media supplemented with 0.8 mm oleic acid complexed to 3% fatty acid-free BSA for 2 h. Conditioned media were cleared by low speed centrifugation and concentrated to 500 μl in Amicon Ultra-4 centrifugal filter units with Ultracel-30 membranes (Millipore). ApoB was immunoprecipitated by standard methodology (59, 60), using saturating concentrations of goat polyclonal antibody against human apoB (AB742; Chemicon International), as described (13, 59, 60). The radioactivity in apoB48 and apo100 was quantified by phosphorimager analysis. Three samples were collected per stable cell line per experiment.
For total protein secretion measurements, cells were incubated in methionine-free medium for 45 min at 37 °C and labeled for 1 h with 125 Ci/ml l-[35S]methionine (ICN Flow). After removal of label, incubations were continued in serum-free medium for 2 h. Cell media were harvested, mixed with protease inhibitors (Calbiochem), and cleared by low speed centrifugation. Proteins were precipitated by incubation with TCA for 15 min on ice. Precipitates were washed in ice-cold acetone and resuspended in I.P. buffer containing protease inhibitors (Calbiochem).
Densities of secreted 35S-labeled apoB lipoproteins were determined by gradient ultracentrifugation. The sucrose gradients were formed as described (13), except that the gradients were ultracentrifuged at 35,400 rpm in a Beckman SW41 rotor and unloaded in 12 fractions. ApoB was recovered from each fraction by immunoprecipitation and size-fractionated by SDS-PAGE, and the amount of radioactivity in apoB48 and full-length apoB100 was quantified by phosphorimager analysis. The density profile of the gradient was determined using an Abbe refractometer.
Immunoblot Analysis of Endogenous Protein Expression
Cell lysates were generated using RIPA buffer (Sigma) and a mixture of protease inhibitors (Roche Applied Science). Proteins were size-fractionated on either 10 or 4–12% gradient NuPage BisTris precast gels (Invitrogen) and transferred to nitrocellulose membranes (Whatman). Membranes were blocked in 5% (w/v) nonfat milk powder before probing with the specified antibodies. The antibodies used were as follows: rabbit polyclonal anti-Hmgcs1 antibody (Proteintech); anti-Lss antibody (Proteintech); anti-Dhcr7 antibody (Abcam); anti-ACSL5 (Proteintech); anti-Sec13 antibody (Abcam); anti-Sec31a antibody, anti-Sar1a antibody, anti-Sar1b antibody, and anti-Srebf2 antibody (Proteintech); anti-ATF6 (Abcam); and mouse monoclonal β-actin or β-tubulin antibodies (Sigma and Abcam). Immunoreactive products were visualized and quantified after labeling with appropriate fluorescent dye-labeled secondary antibodies (LI-COR Biosciences) using the Odyssey imaging system (LI-COR). Apparent molecular masses were estimated using the Novex Sharp Protein Standard (Invitrogen) and the band sizing application in Odyssey software (LI-COR Biosciences).
RT-qPCR Analyses in Cell Lines
Pre-designed gene primers were purchased from Qiagen (Quantitect primer assay), unless stated otherwise (supplemental Table S1). Total RNA was isolated from cell lysates using the RNeasyPlus kit (Qiagen). Cells were homogenized using QIAshredder columns (Qiagen). RNA samples were quantified by a NanoDrop 1000 spectrophotometer (Thermo Scientific). RT-qPCRs were performed in triplicate (with a no RT control) with SYBR Green kits from Qiagen or Stratagene, as recommended by the manufacturers. Data were analyzed with either SDS 2.3 software (Applied Biosystems) or MxPro (Stratagene). Gene expression levels, normalized to Ppia, were calculated by the relative 2−ΔΔCt method (61).
RNA Analyses of Human Small Intestine Biopsies and Liver Samples
After an overnight fast, gastroduodenoscopy was performed, and small intestinal (jejunal) biopsies were taken near the Treitz' ligament in 17 obese male subjects with no history of disease or any medication use (i.e. treatment-naive). To minimize variability in sample processing and handling, biopsy samples were collected immediately and snap-frozen in liquid nitrogen. All subsequent sample processing was performed by the same individual using the same standardized protocol. Tissue was homogenized in MagNA Pure LC RNA Isolation Tissue Lysis Buffer (Roche Applied Science), using a MagNALyser instrument (Roche Applied Science), and using parameters that were optimized on comparable samples prior to commencing the study. Total RNA was prepared using TRIzol according to the manufacturer's manual (Invitrogen), quantified by a NanoDrop 1000 spectrophotometer (Thermo Scientific), and reverse-transcribed into cDNA using a cDNA synthesis kit from Bio-Rad. RT-qPCR was performed in triplicate with 50 ng of cDNA, 200 nm of each primer, and 7.5 μl of SYBR Green mix (CE Biotech). PCRs were performed on a CFX system (Bio-Rad). Gene expression levels normalized to 36B4 were calculated by the relative 2−ΔΔCt method (61). The study was approved by the Research Ethics Committee at Amsterdam Medical Center, Amsterdam, The Netherlands, and all participants gave written informed consent.
RT-qPCR on human liver RNA (First Choice Human Total RNA, Applied Biosytems) was performed with a SYBR Green kit (Qiagen). Correlations between mRNA levels in human liver samples (62, 63) were computed using R2, a Genomics Analysis Visualization Platform. p values were corrected for multiple testing using the formula implemented in R2: t = R/sqrt(1 − R^2)/(n − 2), where R is the correlation coefficient, and n denotes the number of samples.
Microarray Gene Analyses
A minimum of three technical replicates were used for each of two Sar1B:H79G-FLAG cell lines, two control cell lines, two Sar1B-FLAG cell lines, and one Sar1A-FLAG cell line. Cells were cultured in complete DMEM containing blasticidin (10 μg/ml; Sigma) and Zeocin (250 μg/ml; Invitrogen). Twenty four hours prior to harvesting, 1 μg/ml tetracycline was added to the culture media. Total RNA was extracted with the RNeasy Plus mini kit (Qiagen). RNA quality was verified (e.g. flat baseline between 18 S and 28 S ribosomal RNA peaks; RIN score >7) using the RNA 6000 Ladder (Ambion), RNA 6000 NanoChip (Agilent), plus an Agilent 2100 bioanalyzer (Agilent Technologies, Germany). cDNA was synthesized from total RNA using the one-cycle eukaryotic target labeling assay kit (Invitrogen and Affymetrix). Double-stranded cDNA was purified with the GeneChip® sample cleanup module. Biotin-labeled cRNA was prepared using the BioArray High Yield RNA transcript labeling kit (Enzo) and purified with the in vitro transcription cRNA GeneChip® sample cleanup module. RNA quality was verified with the Agilent Bioanalyzer 2100 system and by hybridization to test arrays (Affymetrix). Biotinylated cRNA (20 μg) was hybridized to GeneChip rat genome 230 2.0 arrays (Affymetrix) containing 31,099 probe sets. All cRNA samples were prepared using the same standard protocol.
Data analyses were performed through the CARMAweb (Comprehensive R-based Microarray Analysis web service). Raw intensity values were normalized using the Loess normalization, Affymetrix MAS 5 and summarization algorithms. Differences in probe-set values were determined on the normalized data using the significance analysis of microarray (SAM) algorithm (64) at specified false discovery rates (FDR). These were determined using the “Benjamini and Hochberg” correction, with cutoff values for significance determined by a tuning parameter, the “Δ value.” The number of Δ values was set at 50. Data are available at NCBI Gene Expression Omnibus (GEO accession number GSE52969). The differentially expressed probe sets were examined for enrichment of functionally related Gene Ontology terms using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov) (65, 66).
Lipid Synthesis and Secretion
Cells were seeded at a density of 1 × 104 cells/cm2 in 9-cm plates and grown for 2 days in DMEM containing 20% FCS, 1% penicillin/streptomycin, and 1 mm glutamine at 37 °C under 5% CO2. Lipid synthesis was measured over the final 5 h of the incubation by the addition of 2 mm [14C]acetate (sodium salt; Sigma) (0.8 μCi/ml). Cell media were aspirated for analysis of lipid secretion. Cells were washed in PBS and harvested by trypsinization and centrifugation. Cell pellets were washed in PBS and lipids extracted by addition of 4 ml of chloroform/methanol (2:1). Water (1 ml) was added to the extract and mixed by vortexing. The suspension was centrifuged for 5 min, and the lower solvent layer was taken for analysis. The upper aqueous layer was re-extracted with a further 4 ml of chloroform/methanol, and the two extractions were combined. This was dried down under N2 and resuspended in a minimal volume of hexane (30 μl). Cell medium (4 ml) was extracted as above, using 16 ml of chloroform/methanol (2:1). Lipids were separated by thin layer chromatography on Silica Gel 60 plates (VWR) using petroleum ether/diethyl ether/acetic acid (60:40:1) as the mobile phase and visualized by iodine staining. Each run included standards of cholesterol, fatty acids, triglycerides and cholesterol esters. Lipid synthesis was measured by scraping of spots into vials followed by liquid scintillation counting. Plates were also visualized by autoradiography to determine that no major lipid product was missed.
Statistics
All data are expressed as means ± S.E. Statistical analyses were performed using unpaired Student's t test or one-way analysis of variance followed by the Bonferroni post hoc test using PASW Statistics version 18.0 software (SPSS, IBM), where appropriate. Figures were generated using Graphpad Prism 4.
RESULTS
Characterization of the Effects of Sar1 Isoforms on ApoB Lipoprotein Secretion
To establish whether Sar1B promotes only the secretion of apoB48-containing lipoproteins, we stably transfected McArdle-RH7777 cells that can secrete both apoB48 and apoB100 lipoproteins with constructs encoding Sar1A-FLAG, Sar1B-FLAG, or constitutively active Sar1-FLAG (i.e. Sar1A:H79G and Sar1B:H79G) isoforms that cannot hydrolyze GTP to support completion of COPII vesicle assembly (67). Despite several attempts, we failed to stably transfect McArdle-RH7777 cells with Sar1A:H79G, potentially because Sar1a mRNA is the predominant species expressed in these cells (11.6-fold higher than Sar1b). The Sar1A cell lines contained 5.26 ± 0.48- and 2.41 ± 0.48-fold, respectively, more recombinant Sar1 than the Sar1B and Sar1B:H79G cell lines.
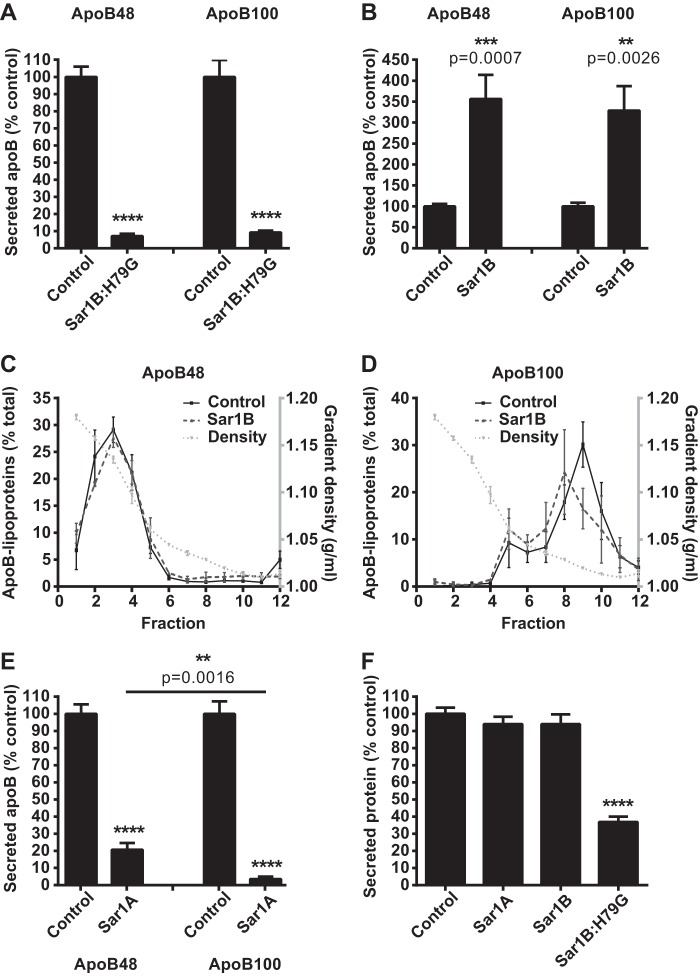
Sar1B:H79G overexpression virtually abolished the secretion of both apoB48 and apoB100 lipoproteins (Fig. 1A). By contrast, Sar1B overexpression increased their secretion by >3-fold (Fig. 1B). Their densities were not decreased (Fig. 1, C and D) with both the apoB48- and apoB100-containing lipoproteins exhibiting their expected densities (13). Sar1A overexpression markedly reduced apoB lipoprotein secretion (Fig. 1E); however, the effect was more pronounced for apoB100 than apoB48 (secretion 3.49 ± 1.5% (apoB100) and 20.65 ± 3.97% (apoB48) of control cells, respectively; p = 0.0016 for difference). Total protein secretion was also decreased in the Sar1B:H79G cell lines (Fig. 1F), but this reduction was significantly less than that observed for apoB48/100 (Fig. 1A). Collectively, these data show that Sar1B promotes secretion of both apoB48 and apoB100 lipoproteins, whereas Sar1A overexpression has the opposite effect, preferentially blocking the secretion of the more lipid-laden particles.
FIGURE 1.
Sar1A and Sar1B have opposite effects on apoB48- and apoB100-containing lipoprotein secretion. A and B, McArdle-RH7777 cells were labeled with l-[35S]methionine for 60 min and chased for 120 min. The l-35S-labeled apoB in the cell media was immunoprecipitated, analyzed by SDS-PAGE, and visualized by phosphorimaging. Data (mean ± S.E.) are from four independent experiments.****, p < 0.0001 versus cells with empty vector control. ***, p < 0.001 versus cells with empty vector control. **, p < 0.01 versus cells with empty vector control. C and D, densities of fractions containing secreted l-35S-labeled apoB48-containing (C) and l-35S-labeled apoB100-containing (D) lipoproteins were determined gravimetrically. Data (mean ± S.E.) are from three independent experiments. E, secreted l-35S-labeled apoB from two independent Sar1A-FLAG cell lines, quantified as in A and B. ****, p < 0.0001 versus cells with empty vector control. **, p < 0.01 versus apoB100 lipoproteins. F, total l-[35S]methionine in secreted protein fraction. Data (mean ± S.E.) are from five independent experiments. ****, p < 0.0001 versus cells with empty vector control.
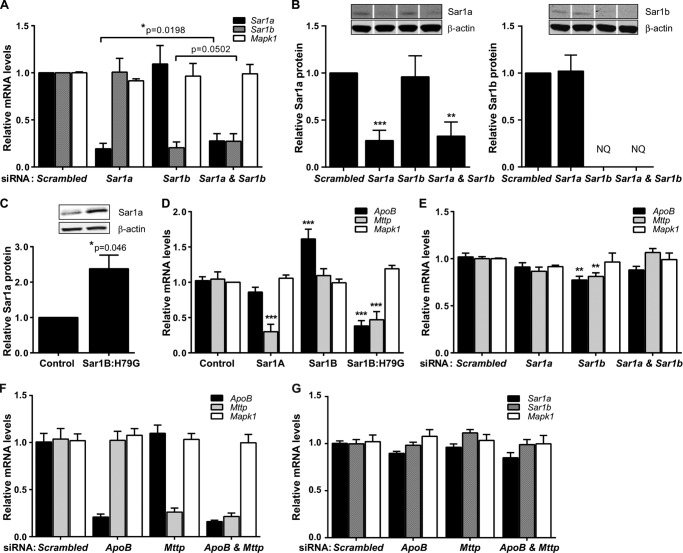
To determine whether Sar1 also had isoform-specific effects on the expression of ApoB and Mttp, we measured the levels of their RNAs by RT-qPCR in our McArdle-RH7777 cell lines plus McArdle-RH7777 cells transfected with Sar1 siRNAs. Individual knockdown of the Sar1 isoforms were isoform-specific, and there were no increases in the mRNA (Fig. 2A) or protein (Fig. 2B) levels of the nontargeted isoform. The amounts of Sar1a protein in cells transfected with Sar1a and Sar1a plus Sar1b siRNAs were 28.16 ± 6.36 and 32.88 ± 8.70%, respectively, of that in cells transfected with the scrambled siRNA control (Fig. 2B). Sar1b knockdown also substantially reduced in Sar1b protein and in the majority of samples to undetectable levels (Fig. 2B). This was due to the Western blot analysis returning weaker signals for Sar1b than Sar1a, consistent with RT-qPCR data (Sar1b mRNA 11.6-fold lower than Sar1a mRNA), plus the efficiency of Sar1b knockdown in reducing Sar1b protein (≥70% in the rare knockdown sample where it could be quantified).
FIGURE 2.
Effect of constitutively active Sar1B and native Sar1 expression on ApoB and Mttp expression in McArdle-RH7777 cells. A and B, isoform-specific effects of Sar1a and Sar1b knockdown on transcript (A) and protein (B) analyzed by RT-qPCR and quantitative Western blot analysis. NQ, not quantified due to undetectable levels of Sar1b in some cell lysates. White line separators indicate that noncontiguous lanes from the same gel are shown. A, *, p < 0.05 versus Sar1a + Sar1b knockdown. B, ***, p < 0.001 versus scrambled control siRNA; **, p < 0.01 versus scrambled control siRNA. C, Sar1a protein in stably overexpressing Sar1B:H79G and empty vector control cells. *, p < 0.05 versus cells with empty vector control. D, Sar1 isoform-specific effects on ApoB and Mttp mRNA levels in overexpressing cell lines, analyzed by RT-qPCR. ***, p < 0.001 versus cells with empty vector control. E, Sar1b knockdown specifically reduces ApoB and Mttp mRNA, analyzed by RT-qPCR. **, p < 0.01 versus scrambled control siRNA. F and G, ApoB and Mttp knockdown (F) does not decrease Sar1a and Sar1b mRNA (G), analyzed by RT-qPCR. A–G, data (mean ± S.E.) are from at least three independent experiments.
Although constitutively active Sar1B increased Sar1a mRNA (4.22 ± 0.29-fold compared with control cells) and protein (Fig. 2C), it reduced ApoB and Mttp mRNA (Fig. 2D). Levels of the control gene, Mapk1, were not altered (Fig. 2D). In the complementary analyses, native Sar1B overexpression increased ApoB mRNA (Fig. 2D), while having no effect on Mttp (Fig. 2D) and Sar1a (1.27 ± 0.03-fold, nonsignificant) mRNAs; and Sar1b knockdown reduced ApoB and Mttp mRNA (Fig. 2E) and had no significant effect on Sar1a mRNA (Fig. 2A). In the control analysis, ApoB and Mttp knockdown (Fig. 2F) did not reduce Sar1b mRNA (Fig. 2G). Interestingly, we found that Sar1A overexpression decreased Mttp mRNA levels, although Sar1a knockdown did not increase Mttp mRNA (Fig. 2E). However, in the double knockdown experiment, decreased Sar1a did prevent the fall in Mttp (and apoB) associated with Sar1b knockdown (Fig. 2E). Thus, collectively these data indicate that Sar1 has isoform-specific effects on ApoB and Mttp mRNA levels. Moreover, because a higher level of MTTP activity is required for producing apoB100 than apoB48 lipoproteins (15), the observed changes in ApoB and Mttp expression may form part of the adaptive responses to Sar1-induced changes in apoB lipoprotein secretion.
Because clinical studies have indicated that SAR1B mutations affect chylomicron secretion and, despite intestinal fat malabsorption, some children with CMRD develop fatty liver (2, 16–18), we examined relative SAR1A and SAR1B mRNA levels in human intestinal biopsies and liver samples. In addition, we examined the correlation between SAR1, APOB, and MTTP mRNA in publically available human liver gene expression datasets.
We found that SAR1B mRNA levels were 239 ± 84- and 3-fold, respectively, higher than SAR1A in intestinal mucosal biopsies (17 donors) and a pooled (three donors) liver sample. By contrast, the predominant SEC23 (A) and SEC31 (A) isoforms expressed in these two tissues displayed more comparable mean fold differences; in the intestinal biopsies, SEC23A and SEC31A were 5.0 ± 0.9- and 43 ± 12-fold higher than their respective B isoforms; and in the pooled liver sample, the corresponding values were 3.2 and 29.4.
We also found positive correlations between hepatic SAR1B, but not SAR1A, mRNA levels and APOB/MTTP mRNA. Thus, in a large dataset comprising 427 individual liver samples, the correlation coefficient (R) between SAR1B and APOB mRNA was 0.301 (p = 8.3 × 10−9), and in the second dataset, comprising 75 samples, this result was replicated (R = 0.292, p = 0.01). Likewise, R values between SAR1B and MTTP mRNA were 0.268 (p = 2.5 × 10−8) in dataset 1 and 0.346 (p = 2.4 × 10−3) in dataset 2. SAR1A displayed no positive correlation with either APOB or MTTP mRNA levels; R values for the SAR1A/APOB correlations were 0.003 (dataset 1, p = NS) and −0.012 (dataset 2, p = NS) and between SAR1A and MTTP, −0.179 (p = 2.3 × 10−4) and −0.019 (p = NS). Notably, the weak inverse correlation between SAR1A and MTTP mRNA levels in dataset 1 is consistent with the effect of Sar1A overexpression on Mttp mRNA levels in McArdle-RH7777 cells (Fig. 2D), plus the Mttp mRNA findings in the set of Sar1 knockdown experiments (Fig. 2E).
Thus, collectively our results (e.g. Fig. 2, D and E) and the previous characterization of the COPII vesicle assembly process (51, 54, 56, 68) are consistent with a model whereby the expression of APOB and MTTP, which are obligatory for apoB lipoprotein production in the ER lumen (12, 69, 70), are modulated by the expression of Sar1B, which facilitates the export of nascent apoB lipoproteins (and potentially other associated cargo) out of the ER in COPII-coated vesicles.
Characterizing Sar1 Effects on Gene Expression and de Novo Cholesterol Biosynthesis
In view of the lipid transport functions of apoB, we determined whether constitutively active Sar1B altered the expression of genes regulating triglyceride, fatty acid, and cholesterol homeostasis. To this end, we performed a microarray RNA analysis on the McArdle-RH7777 cell lines and validated the robustness of the results by measuring Sec mRNA levels via RT-qPCR in an independent experiment (supplemental Table S2). In addition, we confirmed that the respective 2.73 ± 0.15- and 2.90 ± 0.47-fold increases in Sec13 and Sec31a mRNA associated with Sar1B:H79G overexpression (supplemental Table S2) were congruent with estimated rises in Sec13 (1.7 ± 0.20-fold) and Sec31a (4.72 ± 0.94-fold) protein.
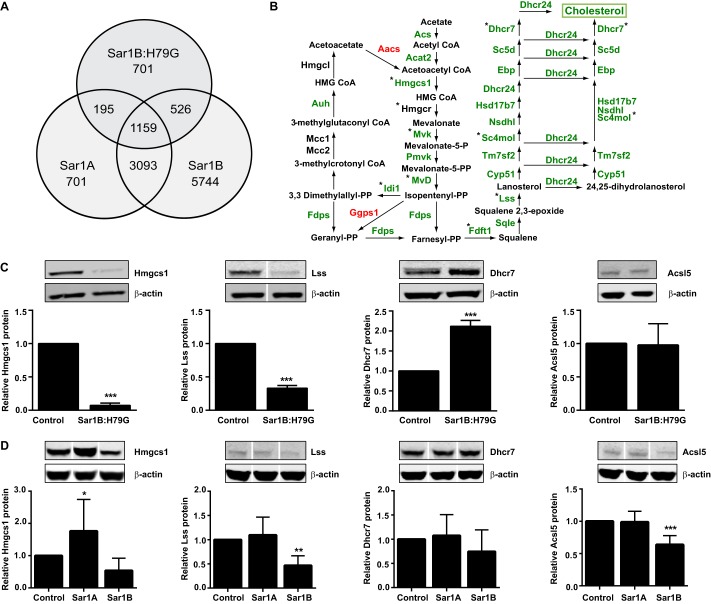
We identified 701 probe sets that had altered expression values (FDR <0.000215) in the Sar1B:H79G cell lines only (Fig. 3A). In the control analyses (i.e. native Sar1 overexpression), fewer probe sets were uniquely differentially expressed as follows: three in the Sar1A- and 70 in the Sar1B-overexpressing cell lines (supplemental Fig. S1). The Sar1B:H79G probe set contained an over-representation of genes assigned the Gene Ontology terms “Cholesterol and Lipid Biosynthetic and Metabolic Processes” (Table 1), although neither the Sar1A nor Sar1B datasets did (supplemental Tables S3 and S4). We therefore collated the expression values for genes active in fatty acid, triglyceride, and cholesterol metabolism, and verified the mRNA levels of representative genes in an independent experiment by RT-qPCR (supplemental Table S5).
FIGURE 3.
Sar1B:H79G decreases cholesterol biosynthesis gene expression in McArdle-RH7777 cells, independently of reduced Apob and Mttp expression. A, expression values of 701 probe sets were significantly altered (FDR, 0.000215; Δ 6.8) by Sar1B:H79G but not Sar1A (FDR 0.009, Δ1.9) or Sar1B (FDR 0.015, Δ 2.0) overexpression. B, green labeling depicts reduced mRNA levels in stably overexpressing Sar1B:H79G cells. *, expression values are verified by RT-qPCR. Note: in stably overexpressing Sar1A and Sar1B cells, mRNA levels of these genes were not reduced (supplemental Table S5). C and D, relative protein levels of three representative enzymes on the cholesterol biosynthetic pathway (Hmgcs1, Lss, Dhcr7) and a control enzyme, Acsl5, in stably transfected cells overexpressing Sar1B:H79G (C), Sar1A, and Sar1B (D). C and D data (mean ± S.E.) are from at least three independent experiments. ***, p < 0.001 versus cells with empty vector control; **, p < 0.01 versus cells with empty vector control. White line separators indicate that noncontiguous lanes from the same gel are shown.
TABLE 1.
Enrichment of gene ontology biological processes terms in the Sar1B:H79G uniquely differentially expressed gene-probe list
The uniquely differentially expressed Sar1B:H79G dataset (see also Fig. 3A) was analysed for enrichment of functionally related Gene Ontology (GO) terms using DAVID (65, 66).
| Cluster description | GO terms in cluster | Differentially expressed genes (n) | Enrichment score | p value range |
|---|---|---|---|---|
| Regulation of molecular function and catalytic activity | 0065009; 0050790; 0051336 | 32 | 3.70 | 2.1 × 10−5-1.5 × 10−3 |
| Cholesterol and lipid biosynthetic and metabolic processes | 0006695; 0008203; 0016126; 0016125; 0044255; 0006629; 0008610; 0006694; 0008202 | 32 | 2.16 | 7.3 × 10−4-8.6 × 10−3 |
| Amine catabolic processes | 0009310; 0044270; 0009065; 0006527; 0009063; 0009064 | 9 | 1.59 | 3.3 × 10−3-2.8 × 10−2 |
| Regulation of GTPase activity | 0043087 | 7 | 1.54 | 6.7 × 10−3 |
| Response to nutrient levels and extracellular stimulus | 0031667; 0009991 | 10 | 1.49 | 8.1 × 10−3-1.0 × 10−2 |
| Regulation of hydrolase activity | 0051336; 0043087; 0032313; 0032483; 0032482; 0032318 | 13 | 1.38 | 1.5 × 10−3-4.5 × 10−2 |
| Response to glucocorticoid and corticosteroid stimulus | 0051384; 0031960 | 5 | 1.3 | 3.0 × 10−2-4.4 × 10−2 |
| Amine catabolic processes | 0009310; 0044270; 0042133; 0006575; 0042219; 0042135 | 16 | 1.28 | 3.3 × 10−3-3.4 × 10−2 |
| Negative regulation of catalytic activity | 0043086; 0033673; 0006469; 0051348 | 9 | 1.21 | 3.4 × 10−3-3.0 × 10−2 |
| Neurogenesis | 0022008; 0048699; 0030182; 0048812; 0048667; 0007409 | 21 | 1.18 | 1.0 × 10−2-4.0 × 10−2 |
| Response to stress | 0006950; 0009605 | 51 | 1.04 | 1.5 × 10−2-1.7 × 10−2 |
| Response to endogenous stimulus | 0009719 | 21 | 0.90 | 1.2 × 10−2 |
| Protein import into nucleus | 0006606; 0051170 | 7 | 0.87 | 2.5 × 10−2-2.8 × 10−2 |
| Alcohol metabolic process | 0006066 | 23 | 0.76 | 3.1 × 10−4 |
Despite reduced secretion of apoB lipoproteins from the Sar1B:H79G and Sar1A cell lines (Fig. 1, A and E), and increased apoB-secretion from Sar1B cells (Fig. 1B), we found that the levels of most transcripts encoding proteins active in fatty acid and triglyceride metabolism were unaltered by increases in Sar1A expression in the Sar1A and Sar1B:H79G (Fig. 2C) cell lines and by Sar1B overexpression (supplemental Table S5). By contrast, constitutively active Sar1B decreased the levels of most mRNAs encoding enzymes on the cholesterol biosynthetic pathway (Fig. 3B and supplemental Table S5), whereas overexpression of native Sar1A and -B did not (supplemental Table S5).
Western blot analysis of Sar1B:H79G cell lysates revealed markedly reduced levels of Hmgcs1 and Lss protein (Fig. 3C, 1st and 2nd panels), whereas Dhcr7, which converts 7-dehydrocholesterol to cholesterol and 7-dehydrodesmostrel to 24-dehydrocholesterol (Fig. 3B), was increased >2-fold (Fig. 3C, 3rd panel). In a control analysis, Sar1B:H79G did not alter RNA (supplemental Table S5) or protein levels of Acsl5 (Fig. 3C, 4th panel), a key regulator of fatty acid metabolism (71). Furthermore, Sar1A overexpression, which also markedly impaired apoB lipoprotein secretion (Fig. 1E), did not reduce Hmgcs1 and Lss protein (Fig. 3D) or increase Dhcr7 protein (Fig. 3D). Rather Hmgcs1 protein was increased (Fig. 3D, 1st panel).
Sar1B overexpression had a variable impact on Hmgcs1 protein levels and only significantly reduced Lss (Fig. 3D). However, no increase in Dhcr7 protein was observed, and in marked contrast to Sar1A and Sar1B:H79G overexpression, Ascl5 protein was reduced (Fig. 3D). Thus, these data indicate that overexpression of Sar1A, Sar1B, and Sar1B:H79G have distinct effects on Hmgcs1, Dhcr7, and Acsl5 protein levels and that only Sar1B:H79G significantly reduced levels of mRNA encoding cholesterol-biosynthetic enzymes (supplemental Table S5).
In a further control experiment, we found that knockdown of ApoB and Mttp did not significantly reduce mRNA levels of cholesterol pathway-encoding enzymes (supplemental Fig. S2). Thus, these data substantiate the evidence from the Sar1A gene expression (supplemental Table S5) and apoB secretion results (Fig. 1E) that partial ablation of cholesterol/cholesteryl ester secretion via the apoB-containing lipoprotein axis (40, 69, 72, 73) was not sufficient by itself to explain the Sar1B:H79G-mediated reductions in the levels of transcripts encoding cholesterol biosynthetic enzymes (Fig. 3B and supplemental Table S5).
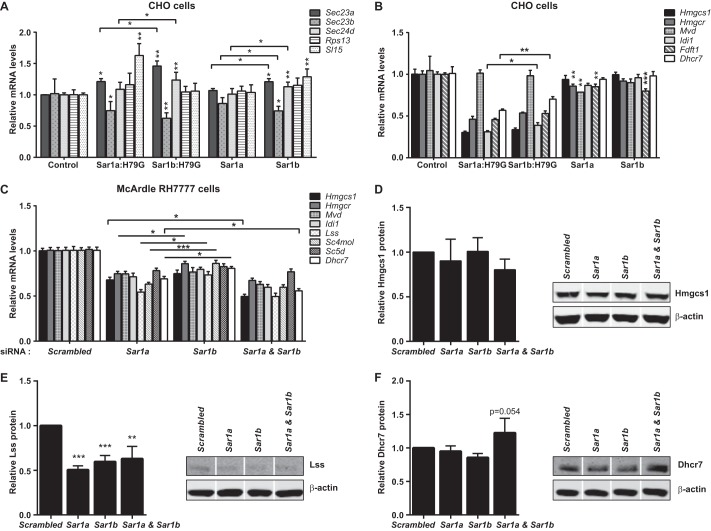
CHO cells do not have the machinery to produce apoB-containing lipoproteins (74). We therefore used them to explore further the impact of native and constitutively active Sar1 overexpression on the levels of mRNAs encoding cholesterol biosynthetic enzymes. Sar1A mRNA levels were respectively increased by 18.68 ± 2.04- and 22.13 ± 1.12-fold in the Sar1A and Sar1A:H79G cell lines; and the corresponding Sar1B values for the Sar1B and Sar1B:H79G cell lines were 21.17 ± 2.87 and 38.08 ± 3.71. Sec and control gene expression was much more modestly affected (Fig. 4A). In these non-apoB lipoprotein-producing cell lines, the overexpression of native Sar1A and Sar1B reduced cholesterol biosynthesis gene expression, but to a lesser extent than the control genes (Fig. 4, A and B). By contrast, constitutively active Sar1A or Sar1B overexpression markedly reduced mRNA levels of the cholesterol gene set (Fig. 4B). Hence, these data support the proposition that the reduced levels of most transcripts encoding enzymes for cholesterol biosynthesis in the McArdle-RH7777 cell lines expressing Sar1B:H79G are partially separable from the apoB lipoprotein secretion defect.
FIGURE 4.
No Sar1 isoform-specific effects on cholesterol biosynthesis gene expression in non-apoB and apoB lipoprotein-secreting cells. A and B, relative mRNA levels of COPII and control genes (A) and representative cholesterol genes (B) in CHO cells stably overexpressing constitutively active Sar1A or Sar1B and native Sar1A or Sar1B, analyzed by RT-qPCR. A, *, p < 0.05 versus cells with empty vector control or cells expressing Sar1b:H79G or Sar1b. **, p < 0.01 versus cells with empty vector control. B, *, p < 0.05 versus cells expressing Sar1b:H79G; **, p < 0.01 versus cells with empty vector control or cells expressing Sar1b:H79G; ***, p < 0.001 versus cells with empty vector control. C, Sar1a and Sar1b knockdown in McArdle-RH7777 cells reduce levels of representative mRNA encoding cholesterol biosynthesis enzymes. *, p < 0.05 versus Sar1b (with or without Sar1a) siRNA; ***, p < 0.001 versus Sar1b siRNA. D–F, relative protein levels of Hmgcs1 (D), Lss (E), and Dhcr7 (F) in McArdle-RH7777 cells transfected with Sar1a and Sar1b siRNAs. White line separators indicate that noncontiguous lanes from the same gel are shown. E, ***, p < 0.001 versus scrambled control siRNA; **, p < 0.01 versus scrambled control siRNA. A–F, data (mean ± S.E.) are from three independent experiments.
The gene expression data from CHO cell line experiments (Fig. 4B) also suggested that both Sar1 isoforms modulate the levels of mRNAs encoding cholesterol biosynthetic enzymes. We examined this further in McArdle-RH7777 cells by determining the effects of Sar1a and Sar1b knockdown on mRNA levels of eight representative members of this gene set. All were decreased (Fig. 4C). Sar1b knockdown generally produced smaller reductions than knockdown of Sar1a, the predominant species in McArdle-RH7777 cells. In the double knockdown, Hmgcs1 and Dhcr7 mRNA levels were significantly lower than in the individual knockdowns (Fig. 4C). However, Hmgcs1 protein levels were not significantly decreased by either individual or combined knockdown of Sar1a and Sar1b (Fig. 4D). In comparison, Lss protein was significantly reduced by both individual and combined Sar1a and Sar1b knockdown (Fig. 4E). Combined (not individual) Sar1a and Sar1b knockdown also produced a nonsignificant rise in Dhcr7 (Fig. 4F, p = 0.054 for difference from scrambled siRNA control). Thus, collectively, these data indicate that both Sar1A and Sar1B modulate the levels of mRNAs encoding cholesterol biosynthetic enzymes, supporting the CHO cell findings (Fig. 4B). Substantiating this conclusion, we also found that in human liver samples, SAR1A and SAR1B mRNA levels exhibited no isoform-specific correlations with 20 of the 21 mRNA species encoding cholesterol biosynthetic enzymes (supplemental Table S6). The only isoform-specific correlation was between that of SAR1B and SC5DL (sterol-C5-desaturase), and both of these genes had expression values that exhibited positive correlation with APOB and MTTP mRNA levels (supplemental Table S6).
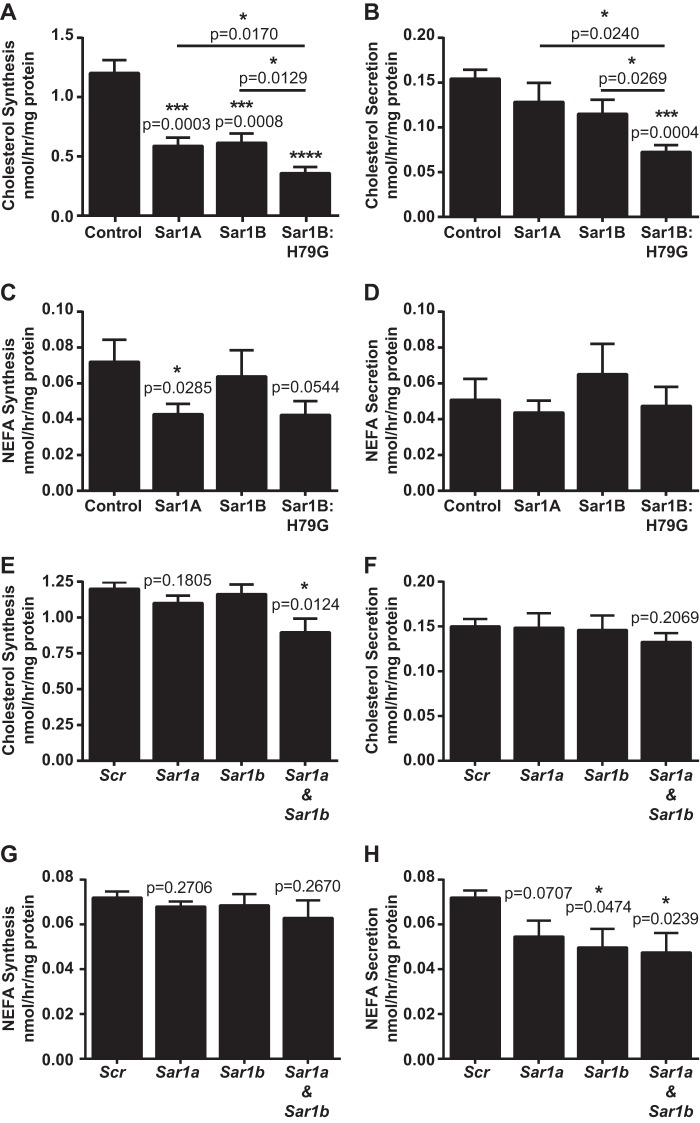
Next, we wanted to establish whether the reductions in “cholesterol” gene expression (Figs. 3B and 4C) and associated protein changes (Figs. 3C and 4, D–F) translated into reduced de novo cholesterol synthesis. As estimated from the incorporation of [14C]acetate into cholesterol, Sar1B:H79G overexpression markedly reduced the rate of de novo cholesterol synthesis (Fig. 5A). Additionally, [14C]cholesterol secretion was reduced (Fig. 5B) but to a lesser extent than de novo cholesterol synthesis. The estimated rate of incorporation of [14C]acetate into nonesterified fatty acids (NEFA) was also reduced (Fig. 5C), but this was to a much lesser degree than de novo cholesterol synthesis, and there was no corresponding reduction in 14C-labeled NEFA secretion (Fig. 5D). Consistent with comparable transcription profiles (supplemental Table S5), the Sar1A and Sar1B cell lines displayed similar rates of de novo cholesterol synthesis (Fig. 5A), both of which were significantly higher than that of the Sar1B:H79G cell lines.
FIGURE 5.
Sar1B:H79G and combined Sar1a and Sar1b deficiency reduce de novo cholesterol synthesis to greater extents than cholesterol secretion. A–H, McArdle-RH7777 cells stably overexpressing specified recombinant Sar1 protein or control McArdle-RH7777 cells transfected with specified siRNA were labeled with 2 mm [14C]acetate for 5 h. A and B, [14C]cholesterol in the cell (A) and media (B) of specified cell lines. C and D, [14C]NEFA contents in the cell (C) and media (D) of specified cell lines. E and F, [14C]cholesterol contents in the cell (E) and media (F) of McArdle-RH7777 cells transfected with specified siRNAs. G and H, [14C]NEFA contents in the cell (G) and media (H) of cells transfected with specified siRNAs. Data (mean ± S.E.) are from three (A–D) or four (E–H) independent experiments, Scr, scrambled siRNA. A and B, ***, p < 0.001 versus cells with empty vector control; ****, p < 0.0001 versus cells with empty vector control; *, p < 0.05 versus cells expressing Sar1B:H79G. C, *, p < 0.05 versus cells with empty vector control. E and H, *, p < 0.5 versus scrambled control siRNA.
Individual knockdown of Sar1a and Sar1b did not reduce the estimated rates of de novo cholesterol synthesis (Fig. 5E). By contrast, combined Sar1a and Sar1b knockdown had a significant impact reducing cellular [14C]cholesterol by 25.33 ± 8.04% (Fig. 5E), and this was paired with a small, albeit nonsignificant, reduction in the estimated rate of [14C]cholesterol secretion (Fig. 5F). The estimated rate of incorporation of [14C]acetate into NEFA was also reduced by combined Sar1a and Sar1b knockdown but by no more than individual Sar1a and Sar1b knockdowns (Fig. 5, G and H). Thus, these data, combined with the results in Figs. 3, B–D, and 4, B–F, and previous findings (52–55), indicate that an insufficiency of Sar1 to support pre-budding complex assembly or the sequestration of certain cargo into pre-budding COPII complexes containing constitutively active Sar1 reduce cholesterol biosynthesis.
Because cholesterol synthesis may both affect and be affected by the processing of Srebp2 in the Golgi apparatus, and COPII vesicles transport the ∼123-kDa ER membrane-bound precursor Srepb2 polypeptide to the Golgi (44, 45), we examined the effects of Sar1B:H79G overexpression and Sar1a and Sar1b knockdown, on Srepb2 expression. Additionally, we determined cellular levels of ATF6, a non-sterol-dependent transcription factor. The ATF6 precursor resides in the ER membrane (75) and is activated by the same proteases as Srebp2 in the Golgi (76, 77) to specifically regulate ER stress-inducible genes (78).
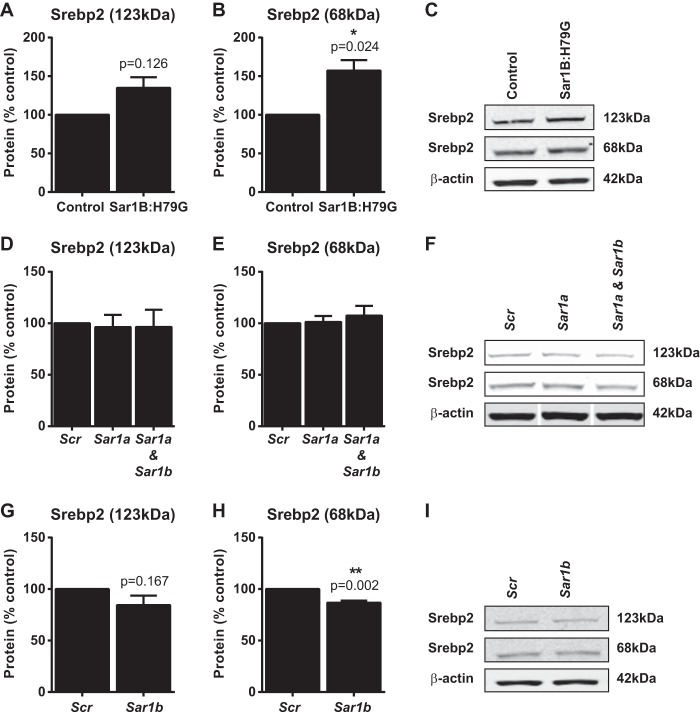
We found that Sar1B:H79G overexpression led to a small, but nonsignificant, rise in the level of precursor Srebp2 (Fig. 6A) and a significant increase in the level of the 68-kDa processed polypeptide (Fig. 6, B and C). Levels of the control protein ATF6 were not altered (supplemental Fig. S3). Thus, these results indicate that there was sufficient endogenous Sar1 in the Sar1B:H79G cell lines (Fig. 2C) to support the ER export of both Srebp2 and ATF6 precursors.
FIGURE 6.
Constitutively active Sar1B increases Srebp2 levels, whereas Sar1b deficiency decreases Srepb2 processing in McArdle-RH7777 cells. A–C, Srebp2 protein in stably overexpressing Sar1B:H79G and empty vector control cells. D–F, Sar1a and Sar1a+ Sar1b knockdown do not alter the levels of the 123-kDa precursor (D and F) or of the 68-kDa processed (E and F) Srebp2 protein. G–I, Sar1b knockdown decreases (H and I) the level of 68-kDa processed Srebp2 protein. A–I, data (mean ± S.E.) are from a minimum of three independent experiments. B, *, p < 0.05 versus cells with empty vector control. H, **, p < 0.01 versus scrambled control siRNA. In Western blot analyses, white line separators indicate that noncontiguous lanes from the same gel are shown. D–I, Scr, scrambled siRNA.
Sar1a knockdown did not reduce either Srebp2 protein levels (Fig. 6, D–F) or Scap and Srebp2 mRNA expression (supplemental Table S7). Likewise, combined Sar1a and Sar1b knockdown did not decrease Srebp2 protein levels or its processing (Fig. 6, D–F). By contrast, knockdown of Sar1b (minority Sar1 species in McArdle-RH7777 cells), which had no impact on de novo cholesterol synthesis (Fig. 5E), significantly reduced (by 13.2 ± 1.93% versus scrambled, p = 0.002 for difference) the level of processed Srebp2 (Fig. 6, G–I) and Scap mRNA (supplemental Table S7). In the control analysis, Sar1b knockdown did not reduce the amount of the non-sterol-responsive transcription factor ATF6 (supplemental Fig. S3).
DISCUSSION
This study was designed to examine the interplay between apoB-containing lipoprotein secretion and de novo cholesterol synthesis and their reliance on the common and individual activities of the two Sar1 isoforms. We provide strong evidence that Sar1B promotes the secretion of triglyceride-rich, apoB-containing lipoproteins from the liver, which would neatly explain the counter-intuitive observation that some CMRD children develop hepatic steatosis, despite severe intestinal fat malabsorption (16–18). Moreover, we show that Sar1A antagonizes Sar1B's lipoprotein secretion-promoting activity. We also establish that SAR1B is the predominantly expressed isoform in human jejunum and liver, two organs that substantially contribute to circulating levels of cholesterol (12, 79, 80), and that Sar1 deficiency, as well as constitutively active Sar1, decrease the levels of transcripts encoding cholesterol biosynthetic enzymes and de novo cholesterol synthesis. The results, which help explain the unique nature of the very severe hypocholesterolemia phenotype (i.e. reduced levels of all cholesterol-carrying lipoproteins) that features in CMRD (16, 18, 81), indicate that Sar1B makes a substantial contribution to the regulatory processes controlling ER and blood cholesterol levels.
As touched upon in the Introduction, there is a close spatial and temporal association between apoB and the ER membrane, extending from its point of entry to its co-translational assembly into a nascent lipoprotein (82). This transformation is promoted by triglyceride and cholesterol synthesis (72, 82–88), plus the conversion of membrane-bound cholesterol to cholesteryl esters (40, 69, 73, 83, 89), whereas the transport of newly assembled apoB lipoproteins from their sites of synthesis in the ER to the Golgi apparatus is mediated by the COPII transport machinery.
With the results from this study, we can now conclude that only Sar1B, and not Sar1A, can secure the efficient secretion of nascent, triglyceride-rich, apoB-containing lipoproteins and that it does so whether they are assembled around apoB48 or apoB100 and in the liver or intestine. Indeed, we found that Sar1A antagonizes Sar1B's apoB lipoprotein secretion-promoting activity, and this antagonism was more pronounced for larger and more lipid-laden apoB lipoproteins. Thus, these data suggest that Sar1B may have a unique capacity to promote the production of COPII vesicles of the size required to secure the ER export of triglyceride-rich, apoB-containing lipoproteins. This proposition fits well with existing knowledge. First, a few abnormally small chylomicrons (diameter 63 ± 19 nm) have been found within the abluminal space of CMRD patients' enterocytes (90), implying that chylomicrons (which are assembled around apoB48) exit the ER in standard sized COPII vesicles so long as they remain small. Second, in both of the two unrelated CMRD patients investigated, VLDL (which is assembled around apoB100) was markedly reduced (91). Third, in rat hepatocytes, VLDL formed around either apoB48 or apoB100 buds from the ER in larger sized COPII vesicles than do protein cargo (92). Moreover, a significant proportion of the apoB100 lipoproteins are found in larger sized COPII vesicles than apoB48 particles (92), likely reflecting that apoB100 has more neutral lipid-binding motifs than apoB48. Indeed, assembling apoB100 lipoproteins not only require more lipid than apoB48 to attain a soluble conformation, they will, with all other things being equal (e.g. lipid availability), bind considerably more lipid than apoB48 (12, 93, 94).
Although early investigations on CMRD patient samples revealed that both intestinal and hepatic apoB-containing lipoproteins were produced in this condition (81, 90), a re-appraisal of these data is long overdue given the new information on Sar1 (53–55) and the results reported herein. Thus, the totality of the data indicates that the removal of apoB lipoproteins from their sites of synthesis in the ER lumen may facilitate the apoB lipoprotein assembly process itself. We note, for example, that Sar1B's lipoprotein secretion-promoting activity was associated with a marked increase in ApoB expression (Fig. 2D) but not of genes regulating triglyceride synthesis (supplemental Table S5) and that Sar1B promoted the production of increased numbers of apoB lipoproteins rather than of more lipid-laden ones (Fig. 1, C and D). In comparison, Sar1B mutations in CMRD culminate in the production of abnormally large chylomicrons (90) and a lipid phenotype resembling that observed in the recessively inherited lipoprotein production disorder, abetalipoproteinemia; specifically, enterocytes from CMRD patients contain large cytosolic lipid droplets, even in the fasting state (90). Thus, loss of Sar1B's lipoprotein secretion-promoting activity appears to impair apoB lipoprotein production, albeit to a lesser extent than the impairment that occurs in abetalipoproteinemia.
Tantalizing clues as to how Sar1B could promote the assembly of specialist pre-budding complexes for mediating the onward transport of nascent apoB lipoproteins have emerged from two in vitro studies (56, 95). The first showed that Sar1B relaxes its host membrane to a greater extent that Sar1A, implying the former may promote assembly of large radius pre-budding complexes more readily than the latter, all other factors being equal (95). The second, incidentally found during study of the pathogenic Sec23A-F382L mutation (56), is that Sar1B-Sec23-Sec24 pre-budding complexes have lower affinity for Sec13/Sec31A than equivalent Sar1A-Sec23-Sec24 complexes, reducing Sec23-mediated hydrolysis of GTP on Sar1. The implication, which seems entirely consistent with their study's morphological analyses, is that Sar1B serves to delay, relative to Sar1A, COPII coat disassembly, thereby tilting the balance toward continued COPII vesicle expansion. In fact, a prolonged COPII vesicle assembly process could permit elongating apoB polypeptides to commandeer its ER-to-Golgi transport vesicle early on in the lipoprotein production process (∼10–15 min for apoB100 (13)), well before it undergoes the huge increase in size that occurs during transformation of hepatic and intestinal apoB into lipoproteins.
We found that relative to SAR1A, SAR1B mRNA levels are much higher in the upper gastrointestinal mucosa than in liver. Nonetheless, it is apparent that SAR1B contributes in a major way to hepatic lipid metabolism given the combination of our apoB lipoprotein studies (e.g. Figs. 1B and 2, D and E), the finding of hepatic steatosis in some children with CMRD (16, 17, 91), the highly significant positive correlations between SAR1B, APOB, and MTTP mRNA in human liver samples, and the finding that partial Sar1b knockdown reduced Srebp2 processing (Fig. 6H) paired with the expected corresponding reduction in levels of mRNAs encoding enzymes active in cholesterol biosynthesis (Fig. 4C).
We found that Sar1 had no isoform-specific effects on the expression of genes regulating triglyceride or fatty acid metabolism (supplemental Table S5). Moreover, even though constitutively active Sar1B blocked apoB lipoprotein secretion (Fig. 1A), it did not reduce the expression of most members of these two gene sets, suggesting that the intracellular transport of Srebp1, the best characterized transcription factor regulating fatty acid and triglyceride gene expression (47, 96–99), was maintained in our Sar1B:H79G cell lines. Indeed, because our stable cell lines overexpressing Sar1B:H79G induced a compensatory rise of endogenous Sar1A (Fig. 2C), we very likely created the means for allowing the onward transport of some ER cargo, while facilitating detection of Sar1B-specific phenotypes.
Although we found that native Sar1A and Sar1B overexpression had no, or minimal, effect on the levels of mRNA encoding cholesterol biosynthetic enzymes (supplemental Table S5 and Fig. 4B), they did reduce de novo cholesterol biosynthesis, albeit to a much lesser extent than Sar1B:H79G overexpression. It seems particularly pertinent that Sar1A overexpression, which markedly decreased apoB lipoprotein secretion (Fig. 1E), was associated with increased Hmgcs1 protein levels, and Sar1B overexpression was associated with reduced Lss protein. Conceivably, this counter-intuitive rise in Hmgcs1 protein (but not mRNA) could reflect a response to the diversion of acetyl-CoA (Fig. 3B) away from the cholesterol biosynthesis pathway toward an alternative metabolic pathway(s), for example, pathways promoting the acetylation of ER resident and ER-transiting proteins (100) and/or histone tails (101). In comparison, the reduction in Lss protein associated with Sar1B overexpression, however it is mediated, may reflect part of an adaptive response that serves to reduce flux through the cholesterol biosynthetic pathway.
The observation that Sar1B:H79G overexpression reduced the levels of most transcripts encoding cholesterol biosynthetic enzymes (Fig. 3B and supplemental Table S5) and that this reduction was paired with increased, rather than decreased, Dhcr7 protein levels is of interest (Fig. 3C). Dhcr7 deficiency in embryonic mouse brains leads to a compensatory rise in Hmgcs1 protein (102). This reciprocal relationship also develops on Sar1B:H79G overexpression, despite it having comparable effects on Hmgcs1 and Dhcr7 mRNA levels (supplemental Table S5). Likewise, the expected reciprocal relationship between reduced de novo cholesterol synthesis and increased Srepb2 processing (Fig. 6, B and C) was sustained. However, this increased processing was insufficient to secure the expected corresponding increases in cholesterol biosynthetic gene expression, as judged by mRNA measurements.
There are several possible explanations that could account for the effect of the Sar1B:H79G mutation on the levels of mRNA encoding cholesterol metabolism genes. For instance, it could reflect reduced Srebp2-mediated activation of these genes, for example, as a result of dimerization of Srebp2 with Srebp1c and/or another transcriptional co-factor (103, 104). Next, the processes regulating stabilization of one or more of these transcripts may be compromised (105). Alternatively, it could reflect their enhanced degradation, for example, via an miRNA-mediated (106) or IRE1α-mediated (107) mechanism(s). What is clear is that the incorporation of constitutively active Sar1B into pre-budding complexes would sequester certain ER cargo as a result of delaying the completion of COPII vesicle assembly and thus the fission of COPII vesicles, along with their selected cargo, from the ER membrane (51, 52, 55). Furthermore, reducing cholesterol secretion via the apoB lipoprotein pathway does not adequately explain the reduced mRNA levels associated with expression of constitutively active Sar1 (e.g. supplemental Fig. S2).
We also found that reducing the level of Sar1a protein decreased the levels of mRNA encoding cholesterol biosynthetic enzymes (Fig. 4C), but despite this, estimated rates of de novo cholesterol synthesis and secretion were not reduced. Consistent with this, Srebp2 processing was not increased. Thus, our results indicate that in our cell system there was sufficient Sar1 (i.e. residual Sar1a protein and Sar1b) to initiate the assembly of pre-budding COPII complexes for ER export of Srebp2 and that factors other than reduced Srebp2 processing underlie the reduced mRNA levels of the cholesterol gene set. By contrast, the reduced processing of Srebp2 associated with knockdown of Sar1b (minority species) may explain some of the reduction in the levels of mRNA encoding cholesterol biosynthetic enzymes. Hence, the observed cholesterol/Srebp2/mRNA differences between the Sar1B:H79G overexpressing cells and cells deficient for Sar1a or Sar1b indicate that the processes regulating de novo cholesterol synthesis in apoB lipoprotein-producing cells may be somewhat more complex than in other cell types. Furthermore, modulation of Sar1A and Sar1B levels in such cells affects processes regulating the accessibility of de novo synthesized cholesterol within the ER membrane (108, 109), its subsequent transfer to and utilization in the plasma membrane (110, 111), in addition to its use in the production of apoB- and apoA1-containing lipoproteins (38–40, 112).
The observation that Sar1 knockdown reduced de novo cholesterol synthesis by ∼25% is likely to be clinically relevant, especially given that this phenotype developed in cells that retained ∼30% of their total Sar1 (i.e. a and b) protein. It is also very striking that the severe hypocholesterolemia of CMRD differs from abetalipoproteinemia (1, 113) and familial hypobetalipoproteinemia (14) in that it is characterized by very low levels of high density lipoprotein (HDL) cholesterol, and indeed, in some cases, absence of this so-called good cholesterol-carrying lipoprotein particle (16, 18, 81, 114). HDL is formed through the efflux of plasma membrane cholesterol onto lipid-free apoAI, and cellular cholesterol levels are a major determinant of this efflux from the plasma membrane (114, 115). Therefore, it must be acknowledged that SAR1B mutations in CMRD very likely contribute to the low HDL cholesterol phenotype through an overall reduction in whole-body de novo cholesterol synthesis and that this reduction may, to a large extent, stem from those cells that have relatively low Sar1A levels or a high requirement for de novo synthesized cholesterol, or a combination of both.
With respect to understanding the role of hepatic de novo cholesterol synthesis in regulating apoB-containing lipoprotein secretion, in vivo data are rather sparse. One study found that liver-specific knockdown of HMGCR in 3- and 4-week-old mice significantly reduced plasma apoB, plus total and LDL cholesterol levels (116). Conversely, another showed that hepatic overexpression of Fdft1, which produces the first specific intermediate in the cholesterol biosynthetic pathway (Fig. 3B), increased de novo cholesterol synthesis and the production of cholesterol-enriched VLDL, culminating in markedly raised plasma levels of total and LDL cholesterol (87). Because the post-squalene cholesterol reactions (Fig. 3B) occur on the ER membrane (24–28), it is plausible that reduced de novo cholesterol synthesis may decrease ACAT activity. Indeed, Parini et al. (38) have shown that culturing cells in cholesterol-enriched media increases the enzymatic activity of ACAT2, the major cholesterol-esterifying enzyme in hepatocytes (39), and Temel et al. (40) demonstrated that ACAT2 stimulates cholesteryl ester secretion in apoB lipoproteins. Furthermore, it has been demonstrated that MTTP increases ACAT's activity by transferring its reaction products (i.e. cholesteryl esters) onto assembling apoB lipoproteins, thereby removing product inhibition (69). Thus, in light of these considerations, and the data presented here, it seems to make sense that in CMRD, VLDL, and LDL in the patients contain, as percentage of total cholesterol, significantly less cholesteryl ester than control samples (81), whereas their VLDL carries a larger than usual triglyceride load (16, 81, 90). More fundamentally, the cholesterol/cholesteryl ester and apoB/MTTP pathways work with each other, as well as the COPII and Srepb2/Scap machineries, to maintain the cholesterol content of the ER membrane at a concentration(s) compatible with this cellular compartment's biosynthetic organizing functions (117).
Acknowledgments
We thank Drs. A. F. Dean and J. Kowalczyk for helpful discussions.
This work was supported by funding from the British Heart Foundation Grants PG/04/010/16596 and FS/12/25/29569 and Medical Research Council plus a ZONMW/NWO VENI award (to M. N.). This work forms part of the research areas contributing to the translational research portfolio of the Cardiovascular Biomedical Research Unit at Barts, which is supported and funded by the National Institute for Health Research.

This article contains supplemental Figs. S1–S3 and Tables S1–S7.
- MTTP
- microsomal triglyceride transfer protein
- ER
- endoplasmic reticulum
- apo
- apolipoprotein
- COPII
- coat protein II
- CMRD
- chylomicron retention disease
- ACAT
- acyl-CoA:cholesterol acyltransferase
- SREBP
- sterol regulatory element-binding protein
- FDR
- false detection rate
- NEFA
- nonesterified fatty acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qPCR
- quantitative PCR.
REFERENCES
- 1. Berriot-Varoqueaux N., Aggerbeck L. P., Samson-Bouma M., Wetterau J. R. (2000) The role of the microsomal triglyceride transfer protein in abetalipoproteinemia. Annu. Rev. Nutr. 20, 663–697 [DOI] [PubMed] [Google Scholar]
- 2. Jones B., Jones E. L., Bonney S. A., Patel H. N., Mensenkamp A. R., Eichenbaum-Voline S., Rudling M., Myrdal U., Annesi G., Naik S., Meadows N., Quattrone A., Islam S. A., Naoumova R. P., Angelin B., Infante R., Levy E., Roy C. C., Freemont P. S., Scott J., Shoulders C. C. (2003) Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat. Genet. 34, 29–31 [DOI] [PubMed] [Google Scholar]
- 3. Porter F. D., Herman G. E. (2011) Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 52, 6–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patterson M. C., Hendriksz C. J., Walterfang M., Sedel F., Vanier M. T., Wijburg F. (2012) Recommendations for the diagnosis and management of Niemann-Pick disease type C: an update. Mol. Genet. Metab. 106, 330–344 [DOI] [PubMed] [Google Scholar]
- 5. Schmidt H. H., Tietge U. J., Buettner J., Barg-Hock H., Offner G., Schweitzer S., Dedoussis G. V., Rodeck B., Kallfelz H. C., Schlitt H. J., Oldhafer K., Klempnauer J. (2008) Liver transplantation in a subject with familial hypercholesterolemia carrying the homozygous p.W577R LDL-receptor gene mutation. Clin. Transplant. 22, 180–184 [DOI] [PubMed] [Google Scholar]
- 6. Palacio C. H., Harring T. R., Nguyen N. T., Goss J. A., O'Mahony C. A. (2011) Homozygous familial hypercholesterolemia: case series and review of the literature. Case Rep. Transplant. 2011, 154908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bansal S., Buring J. E., Rifai N., Mora S., Sacks F. M., Ridker P. M. (2007) Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. JAMA 298, 309–316 [DOI] [PubMed] [Google Scholar]
- 8. Nordestgaard B. G., Benn M., Schnohr P., Tybjaerg-Hansen A. (2007) Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA 298, 299–308 [DOI] [PubMed] [Google Scholar]
- 9. Nagasawa S. Y., Okamura T., Iso H., Tamakoshi A., Yamada M., Watanabe M., Murakami Y., Miura K., Ueshima H. (2012) Relation between serum total cholesterol level and cardiovascular disease stratified by sex and age group: a pooled analysis of 65,594 individuals from 10 cohort studies in Japan. J. Am. Heart. Assoc. 1, e001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leamy A. K., Egnatchik R. A., Young J. D. (2013) Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Prog. Lipid Res. 52, 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strand B. H., Langballe E. M., Hjellvik V., Handal M., Næss O., Knudsen G. P., Refsum H., Tambs K., Nafstad P., Schirmer H., Bergem A. L., Selmer R., Engedal K., Magnus P., Bjertness E., and GENIDEM-Group (2013) Midlife vascular risk factors and their association with dementia deaths: results from a Norwegian prospective study followed up for 35 years. J. Neurol. Sci. 324, 124–130 [DOI] [PubMed] [Google Scholar]
- 12. Calandra S., Tarugi P., Speedy H. E., Dean A. F., Bertolini S., Shoulders C. C. (2011) Mechanisms and genetic determinants regulating sterol absorption, circulating LDL levels, and sterol elimination: implications for classification and disease risk. J. Lipid Res. 52, 1885–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borén J., Rustaeus S., Olofsson S. O. (1994) Studies on the assembly of apolipoprotein B-100- and B-48-containing very low density lipoproteins in McA-RH7777 cells. J. Biol. Chem. 269, 25879–25888 [PubMed] [Google Scholar]
- 14. Welty F. K., Ordovas J., Schaefer E. J., Wilson P. W., Young S. G. (1995) Identification and molecular analysis of two apoB gene mutations causing low plasma cholesterol levels. Circulation 92, 2036–2040 [DOI] [PubMed] [Google Scholar]
- 15. Nicodeme E., Benoist F., McLeod R., Yao Z., Scott J., Shoulders C. C., Grand-Perret T. (1999) Identification of domains in apolipoprotein B100 that confer a high requirement for the microsomal triglyceride transfer protein. J. Biol. Chem. 274, 1986–1993 [DOI] [PubMed] [Google Scholar]
- 16. Nemeth A., Myrdal U., Veress B., Rudling M., Berglund L., Angelin B. (1995) Studies on lipoprotein metabolism in a family with jejunal chylomicron retention. Eur. J. Clin. Invest. 25, 271–280 [DOI] [PubMed] [Google Scholar]
- 17. Charcosset M., Sassolas A., Peretti N., Roy C. C., Deslandres C., Sinnett D., Levy E., Lachaux A. (2008) Anderson or chylomicron retention disease: molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Mol. Genet. Metab. 93, 74–84 [DOI] [PubMed] [Google Scholar]
- 18. Peretti N., Sassolas A., Roy C. C., Deslandres C., Charcosset M., Castagnetti J., Pugnet-Chardon L., Moulin P., Labarge S., Bouthillier L., Lachaux A., Levy E. (2010) Guidelines for the diagnosis and management of chylomicron retention disease based on a review of the literature and the experience of two centers. Orphanet. J. Rare Dis. 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sudhop T., Lütjohann D., Kodal A., Igel M., Tribble D. L., Shah S., Perevozskaya I., von Bergmann K. (2002) Inhibition of intestinal cholesterol absorption by ezetimibe in humans. Circulation 106, 1943–1948 [DOI] [PubMed] [Google Scholar]
- 20. Sudhop T., Reber M., Tribble D., Sapre A., Taggart W., Gibbons P., Musliner T., von Bergmann K., Lütjohann D. (2009) Changes in cholesterol absorption and cholesterol synthesis caused by ezetimibe and/or simvastatin in men. J. Lipid Res. 50, 2117–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones P. J., Lichtenstein A. H., Schaefer E. J. (1994) Interaction of dietary fat saturation and cholesterol level on cholesterol synthesis measured using deuterium incorporation. J. Lipid Res. 35, 1093–1101 [PubMed] [Google Scholar]
- 22. Haggarty P., Shetty P., Thangam S., Kumar S., Kurpad A., Ashton J., Milne E., Earl C. (2000) Free and esterified fatty acid and cholesterol synthesis in adult males and its effect on the doubly-labelled water method. Br. J. Nutr. 83, 227–234 [DOI] [PubMed] [Google Scholar]
- 23. Dietschy J. M., Turley S. D., Spady D. K. (1993) Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J. Lipid Res. 34, 1637–1659 [PubMed] [Google Scholar]
- 24. Stamellos K. D., Shackelford J. E., Shechter I., Jiang G., Conrad D., Keller G. A., Krisans S. K. (1993) Subcellular localization of squalene synthase in rat hepatic cells. Biochemical and immunochemical evidence. J. Biol. Chem. 268, 12825–12836 [PubMed] [Google Scholar]
- 25. Gaylor J. L. (2002) Membrane-bound enzymes of cholesterol synthesis from lanosterol. Biochem. Biophys. Res. Commun. 292, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 26. Caldas H., Herman G. E. (2003) NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum. Mol. Genet. 12, 2981–2991 [DOI] [PubMed] [Google Scholar]
- 27. Marijanovic Z., Laubner D., Moller G., Gege C., Husen B., Adamski J., Breitling R. (2003) Closing the gap: identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Mol. Endocrinol. 17, 1715–1725 [DOI] [PubMed] [Google Scholar]
- 28. Lu X., Li Y., Liu J., Cao X., Wang X., Wang D., Seo H., Gao B. (2012) The membrane topological analysis of 3β-hydroxysteroid-Δ24 reductase (DHCR24) on endoplasmic reticulum. J. Mol. Endocrinol. 48, 1–9 [DOI] [PubMed] [Google Scholar]
- 29. Jo Y., Debose-Boyd R. A. (2010) Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit. Rev. Biochem. Mol. Biol. 45, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baigent C., Landray M. J., Reith C., Emberson J., Wheeler D. C., Tomson C., Wanner C., Krane V., Cass A., Craig J., Neal B., Jiang L., Hooi L. S., Levin A., Agodoa L., Gaziano M., Kasiske B., Walker R., Massy Z. A., Feldt-Rasmussen B., Krairittichai U., Ophascharoensuk V., Fellström B., Holdaas H., Tesar V., Wiecek A., Grobbee D., de Zeeuw D., Grönhagen-Riska C., Dasgupta T., Lewis D., Herrington W., Mafham M., Majoni W., Wallendszus K., Grimm R., Pedersen T., Tobert J., Armitage J., Baxter A., Bray C., Chen Y., Chen Z., Hill M., Knott C., Parish S., Simpson D., Sleight P., Young A., Collins R., and SHARP Investigators (2011) The effects of lowering LDL cholesterol with simvastatin plus ezetimibe in patients with chronic kidney disease (Study of Heart and Renal Protection): a randomised placebo-controlled trial. Lancet 377, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor F., Huffman M. D., Macedo A. F., Moore T. H., Burke M., Davey Smith G., Ward K., Ebrahim S. (2013) Statins for the primary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 1, CD004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Calkin A. C., Tontonoz P. (2012) Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berr F., Eckel R. H., Kern F., Jr. (1986) Contraceptive steroids increase hepatic uptake of chylomicron remnants in healthy young women. J. Lipid Res. 27, 645–651 [PubMed] [Google Scholar]
- 34. Welty F. K., Lichtenstein A. H., Barrett P. H., Dolnikowski G. G., Schaefer E. J. (2004) Interrelationships between human apolipoprotein A-I and apolipoproteins B-48 and B-100 kinetics using stable isotopes. Arterioscler. Thromb. Vasc. Biol. 24, 1703–1707 [DOI] [PubMed] [Google Scholar]
- 35. Wojtanik K. M., Liscum L. (2003) The transport of low density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J. Biol. Chem. 278, 14850–14856 [DOI] [PubMed] [Google Scholar]
- 36. Urano Y., Watanabe H., Murphy S. R., Shibuya Y., Geng Y., Peden A. A., Chang C. C., Chang T. Y. (2008) Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc. Natl. Acad. Sci. U.S.A. 105, 16513–16518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Kant R., Zondervan I., Janssen L., Neefjes J. (2013) Cholesterol-binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J. Lipid Res. 54, 2153–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parini P., Davis M., Lada A. T., Erickson S. K., Wright T. L., Gustafsson U., Sahlin S., Einarsson C., Eriksson M., Angelin B., Tomoda H., Omura S., Willingham M. C., Rudel L. L. (2004) ACAT2 is localized to hepatocytes and is the major cholesterol-esterifying enzyme in human liver. Circulation 110, 2017–2023 [DOI] [PubMed] [Google Scholar]
- 39. Pramfalk C., Angelin B., Eriksson M., Parini P. (2007) Cholesterol regulates ACAT2 gene expression and enzyme activity in human hepatoma cells. Biochem. Biophys. Res. Commun. 364, 402–409 [DOI] [PubMed] [Google Scholar]
- 40. Temel R. E., Hou L., Rudel L. L., Shelness G. S. (2007) ACAT2 stimulates cholesteryl ester secretion in apoB-containing lipoproteins. J. Lipid Res. 48, 1618–1627 [DOI] [PubMed] [Google Scholar]
- 41. Kristiana I., Yang H., Brown A. J. (2008) Different kinetics of cholesterol delivery to components of the cholesterol homeostatic machinery: implications for cholesterol trafficking to the endoplasmic reticulum. Biochim. Biophys. Acta 1781, 724–730 [DOI] [PubMed] [Google Scholar]
- 42. Espenshade P. J., Li W. P., Yabe D. (2002) Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. U.S.A. 99, 11694–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun L. P., Seemann J., Goldstein J. L., Brown M. S. (2007) Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Motamed M., Zhang Y., Wang M. L., Seemann J., Kwon H. J., Goldstein J. L., Brown M. S. (2011) Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J. Biol. Chem. 286, 18002–18012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown M. S., Goldstein J. L. (2009) Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J. Lipid Res. 50, S15–S27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nagai M., Sakakibara J., Nakamura Y., Gejyo F., Ono T. (2002) SREBP-2 and NF-Y are involved in the transcriptional regulation of squalene epoxidase. Biochem. Biophys. Res. Commun. 295, 74–80 [DOI] [PubMed] [Google Scholar]
- 47. Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bennett M. K., Seo Y. K., Datta S., Shin D. J., Osborne T. F. (2008) Selective binding of sterol regulatory element-binding protein isoforms and co-regulatory proteins to promoters for lipid metabolic genes in liver. J. Biol. Chem. 283, 15628–15637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishimoto K., Tachibana K., Hanano I., Yamasaki D., Nakamura H., Kawai M., Urano Y., Tanaka T., Hamakubo T., Sakai J., Kodama T., Doi T. (2010) Sterol-regulatory-element-binding protein 2 and nuclear factor Y control human farnesyl diphosphate synthase expression and affect cell proliferation in hepatoblastoma cells. Biochem. J. 429, 347–357 [DOI] [PubMed] [Google Scholar]
- 50. Schiavoni G., Bennati A. M., Castelli M., Fazia M. A., Beccari T., Servillo G., Roberti R. (2010) Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochim. Biophys. Acta 1801, 587–592 [DOI] [PubMed] [Google Scholar]
- 51. Antonny B., Madden D., Hamamoto S., Orci L., Schekman R. (2001) Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 3, 531–537 [DOI] [PubMed] [Google Scholar]
- 52. Futai E., Hamamoto S., Orci L., Schekman R. (2004) GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J. 23, 4146–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bielli A., Haney C. J., Gabreski G., Watkins S. C., Bannykh S. I., Aridor M. (2005) Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J. Cell Biol. 171, 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Long K. R., Yamamoto Y., Baker A. L., Watkins S. C., Coyne C. B., Conway J. F., Aridor M. (2010) Sar1 assembly regulates membrane constriction and ER export. J. Cell Biol. 190, 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sato K., Nakano A. (2005) Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat. Struct. Mol. Biol. 12, 167–174 [DOI] [PubMed] [Google Scholar]
- 56. Fromme J. C., Ravazzola M., Hamamoto S., Al-Balwi M., Eyaid W., Boyadjiev S. A., Cosson P., Schekman R., Orci L. (2007) The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev. Cell 13, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mann C. J., Anderson T. A., Read J., Chester S. A., Harrison G. B., Köchl S., Ritchie P. J., Bradbury P., Hussain F. S., Amey J., Vanloo B., Rosseneu M., Infante R., Hancock J. M., Levitt D. G., Banaszak L. J., Scott J., Shoulders C. C. (1999) The structure of vitellogenin provides a molecular model for the assembly and secretion of atherogenic lipoproteins. J. Mol. Biol. 285, 391–408 [DOI] [PubMed] [Google Scholar]
- 58. Griffin J. L., Mann C. J., Scott J., Shoulders C. C., Nicholson J. K. (2001) Choline containing metabolites during cell transfection: an insight into magnetic resonance spectroscopy detectable changes. FEBS Lett. 509, 263–266 [DOI] [PubMed] [Google Scholar]
- 59. Leiper J. M., Bayliss J. D., Pease R. J., Brett D. J., Scott J., Shoulders C. C. (1994) Microsomal triglyceride transfer protein, the abetalipoproteinemia gene product, mediates the secretion of apolipoprotein B-containing lipoproteins from heterologous cells. J. Biol. Chem. 269, 21951–21954 [PubMed] [Google Scholar]
- 60. Fisher E. A., Lapierre L. R., Junkins R. D., McLeod R. S. (2008) The AAA-ATPase p97 facilitates degradation of apolipoprotein B by the ubiquitin-proteasome pathway. J. Lipid Res. 49, 2149–2160 [DOI] [PubMed] [Google Scholar]
- 61. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 62. Schadt E. E., Molony C., Chudin E., Hao K., Yang X., Lum P. Y., Kasarskis A., Zhang B., Wang S., Suver C., Zhu J., Millstein J., Sieberts S., Lamb J., GuhaThakurta D., Derry J., Storey J. D., Avila-Campillo I., Kruger M. J., Johnson J. M., Rohl C. A., van Nas A., Mehrabian M., Drake T. A., Lusis A. J., Smith R. C., Guengerich F. P., Strom S. C., Schuetz E., Rushmore T. H., Ulrich R. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wurmbach E., Chen Y. B., Khitrov G., Zhang W., Roayaie S., Schwartz M., Fiel I., Thung S., Mazzaferro V., Bruix J., Bottinger E., Friedman S., Waxman S., Llovet J. M. (2007) Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45, 938–947 [DOI] [PubMed] [Google Scholar]
- 64. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 66. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aridor M., Bannykh S. I., Rowe T., Balch W. E. (1995) Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 131, 875–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Townley A. K., Feng Y., Schmidt K., Carter D. A., Porter R., Verkade P., Stephens D. J. (2008) Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J. Cell Sci. 121, 3025–3034 [DOI] [PubMed] [Google Scholar]
- 69. Iqbal J., Rudel L. L., Hussain M. M. (2008) Microsomal triglyceride transfer protein enhances cellular cholesteryl esterification by relieving product inhibition. J. Biol. Chem. 283, 19967–19980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee R. G., Fu W., Graham M. J., Mullick A. E., Sipe D., Gattis D., Bell T. A., Booten S., Crooke R. M. (2013) Comparison of the pharmacological profiles of murine antisense oligonucleotides targeting apolipoprotein B and microsomal triglyceride transfer protein. J. Lipid Res. 54, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bu S. Y., Mashek D. G. (2010) Hepatic long-chain acyl-CoA synthetase 5 mediates fatty acid channeling between anabolic and catabolic pathways. J. Lipid Res. 51, 3270–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brown A., Wiggins D., Gibbons G. F. (1999) Manipulation of cholesterol and cholesteryl ester synthesis has multiple effects on the metabolism of apolipoprotein B and the secretion of very-low-density lipoprotein by primary hepatocyte cultures. Biochim. Biophys. Acta 1440, 253–265 [DOI] [PubMed] [Google Scholar]
- 73. Alger H. M., Brown J. M., Sawyer J. K., Kelley K. L., Shah R., Wilson M. D., Willingham M. C., Rudel L. L. (2010) Inhibition of acyl-coenzyme A:cholesterol acyltransferase 2 (ACAT2) prevents dietary cholesterol-associated steatosis by enhancing hepatic triglyceride mobilization. J. Biol. Chem. 285, 14267–14274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fleming J. F., Spitsen G. M., Hui T. Y., Olivier L., Du E. Z., Raabe M., Davis R. A. (1999) Chinese hamster ovary cells require the coexpression of microsomal triglyceride transfer protein and cholesterol 7α-hydroxylase for the assembly and secretion of apolipoprotein B-containing lipoproteins. J. Biol. Chem. 274, 9509–9514 [DOI] [PubMed] [Google Scholar]
- 75. Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nadanaka S., Yoshida H., Sato R., Mori K. (2006) Analysis of ATF6 activation in Site-2 protease-deficient Chinese hamster ovary cells. Cell Struct. Funct. 31, 109–116 [DOI] [PubMed] [Google Scholar]
- 77. Ye J., Rawson R. B., Komuro R., Chen X., Davé U. P., Prywes R., Brown M. S., Goldstein J. L. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 78. Sela D., Chen L., Martin-Brown S., Washburn M. P., Florens L., Conaway J. W., Conaway R. C. (2012) Endoplasmic reticulum stress-responsive transcription factor ATF6α directs recruitment of the mediator of RNA polymerase II transcription and multiple histone acetyltransferase complexes. J. Biol. Chem. 287, 23035–23045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nguyen L. B., Shefer S., Salen G., Ness G. C., Tint G. S., Zaki F. G., Rani I. (1990) A molecular defect in hepatic cholesterol biosynthesis in sitosterolemia with xanthomatosis. J. Clin. Invest. 86, 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Repa J. J., Turley S. D., Quan G., Dietschy J. M. (2005) Delineation of molecular changes in intrahepatic cholesterol metabolism resulting from diminished cholesterol absorption. J. Lipid Res. 46, 779–789 [DOI] [PubMed] [Google Scholar]
- 81. Levy E., Marcel Y., Deckelbaum R. J., Milne R., Lepage G., Seidman E., Bendayan M., Roy C. C. (1987) Intestinal apoB synthesis, lipids, and lipoproteins in chylomicron retention disease. J. Lipid Res. 28, 1263–1274 [PubMed] [Google Scholar]
- 82. Borén J., Rustaeus S., Wettesten M., Andersson M., Wiklund A., Olofsson S. O. (1993) Influence of triacylglycerol biosynthesis rate on the assembly of apoB-100-containing lipoproteins in Hep G2 cells. Arterioscler. Thromb. 13, 1743–1754 [DOI] [PubMed] [Google Scholar]
- 83. Cianflone K. M., Yasruel Z., Rodriguez M. A., Vas D., Sniderman A. D. (1990) Regulation of apoB secretion from HepG2 cells: evidence for a critical role for cholesteryl ester synthesis in the response to a fatty acid challenge. J. Lipid Res. 31, 2045–2055 [PubMed] [Google Scholar]
- 84. Tanaka M., Jingami H., Otani H., Cho M., Ueda Y., Arai H., Nagano Y., Doi T., Yokode M., Kita T. (1993) Regulation of apolipoprotein B production and secretion in response to the change of intracellular cholesteryl ester contents in rabbit hepatocytes. J. Biol. Chem. 268, 12713–12718 [PubMed] [Google Scholar]
- 85. Cuchel M., Schaefer E. J., Millar J. S., Jones P. J., Dolnikowski G. G., Vergani C., Lichtenstein A. H. (1997) Lovastatin decreases de novo cholesterol synthesis and LDL Apo B-100 production rates in combined-hyperlipidemic males. Arterioscler. Thromb. Vasc. Biol. 17, 1910–1917 [DOI] [PubMed] [Google Scholar]
- 86. Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X. Z., Kelly S., Chen S., McKay R., Monia B. P., Bhanot S. (2005) Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology 42, 362–371 [DOI] [PubMed] [Google Scholar]
- 87. Okazaki H., Tazoe F., Okazaki S., Isoo N., Tsukamoto K., Sekiya M., Yahagi N., Iizuka Y., Ohashi K., Kitamine T., Tozawa R., Inaba T., Yagyu H., Okazaki M., Shimano H., Shibata N., Arai H., Nagai R. Z., Kadowaki T., Osuga J., Ishibashi S. (2006) Increased cholesterol biosynthesis and hypercholesterolemia in mice overexpressing squalene synthase in the liver. J. Lipid Res. 47, 1950–1958 [DOI] [PubMed] [Google Scholar]
- 88. Liu Y., Millar J. S., Cromley D. A., Graham M., Crooke R., Billheimer J. T., Rader D. J. (2008) Knockdown of acyl-CoA:diacylglycerol acyltransferase 2 with antisense oligonucleotide reduces VLDL TG and ApoB secretion in mice. Biochim. Biophys. Acta 1781, 97–104 [DOI] [PubMed] [Google Scholar]
- 89. Parini P., Gustafsson U., Davis M. A., Larsson L., Einarsson C., Wilson M., Rudling M., Tomoda H., Omura S., Sahlin S., Angelin B., Rudel L. L., Eriksson M. (2008) Cholesterol synthesis inhibition elicits an integrated molecular response in human livers including decreased ACAT2. Arterioscler. Thromb. Vasc. Biol. 28, 1200–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dannoura A. H., Berriot-Varoqueaux N., Amati P., Abadie V., Verthier N., Schmitz J., Wetterau J. R., Samson-Bouma M. E., Aggerbeck L. P. (1999) Anderson's disease: exclusion of apolipoprotein and intracellular lipid transport genes. Arterioscler. Thromb. Vasc. Biol. 19, 2494–2508 [DOI] [PubMed] [Google Scholar]
- 91. Ouguerram K., Zaïr Y., Kasbi-Chadli F., Nazih H., Bligny D., Schmitz J., Aparicio T., Chétiveaux M., Magot T., Aggerbeck L. P., Samson-Bouma M. E., Krempf M. (2012) Low rate of production of apolipoproteins B100 and AI in 2 patients with Anderson disease (chylomicron retention disease). Arterioscler. Thromb. Vasc. Biol. 32, 1520–1525 [DOI] [PubMed] [Google Scholar]
- 92. Siddiqi S. A. (2008) VLDL exits from the endoplasmic reticulum in a specialized vesicle, the VLDL transport vesicle, in rat primary hepatocytes. Biochem. J. 413, 333–342 [DOI] [PubMed] [Google Scholar]
- 93. Borén J., Graham L., Wettesten M., Scott J., White A., Olofsson S. O. (1992) The assembly and secretion of ApoB 100-containing lipoproteins in Hep G2 cells. ApoB 100 is cotranslationally integrated into lipoproteins. J. Biol. Chem. 267, 9858–9867 [PubMed] [Google Scholar]
- 94. Spring D. J., Chen-Liu L. W., Chatterton J. E., Elovson J., Schumaker V. N. (1992) Lipoprotein assembly. Apolipoprotein B size determines lipoprotein core circumference. J. Biol. Chem. 267, 14839–14845 [PubMed] [Google Scholar]
- 95. Loftus A. F., Hsieh V. L., Parthasarathy R. (2012) Modulation of membrane rigidity by the human vesicle trafficking proteins Sar1A and Sar1B. Biochem. Biophys. Res. Commun. 426, 585–589 [DOI] [PubMed] [Google Scholar]
- 96. Guillou H., Martin P. G., Pineau T. (2008) Transcriptional regulation of hepatic fatty acid metabolism. Subcell. Biochem. 49, 3–47 [DOI] [PubMed] [Google Scholar]
- 97. Radonjic M., de Haan J. R., van Erk M. J., van Dijk K. W., van den Berg S. A., de Groot P. J., Müller M., van Ommen B. (2009) Genome-wide mRNA expression analysis of hepatic adaptation to high-fat diets reveals switch from an inflammatory to steatotic transcriptional program. PLoS One 4, e6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Harada N., Fujimoto E., Okuyama M., Sakaue H., Nakaya Y. (2012) Identification and functional characterization of human glycerol-3-phosphate acyltransferase 1 gene promoters. Biochem. Biophys. Res. Commun. 423, 128–133 [DOI] [PubMed] [Google Scholar]
- 99. Knebel B., Haas J., Hartwig S., Jacob S., Köllmer C., Nitzgen U., Muller-Wieland D., Kotzka J. (2012) Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLoS One 7, e31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pehar M., Lehnus M., Karst A., Puglielli L. (2012) Proteomic assessment shows that many endoplasmic reticulum (ER)-resident proteins are targeted by Nϵ-lysine acetylation in the lumen of the organelle and predicts broad biological impact. J. Biol. Chem. 287, 22436–22440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kaelin W. G., Jr., McKnight S. L. (2013) Influence of metabolism on epigenetics and disease. Cell 153, 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jiang X. S., Backlund P. S., Wassif C. A., Yergey A. L., Porter F. D. (2010) Quantitative proteomics analysis of inborn errors of cholesterol synthesis: identification of altered metabolic pathways in DHCR7 and SC5D deficiency. Mol. Cell. Proteomics 9, 1461–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Datta S., Osborne T. F. (2005) Activation domains from both monomers contribute to transcriptional stimulation by sterol regulatory element-binding protein dimers. J. Biol. Chem. 280, 3338–3345 [DOI] [PubMed] [Google Scholar]
- 104. Chatterjee S., Szustakowski J. D., Nanguneri N. R., Mickanin C., Labow M. A., Nohturfft A., Dev K. K., Sivasankaran R. (2009) Identification of novel genes and pathways regulating SREBP transcriptional activity. PLoS One 4, e5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vargas N. B., Brewer B. Y., Rogers T. B., Wilson G. M. (2009) Protein kinase C activation stabilizes LDL receptor mRNA via the JNK pathway in HepG2 cells. J. Lipid Res. 50, 386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Clingman C. C., Ryder S. P. (2013) Metabolite sensing in eukaryotic mRNA biology. Wiley Interdiscip. Rev. RNA 4, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. So J. S., Hur K. Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A. H., Iwawaki T., Glimcher L. H., Lee A. H. (2012) Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sokolov A., Radhakrishnan A. (2010) Accessibility of cholesterol in endoplasmic reticulum membranes and activation of SREBP-2 switch abruptly at a common cholesterol threshold. J. Biol. Chem. 285, 29480–29490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Olsen B. N., Bielska A. A., Lee T., Daily M. D., Covey D. F., Schlesinger P. H., Baker N. A., Ory D. S. (2013) The structural basis of cholesterol accessibility in membranes. Biophys. J. 105, 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Baumann N. A., Sullivan D. P., Ohvo-Rekilä H., Simonot C., Pottekat A., Klaassen Z., Beh C. T., Menon A. K. (2005) Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 44, 5816–5826 [DOI] [PubMed] [Google Scholar]
- 111. Woudenberg J., Rembacz K. P., Hoekstra M., Pellicoro A., van den Heuvel F. A., Heegsma J., van Ijzendoorn S. C., Holzinger A., Imanaka T., Moshage H., Faber K. N. (2010) Lipid rafts are essential for peroxisome biogenesis in HepG2 cells. Hepatology 52, 623–633 [DOI] [PubMed] [Google Scholar]
- 112. Auton M., Bassett G. R., Gillard B. K., Pownall H. J. (2013) Free cholesterol determines reassembled high-density lipoprotein phospholipid phase structure and stability. Biochemistry 52, 4324–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shoulders C. C., Brett D. J., Bayliss J. D., Narcisi T. M., Jarmuz A., Grantham T. T., Leoni P. R., Bhattacharya S., Pease R. J., Cullen P. M. (1993) Abetalipoproteinemia is caused by defects of the gene encoding the 97-kDa subunit of a microsomal triglyceride transfer protein. Hum. Mol. Genet. 2, 2109–2116 [DOI] [PubMed] [Google Scholar]
- 114. Sorci-Thomas M. G., Thomas M. J. (2012) High density lipoprotein biogenesis, cholesterol efflux, and immune cell function. Arterioscler. Thromb. Vasc. Biol. 32, 2561–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fisher E. A., Feig J. E., Hewing B., Hazen S. L., Smith J. D. (2012) High-density lipoprotein function, dysfunction, and reverse cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 32, 2813–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nagashima S., Yagyu H., Ohashi K., Tazoe F., Takahashi M., Ohshiro T., Bayasgalan T., Okada K., Sekiya M., Osuga J., Ishibashi S. (2012) Liver-specific deletion of 3-hydroxy-3-methylglutaryl coenzyme A reductase causes hepatic steatosis and death. Arterioscler. Thromb. Vasc. Biol. 32, 1824–1831 [DOI] [PubMed] [Google Scholar]
- 117. Simons K., Sampaio J. L. (2011) Membrane organization and lipid rafts. Cold Spring Harbor Perspect. Biol. 3, a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]