Key Points
The deletion of pfn1 led to bone marrow failure, loss of quiescence, increased apoptosis and mobilization, and a metabolic switch of HSCs.
Pfn1 partially acts through the axis of pfn1/Gα13/EGR1 to regulate stem cell retention and metabolism in the bone marrow.
Abstract
How stem cells interact with the microenvironment to regulate their cell fates and metabolism is largely unknown. Here we demonstrated that the deletion of the cytoskeleton-modulating protein profilin 1 (pfn1) in hematopoietic stem cell (HSCs) led to bone marrow failure, loss of quiescence, and mobilization and apoptosis of HSCs in vivo. A switch from glycolysis to mitochondrial respiration with increased reactive oxygen species (ROS) level was also observed in HSCs on pfn1 deletion. Importantly, treatment of pfn1-deficient mice with the antioxidant N-acetyl-l-cysteine reversed the ROS level and loss of quiescence of HSCs, suggesting that the metabolism is mechanistically linked to the cell cycle quiescence of stem cells. The actin-binding and proline-binding activities of pfn1 are required for its function in HSCs. Our study provided evidence that pfn1 at least partially acts through the axis of pfn1/Gα13/EGR1 to regulate stem cell retention and metabolism in the bone marrow.
Introduction
In the bone marrow (BM) microenvironment, the fates of hematopoietic stem cells (HSCs) are balanced among quiescence, self-renewal, differentiation, apoptosis, and motility. This balance is achieved through the dynamic interaction of factors including transcription factors, epigenetic regulators, cytoskeleton molecules, and extrinsic cues from the environment. Recently, we demonstrated that HSCs possess a distinct metabolic profile with a preference for glycolysis rather than mitochondrial respiration.1 However, the relationship among cell fates, metabolism, and the microenvironment of HSCs is not well understood.
Profilin 1 (Pfn1) is a ubiquitously expressed member of the profilin family. Pfn1 enhances ADP-to-ATP exchange on G-actin and is capable of adding ATP-bound G-actin to the barbed ends of growing filaments. Pfn1 was therefore thought to play a central role in the regulation of de novo actin polymerization.2 Pfn1 also binds to phosphoinositides, phosphatidylinositol-3,4,5-triphosphate, and a number of proline-rich proteins ranging from those participating in cytoskeletal organization to transcriptional control in cells. Binding of profilins to signaling molecules, such as Arp2/3 complex, Mena, VASP, N-WASP, dynamin I, and others, further implicates them as regulators of diverse activities including proliferation, migration, endocytosis, mRNA splicing, and transcription.3
The pfn1 gene was first identified as a survival essential gene in Drosophila.4 Homozygous pfn1 knockout mouse embryos die as early as the 2-cell stage, indicating its essential role in survival and cell division of embryonic development.2 The roles of pfn1 in motility are not consistent among different cell types. A number of studies indicated that pfn1 stimulates migration of endothelial cells, chondrocytes, human mesenchymal stem cells, and granule neurons.3,5-7 By contrast, pfn1 decreases motility and invasiveness of breast cancer cells in a mouse model8 and is down-regulated in invasive bladder cancer cells compared with noninvasive counterparts.9
The in vivo role of pfn1 in tissue-specific stem cells has not been reported. The availability of the pfn1flox/flox mice provided us an opportunity to clarify the function of pfn1 in different tissues and stem cells in the whole animal. We bred HSC-specific Cre-ER mice that express inducible Cre in HSCs10 and pfn1flox/flox mice to inducibly delete pfn1 in HSCs. We used this model to study the roles of pfn1 in hematopoietic development and to investigate the relationships of BM environment and metabolism and cell fates of HSCs. We showed that, different from its roles in many other types of cells,3,5-7 pfn1 is essential for the retention and quiescence of HSCs in the BM. Pfn1 also maintains glycolysis to directly control HSC quiescence, indicating that the unique metabolic property of HSCs is a determinant of quiescence of these stem cells.
Methods
Mice
C57 BL/6 CD45.2 and CD45.1 mice were purchased from the National Cancer Institute and from the University of Texas Southwestern Medical Center animal breeding core facility. To obtain an HSC-specific deletion of pfn1, C57BL/6 mice carrying the loxP-flanked pfn1 gene3 were crossed with transgenic C57BL/6 mice expressing the tamoxifen-inducible Cre recombinase under the control of stem cell leukemia (Scl) HSC enhancer10 to produce Sclpfn1 mice (supplemental Table 1, available on the Blood Web site). For induction of Cre-ER recombinase, mice received intraperitoneal tamoxifen (1 mg/0.1 mL of corn oil; Sigma-Aldrich) injections as previously described.10 Mice were maintained at the University of Texas Southwestern Medical Center animal facility. All animal experiments were performed with the approval of University of Texas Southwestern Committee on Animal Care.
Flow cytometry, mouse HSC culture, competitive reconstitution analysis, and homing assay
The isolation of Lin−Sca-1+Kit+Flk2−CD34− cells (long-term HSCs [LT-HSCs]), analysis of repopulation of mouse HSCs, and the Hoechst 33342/pyronin Y staining and bromodeoxyuridine (BrdU) incorporation were performed as previously described.11 Indicated numbers of BM Lin−Sca-1+Kit+Flk2−CD34− cells were cultured in serum-free medium supplemented with stem cell factor, thrombopoietin, and fibroblast growth factor-1 as previously described.12 The competitive reconstitution analysis was conducted as we described previously.13,14 Homing assays were performed as described previously.11,13 Details are included in the supplemental Methods.
Measurement of 13C lactate production, ATP assay, and oxygen consumption analysis
The metabolic assays were performed essentially as we described.1,15 Details are described in the supplemental Methods.
Measurement of reactive oxygen species
The measure of reactive oxygen species (ROS) was performed essentially as described previously.15 Briefly, control and Sclpfn1 Lin− cells were incubated with 1 µM 5-(and-6)-carboxy-2′,7′-Dichlorofluorescein diacetate (carboxy-DCFDA, C-369; Invitrogen) for 30 minutes at 37°C in the dark. Then, cells were stained for HSCs markers Sca-1-phycoerythrin (PE)/Cy5.5, C-Kit-Allophycocyanin, CD34-PE, and Flk2-PE and assayed by flow cytometer. Antibodies were all purchased from BD Biosciences.
Treatment with N-acetyl-l-cysteine
Control and Sclpfn1 mice were treated with tamoxifen for 7 days followed by treatment with N-acetyl-l-cysteine (NAC; Sigma-Aldrich) at a dose of 100 mg/kg body weight for another 8 days. NAC was injected into mice intraperitoneally.
Retrovirus infection and quantitative RT-PCR
The retrovirus infection and real-time reverse transcriptase-polymerase chain reaction (RT-PCR) were performed essentially as previously described,11 and details are included in the supplemental Methods. Real-time RT-PCR primers are described in supplemental Table 2.
Colony assays
The colony-forming unit (CFU)-erythroid, mature burst-forming unit-erythroid, CFU-granulocyte macrophage, CFU-pre-B lymphoid progenitor assays were performed as described previously.13 Details are described in the supplemental Methods.
Transwell experiments
A total of 20 000 fluorescence-activated cell sorter–collected wild-type (WT) and Sclpfn1 Lin−Sca-1+Kit+Flk2−CD34− cells were suspended in 300 μL of Hanks balanced salt solution/0.1% bovine serum albumin in the upper chamber and allowed to migrate across the 5-μm pore inserts (Corning-Costar) toward the lower chamber containing Hanks balanced salt solution/0.1% bovine serum albumin and 160 ng/mL CXCL12 (Peprotech). Transmigrated cells were harvested after 4 hours. The transmigrated cells and the cells that remained in the upper chamber were analyzed by flow cytometery.
Statistical analyses
Data are expressed as mean ± standard error of the mean. Data were analyzed by the Student t-test and were considered statistically significant if P < .05.
Results
Maintenance of BM HSCs requires Pfn1 function
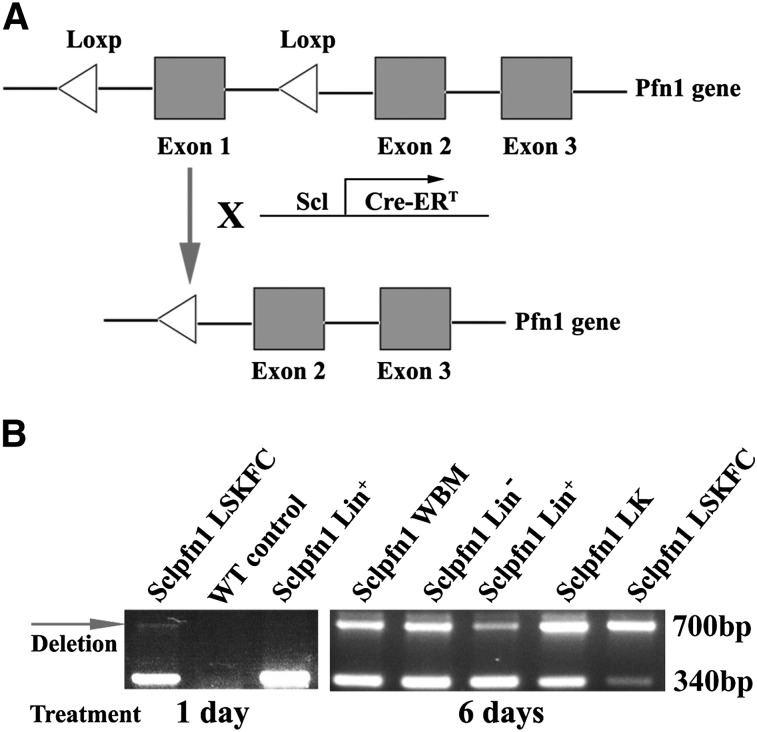
To obtain an inducible loss-of-function model for pfn1 in HSCs, we crossed pfn1fl/fl mice3 with transgenic mice expressing the tamoxifen-inducible Cre recombinase under the control of the Scl HSC enhancer, which deletes floxed genes in HSCs and hematopoietic progenitors on tamoxifen treatment10 (supplemental Table 1). The resultant Scl-Cre-ER/pfn1fl/fl mice (Sclpfn1; Figure 1A) and the control mice (Scl-Cre-ER/ pfn1+/+) were injected with tamoxifen to induce Cre expression in HSCs. Specific primers are designed to distinguish the WT, pfn1fl/fl, and pfn1−/− mice (on tamoxifen treatment; supplemental Figure 1A). Genotyping of BM LT-HSCs as Lin−Sca-1+Kit+CD34−Flk2− (LSKFC) cells and differentiated hematopoietic Lin+ cells of the Sclpfn1 mice revealed that loss of pfn1 had occurred in HSCs, but not in Lin+ cells at 24 hours after treatment of tamoxifen (Figure 1B). This pfn1 loss was dramatically increased in all hematopoietic cells at day 6 after the differentiation of HSCs (Figure 1B).
Figure 1.
Conditional deletion of pfn1 in HSCs. (A) Schematic of pfn1 floxed allele showing deletion of floxed exon 1 following Cre recombinase activity. Use of Scl-Cre-ERT results in specific deletion of the pfn1 gene in HSCs following tamoxifen treatment. (B) Genotype analyses of LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−, LSKFC) and Lin+, Lin−, Lin−Kit+ (LK) cells and whole BM (WBM) cells at 1 and 6 days after tamoxifen injections. Deletion of pfn1 is indicated by the arrow (700 bp).
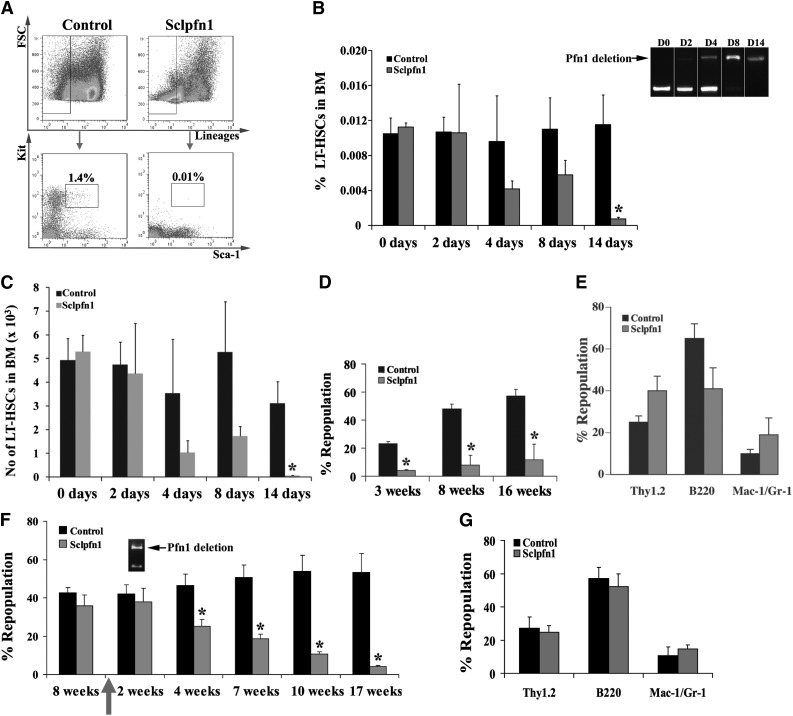
To evaluate the role of pfn1 in HSC maintenance and function, we analyzed the frequency and number of LSK cells and LSKFC cells (as LT-HSCs) in control and Sclpfn1 mice at different time points after tamoxifen treatment (Figure 2A-B). LSKFC cells in control mice were 0.011 ± 0.0038% and 0.012 ± 0.0036% of the total BM cells at days 8 and 14, respectively. In contrast, LSKFC cells accounted for only 0.0058 ± 0.0017% of BM cells at day 8 and 0.0008 ± 0.00021% at day 14 in Sclpfn1 mice (Figure 2B). Total numbers of BM cells in Sclpfn1 mice dramatically decreased from day 8 to day 14, consistent with exhaustion of HSCs (supplemental Figure 1B). Consistently, the total HSC number in BM decreased from 5.3 ± 0.067 × 103 at day 0 to 0.067 ± 0.019 × 103 at day 14 (Figure 2C). Meanwhile, lineage distribution in BM was changed at day 14 in Sclpfn1 mice compared with the WT distribution with increases in percentages of CD3+ and B220+ lymphoid cells and CD71+Ter119+ erythroid cells, but decreases in Mac-1+ and Gr-1+ myeloid cells (supplemental Figure 1C). A colony assay was performed to assess the dynamics of progenitor cells after pfn1 deletion. As shown in supplemental Figure 1D-E, numbers of myeloid progenitor cells, but not erythroid progenitors or B cell progenitors, were dramatically decreased to 40.70 ± 8.01% of the control at 4 days after tamoxifen treatment. The flow cytometry analyses also showed that the common myeloid progenitors decreased dramatically by >60% than the control (supplemental Figure 1H-I). The loss of cellularity of BM and dislocation of HSCs in bone marrow niches in pfn1-null mice were also evident (supplemental Figure 1F-G). Overall, our data in Figure 2 and supplemental Figure 1 indicate that pfn1 deletion results in the exhaustion of HSCs and the fast cycling myeloid progenitors in the mouse BM.
Figure 2.
The maintenance of BM HSCs requires pfn1 function. (A) Representative flow cytometry profile of Lin−Sca-1+Kit+ cells from control and Sclpfn1 mice. (B) The frequency of LSKFC in the Sclpfn1 mouse was dramatically decreased compared with the control mouse (1.4% vs 0.01%) at day 14 after tamoxifen treatment. Deletion of pfn1 over time is indicated by the arrow. (B-C) Quantification of HSC (Lin−Sca-1+Kit+Flk2−CD34−) frequency and number in BM of control and Sclpfn1 mice (n = 5-8) at different time points (*P < .05). (D) Competitive reconstitution analysis of control and Sclpfn1 HSCs treated with tamoxifen for 4 days. BM CD45.2 cells (2 × 105 cells) from donor mice along with 2 × 105 freshly isolated CD45.1 competitor cells were transplanted into lethally irradiated CD45.1 recipient mice. The mice (n = 5 per group) were analyzed for engraftment through 16 weeks after transplant (*P < .05). (E) Multilineage contribution of donor cells in the recipients at 14 weeks after transplant (n = 5). (F) Control and Sclpfn1 donor cells (2 × 105 cells) along with 2 × 105 competitor cells were transplanted into lethally irradiated CD45.1 recipient mice. These primary transplanted mice (n = 5) were treated with tamoxifen at 8 weeks after transplant and were then analyzed for the engraftment from 2 to 17 weeks after tamoxifen treatment. (*P < .05). Deletion of pfn1 at 2 weeks after tamoxifen treatment is indicated by the arrow. (G) Multilineage contribution of donor cells in the recipients at 16 weeks after transplant (n = 5).
To further determine whether the reduction in phenotypic HSCs in Sclpfn1 mice reflected an intrinsic defect, we performed 2 types of competitive transplantation experiments. In the first experiment, we transplanted 2 × 105 BM cells from either tamoxifen-treated Sclpfn1 mice or control animals with 2 × 105 CD45.1 competitors into lethally irradiated CD45.1 recipients. Significant numbers of donor-derived cells were observed in the peripheral blood of mice injected with control HSCs (23.21 ± 1.53%, 47.97 ± 3.47%, and 57.28 ± 4.61% repopulation at 3, 8, and 16 weeks, respectively); however, donor Sclpfn1 cells only contributed to minor chimerism (4.15 ± 0.52%, 8.04 ± 7.04%, and 11.69 ± 11.15% repopulation at 3, 8, and 16 weeks, respectively; Figure 2D). Nevertheless, Sclpfn1 donor cells did repopulate the lymphoid and myeloid lineages (Figure 2E), indicating differentiation was not impaired in pfn1−/− HSCs. Sclpfn1 donor cells treated with tamoxifen repopulated lethally irradiated CD45.1 recipients at barely detectable levels (supplemental Figure 2), which further confirmed that pfn1 plays a crucial role in maintenance of HSC activity.
In the other experiment, we transplanted 2 × 105 Sclpfn1 cells (without tamoxifen treatment) or control BM cells with 2 × 105 CD45.1 competitors into lethally irradiated CD45.1 recipients. Both groups were repopulated at similar efficiencies (42.61 ± 2.74% and 35.77 ± 5.64%, respectively; Figure 2F) at 8 weeks after transplantation. When pfn1 deletion was induced by tamoxifen treatment, levels of the pfn1-deficient HSCs significantly decreased over time from 35.77 ± 5.64% at 4 weeks to 4.21 ± 0.60% at 17 weeks after treatment began (Figure 2F). The repopulation of the control group remained relatively constant (46.54 ± 5.78%, 50.71 ± 6.36%, 54.03 ± 8.07%, and 53.21 ± 9.88% at weeks 4, 7, 10, and 17, respectively). The pfn1−/− donor cells repopulated all lymphoid and myeloid lineages (Figure 2G). Collectively, these results indicate that pfn1 is essential for the repopulating activity of HSCs.
Spontaneous and constitutive mobilization of HSCs in Sclpfn1 mice
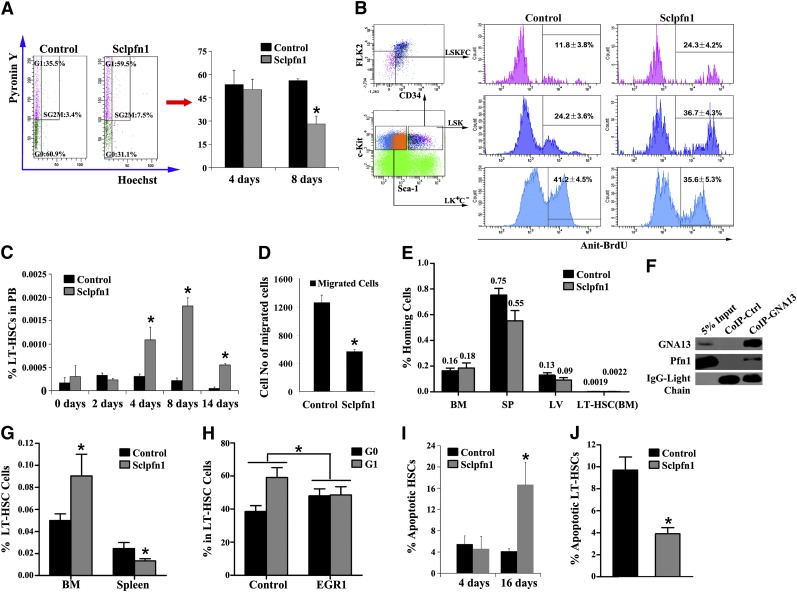
A number of possible mechanisms may account for the decreased HSC frequency and activity observed in Sclpfn1 mice including (1) decreased proliferation of HSCs, (2) increased mobilization, and (3) increased apoptosis. We first determined the impact of pfn1 deficiency on HSC quiescence/proliferation by analyzing the cell cycle profile of pfn1−/− BM HSCs. Hoechst 33342 and pyronin Y staining16 of sorted HSCs (as LSKFC cells) revealed no significant difference between Sclpfn1 and control mice at day 4 after tamoxifen treatment. However, there was an ∼50% decrease in G0 phase of pfn1−/− HSCs compared with WT controls at day 8 after tamoxifen treatment (Figure 3A). This decreased quiescence of pfn1−/− HSCs in BM was further confirmed by BrdU incorporation assays, which showed twofold greater (24.3 ± 4.2% vs 11.8 ± 3.8% in HSCs and 36.7 ± 4.3% vs 24.2 ± 3.6% in LSK cells) incorporation of BrdU into pfn1−/− HSCs or LSK cells compared with control HSCs or LSK cells (Figure 3B). Therefore pfn1−/− HSCs exhibited decreased quiescence in BM.
Figure 3.
Spontaneous and constitutive mobilization of HSCs in Sclpfn1 mice. (A) Sclpfn1 BM HSCs are less quiescent than control HSCs. (Left) LT-HSCs (as Lin−Sca-1+Kit+Flk2−CD34− cells) from a WT mouse, stained with Hoechst 33342 and pyronin Y, were analyzed for cell cycle stage. (Right) The percentages of G0 cells in control and Sclpfn1 mice (n = 3-4) at days 4 and 8 after tamoxifen treatment are shown (*P < .05). (B) BrdU incorporation indicates a decreased cycling in HSCs isolated from control mice compared with Sclpfn1 mice (n = 3; *P < .05). (C) Relative frequency of LT-HSCs in peripheral blood (PB) was analyzed over time in control and Sclpfn1 mice after tamoxifen treatment (n = 3-7; *P < .05). (D) The migrations of LT-HSCs (as Lin−Sca-1+Kit+Flk2−CD34− cells) isolated from control and Sclpfn1 mice 8 days after tamoxifen treatment were compared in a transwell experiment (n = 3; *P < .05). (E) Control and Sclpfn1 HSCs home equivalently to recipient BM. BM from control or Sclpfn1 mice (n = 5) was labeled with 5- and 6-carboxyfluorescein succinimidyl ester (CFSE), and 1 × 107 cells were transplanted into lethally irradiated recipients. After 16 hours, the total percentage of CFSE+ cells in the BM, spleen, and liver and LT-HSCs (CFSE+Lin−Sca-1+Kit+ Flk2−CD34− cells) in BM were determined by flow cytometry. (F) Lysates of WT mouse BM cells were coimmunoprecipitated with anti-Gα13 antibody or control rabbit IgG, and precipitation was then determined by western blotting using anti-Gα13 and anti-pfn1 antibodies (G-H) Rescue of Sclpfn1 LT-HSCs by EGR1. Control or EGR1 overexpressed Sclpfn1 BM cells were transplanted into CD45.1 recipients with competitors. Mice were treated with tamoxifen after 5 weeks. (G) Percentages of donor-derived CD45.2+ LT-HSCs were determined in BM and spleen (n = 3-4, *P < .05). (H) LT-HSCs were sorted from the BM of control or EGR1 rescued mice, and cell cycle was then measured by Hoechst 33342 and pyronin Y staining (n = 3-4, *P < .05). (I) Control and Sclpfn1 Lin−Sca-1+Kit+Flk2−CD34− cells were analyzed for apoptosis by using Annexin V/7-AAD staining (n = 3-5; *P < .05). (J) LT-HSCs from the BM of control or EGR1 rescued mice were cultured for 8 days, and apoptosis was measured (n = 3-4; *P < .05).
Next, we tested whether pfn1 plays a role in retaining HSCs in the BM. To this end, we analyzed LT-HSC frequency and number in the peripheral blood of Sclpfn1 and control mice. Beginning at day 4 after tamoxifen treatment, the frequency of circulating HSCs in Sclpfn1 mice increased about threefold compared with controls (Figure 3C). At days 8 and 14 after treatment with tamoxifen, the frequencies of circulating Sclpfn1 HSCs were dramatically increased by up to 10-fold in Sclpfn1 mice relative to controls. Because the spontaneous mobilization of LT-HSCs started at day 4 after tamoxifen treatment (Figure 3C), concordantly with the decrease of the frequency of HSCs in BM (starting after day 4; Figure 2B), HSC mobilization is likely a cause of the decrease of HSCs in BM in Sclpfn1 mice.
To investigate the mechanistic role of pfn1 in retention of HSCs in the BM, we performed a transwell experiment with control and Sclpfn1 HSCs treated for 8 days with tamoxifen. As shown in Figure 3D, Sclpfn1 HSCs had significantly decreased chemoattraction for SDF-1 compared with control cells, indicating that pfn1 regulates the BM retention of HSCs, possibly through SDF-1–mediated chemotaxis.
Both mobilization and homing are measures of HSC motility. Although Sclpfn1 HSCs had drastically increased mobilization and pfn1 deletion up-regulates CXCR4 expression in LT-HSCs (supplemental Figure 1H,J), the Sclpfn1 HSCs did not show significant difference in homing to BM compared with controls (Figure 3E). This result suggests that pfn1 is essential for the retention but not for the immediate homing of HSCs in the BM niche. Therefore, pfn1 deficiency induces an uncommon uncoupling of homing and mobilization of HSCs in BM.
It was reported that EGR1 regulates HSC migration and retention in the BM niche, and their knockout showed similar phenotypes as pfn1−/− mice.17,18 To test the possible role of pfn1 in regulation of EGR1, we examined EGR1 expression in control and Sclpfn1 HSCs treated with tamoxifen for 3 days using quantitative RT-PCR. Interestingly, we found that the EGR1 level in Sclpfn1 HSCs decreased to 38.99 ± 4.76% of that in controls (supplemental Figure 3A). Another protein, MLL, which was shown to support HSC BM retention,18 was also decreased in Sclpfn1 HSCs (supplemental Figure 3B). To determine how pfn1 regulates HSC BM retention, we demonstrated that Pfn1 was capable of interacting with a guanine nucleotide-binding protein Gα13 (Figure 3F; supplemental Figure 3C), which was known to transactivate the expression of EGR1.19 Importantly, a retroviral expression of EGR1 rescued the defective retention of Sclpfn1 HSCs in the BM but not in the spleen and the decreased quiescence of Sclpfn1 HSCs (Figure 3G-H). These results indicate that Pfn1 interacts with Gα13, which up-regulates EGR1 to maintain HSCs in the BM.
Finally, to determine the role of pfn1 in apoptosis of HSCs, we assessed Annexin V and 7-aminoactinomycin D (7-AAD) staining of LSKFC cells as a function of time after pfn1 deletion. As shown in Figure 3I, apoptosis of HSCs was dramatically increased in Sclpfn1 mice compared with controls at 16 days but not at 4 days after tamoxifen treatment. Furthermore, the increased apoptosis of Sclpfn1 HSCs can also be rescued by EGR1 (Figure 3J). Overall, Pfn1 plays a significant role in the survival of HSCs.
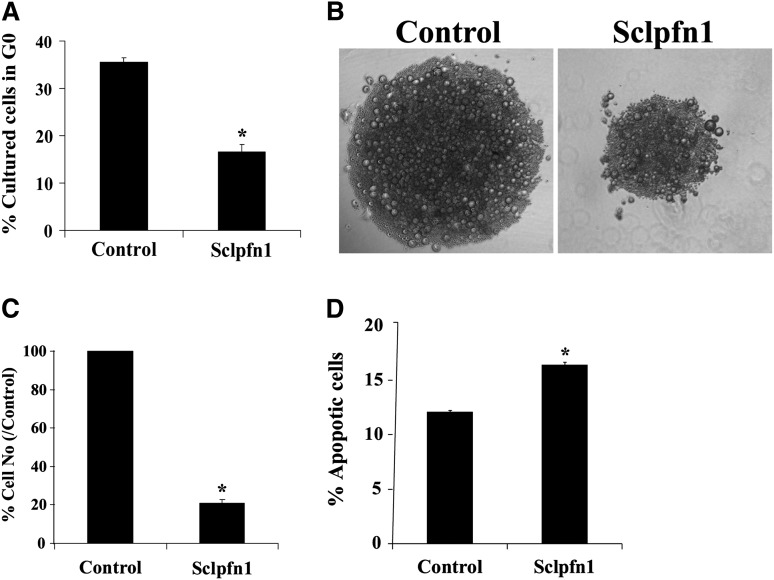
To understand the relationship between apoptosis and BM exhaustion of Sclpfn1 HSCs, we analyzed the growth of pfn1−/− and control HSCs in vitro. Similar to the observed loss of quiescence in vivo, null HSCs had faster cycling in culture than WT HSCs (Figure 4A). This suggests that pfn1 negatively regulates HSC proliferation. The null HSCs expanded much more slowly than their WT counterparts in vitro (Figure 4B-C). To explain these seemingly contradictory observations, we analyzed apoptosis and differentiation of control and pfn1−/− HSCs in culture (Figure 4D; supplemental Figure 4A). We found that null HSCs had significantly increased levels of apoptosis relative to controls (Figure 4D). Similar to results obtained in vivo, pfn1−/− HSCs differentiated normally in culture (supplemental Figure 4A). Overall, these in vivo and in vitro results suggest that pfn1 supports the survival and quiescence of HSCs. The deletion of pfn1 in HSCs led to increased apoptosis, cycling, and mobilization of HSCs, all of which account for the BM failure of the mutant mice.
Figure 4.
Pfn1 supports the survival of HSCs. (A) Cultured HSCs were stained with Hoechst 33342 and pyronin Y and analyzed for cell cycle stage. The percentages of G0 cells in cultured control and Sclpfn1 HSCs are shown (n = 3; *P < .05). (B) LT-HSCs (as Lin−Sca-1+Kit+Flk2−CD34− cells) were isolated from control and Sclpfn1 mice 4 days after tamoxifen treatment and cultured in STF medium for 8 to 10 days. Figures are representative for samples after a 10-day culture. (C) Quantification of control and Sclpfn1 HSCs after 10 days in culture (n = 3; *P < .05). (D) Cultured HSCs from control and Sclpfn1 mice were analyzed for apoptosis using Annexin V/7-AAD staining (n = 3; *P < .05).
Pfn1 supports glycolysis of HSCs
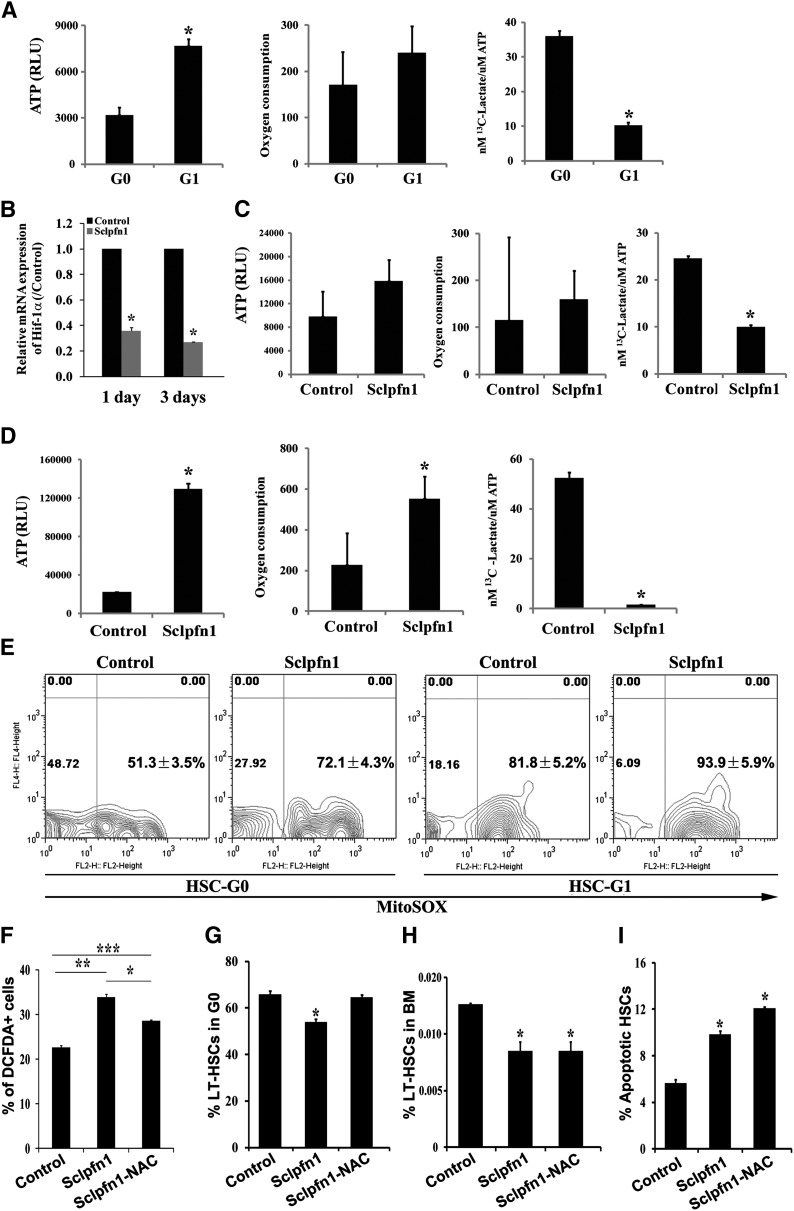
Previous studies suggested that some cytoskeleton molecules, including moesin, γ-actin, and cofilin-1, may be involved in metabolic regulation.20 We recently demonstrated that HSCs have a distinct metabolic profile and prefer to use glycolysis rather than mitochondrial oxidation as their main energy source.1 We thus sought to test whether pfn1, as an important cytoskeleton molecule, plays a role in HSC metabolism. To this end, we first examined whether metabolic properties of HSCs changed over the course of the cell cycle. We collected LT-HSCs at G0 and G1 by fluorescence-activated cell sorter and measured their ATP content, oxygen consumption, and glycolytic flux relative to ATP level. We found that the G0 HSCs, which are quiescent, had 41.10 ± 4.30% ATP content and 3.59 ± 0.16-fold higher glycolytic flux per ATP than did the G1 population (Figure 5A). G0 HSCs also showed a lower level of oxygen consumption on average (Figure 5A). These results, for the first time, demonstrate that HSCs in different stages of the cell cycle have different metabolic preferences.
Figure 5.
Pfn1 regulates metabolism of HSCs. (A) Measurement of ATP content, oxygen consumption, and labeled glycolytic flux per ATP in G0 and G1 fractions of LT-HSCs (as Lin−Sca-1+Kit+Flk2−CD34− cells) (n = 3-6; *P < .05). (B) Hif-1a expression in control and Sclpfn1 HSCs at days 1 and 3 after tamoxifen treatment (n = 3; *P < .05). (C-D) Metabolic profiles (ATP content, oxygen consumption, and glycolytic flux per ATP) in control and Sclpfn1 HSCs at (C) day 2 or (D) day 8 after tamoxifen treatment. Sclpfn1 HSCs are much less glycolytic (n = 3-6; *P < .05). (E) Mitochondrial super-oxide (MitoSOX) was analyzed in G0 and G1 cells from control and Sclpfn1 LT-HSCs (n = 3). (F-I) Control and Sclpfn1 mice were treated with tamoxifen for 7 days followed by NAC treatment of another 8 days. (F) Levels of ROS were determined by analysis for carboxy-DCFDA, and percentage of DCFDA+ LT-HSCs are shown (n = 4-6; */**/***P < .05). (J) Cell cycle status, (H) frequency, and (I) levels of apoptosis of HSCs were analyzed (n = 3; *P < .05).
Because pfn1 deletion in HSCs leads to decrease of G0 that possesses greater glycotic flux, we sought to test whether pfn1 plays a role in regulation of HSC metabolism. We started to measure the expression of hypoxia inducible factor 1 (Hif-1)α, an important regulator of hypoxia and metabolism,1,21 by real-time RT-PCR in control and pfn1−/− HSCs. As shown in Figure 5B, Hif-1α expression was dramatically decreased to 35.91 ± 2.40% or 27.02 ± 0.09% at day 1 or 3, respectively, after tamoxifen treatment in null HSCs. This result is in agreement with our finding that pfn1 up-regulates EGR1 (supplemental Figure 3A; Figure 6E) and the fact that EGR1 directly transactivates Hif-1α 22 (Figure 6D,F). Our results thus suggest that pfn1 may play a role in regulation of glycolysis and mitochondrial oxidation. Indeed, the HSC-specific deletion of pfn1 resulted in decreased glycolytic flux per ATP (40.5 ± 0.94% compared with that of controls) at only 2 days after tamoxifen treatment (Figure 5C). At 8 days after tamoxifen treatment, the Sclpfn1 HSCs had 6.56 ± 1.42-fold increased levels of ATP, a 2.5-fold (2.50 ± 0.12%) increase of oxygen consumption, and a 97.05 ± 0.21% decrease of glycolytic flux per ATP compared with controls (Figure 5D). Consistently, the glycolysis-related enzymes were down-regulated and Kreb cycle–related enzymes PDHA1 and PDHB were up-regulated in Sclpfn1 HSCs (supplemental Figure 5A-B). Interestingly, this metabolic switch on pfn1 deletion is specific to HSCs, because it did not occur in various hematopoietic progenitors (supplemental Figure 5C-E). These results clearly demonstrate that deletion of pfn1 results in a switch from anaerobic glycolytic metabolism to oxidative phosphorylation in HSCs.
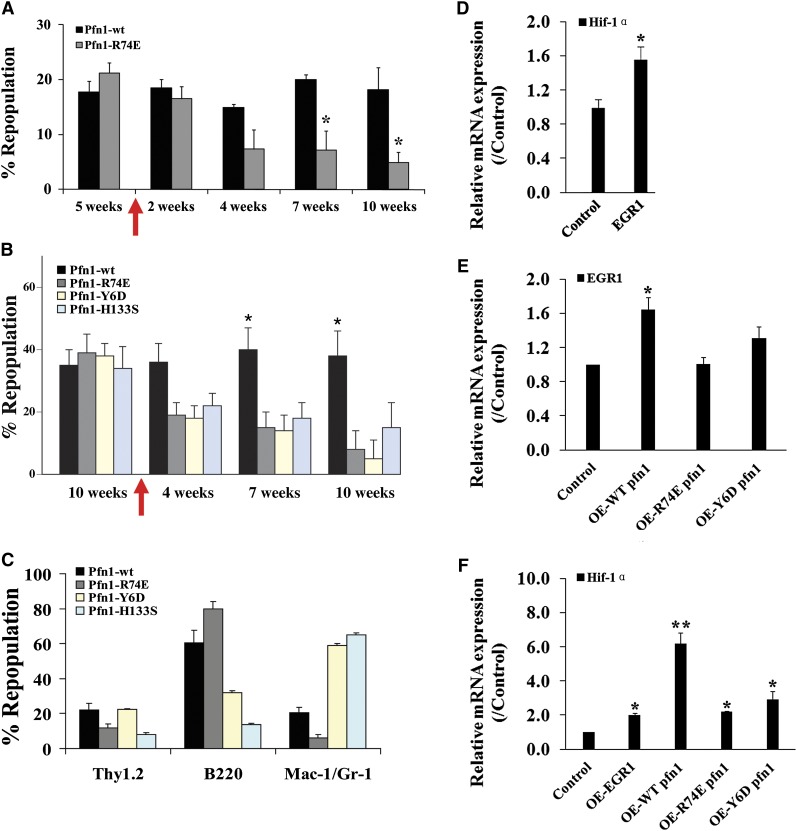
Figure 6.
Actin and poly-proline binding is critical to pfn1’s function in HSCs. (A-B) E16 fetal liver Sclpfn1 Lin− cells were infected with retrovirus encoding WT, actin binding–deficient pfn1, or poly-proline-binding–deficient pfn1 and were then transplanted into lethally irradiated CD45.1 recipients. Five to 10 weeks later, the mice were treated with tamoxifen for 2 weeks. Donor engraftment at indicated time after transplant is shown (n = 5). (C) Multilineage contribution of donor cells in recipients at 10 or 15 weeks after transplant (n = 5). (D) Control or EGR1 retrovirally expressed Sclpfn1 BM cells were transplanted into CD45.1 recipients with competitors. Mice were treated with tamoxifen after 5 weeks. Donor-derived CD45.2+ LSK cells were sorted, and expression of Hif-1α was measured (n = 3). (E) Pfn1 or Pfn1 mutants were retrovially expressed in WT LSK cells, and expression of EGR1 was measured (n = 3). (F) Pfn1, Pfn1 mutants, or EGR1 was retrovially expressed in WT LSK cells, and expression of Hif-1α was measured (n = 3). *P < .05, different from control values; **P < .05, different from mutant pfn1 values.
To identify the possible cause for this metabolic switch in HSCs on pfn1 deletion, we examined the ATP production and oxygen consumption of control and Sclpfn1 HSCs during in vitro culture (supplemental Figure 4B-C). Although the in vitro cultured Sclpfn1 HSCs have increased apoptosis (Figure 4D), unlike the in vivo situation, they did not show a metabolic switch (supplemental Figure 4B-C). In addition, the in vivo metabolic switch happens in pfn1-deleted HSCs much earlier than the apoptosis changes. These results indicate that the altered metabolic profiling of Sclpfn1 HSCs does not result from the apoptosis of Sclpfn1 HSCs. Given that pfn1 up-regulates EGR1 (supplemental Figure 3A) that suppresses mobilization17 and EGR1 transactivates Hif-1α22 (Figure 6D-F) and partially rescues pfn1-null metabolic phenotype (supplemental Figure 3D), we reasoned that the change of metabolic profiling on pfn1 deletion intrinsically links to the mobilization of HSCs.
Mitochondria are considered the major source of ROS in cells and play important roles in aging, degenerative diseases, and HSC dysfunction.23-25 We found that the level of mitochondrial super-oxide (MitoSOX) is much lower in G0 cells than G1 cells in wild-type LT-HSCs, and MitoSOX levels in either G0 or G1 cells of Sclpfn1 LT-HSCs are higher than that of controls, respectively (Figure 5E). Accordingly, the mitochondrial potential in these cells showed a similar trend (supplemental Figure 5F). Our results are consistent with the view that ROS can induce loss of quiescence and apoptosis of HSCs.26 Our previous results indicate that ROS production in HSCs plays an important role in regulation of the metabolic phenotype and cell fates.15 To investigate whether ROS is responsible for the phenotypes of Sclpfn1 HSCs, we treated Sclpfn1 and control mice with the antioxidant NAC for 7 to 8 days and then examined the cell fates of HSCs. As shown in Figure 5F, the level of ROS in Sclpfn1 HSCs was significantly higher than in control cells and was statistically decreased after an 8-day treatment with NAC. Interestingly, NAC treatment reversed the loss of quiescence of the null HSCs (Figure 5G; supplemental Figure 5G) but had no effect on HSC frequency or apoptosis in Sclpfn1 mice (Figure 5H-I; supplemental Figure 5H-I). Taken together, these data indicate that metabolic changes are one of the earliest events in pfn1-deficient HSCs in BM, which accounts for their loss of quiescence.
Actin and poly-proline binding are critical to pfn1’s function in HSCs
Previous studies showed that the actin binding and the poly-proline binding ability of pfn1 are critical for the fission yeast viability and chondrocyte function.3,27 To investigate the relative contributions of actin and nonactin ligands to pfn1 activity in HSCs, we infected Sclpfn1 HSCs with retroviruses encoding WT pfn1, an actin binding–deficient point mutant of pfn1 (R74E), or pfn1 point mutants (Y6D and H133S) with reduced affinities for poly-proline.3 Five weeks after transplantation of the infected HSCs, tamoxifen was used to induce deletion of pfn1 to produce pfn1−/− donor HSCs. As expected, retroviral infection with the WT pfn1, but not the actin binding–deficient mutant or the poly-proline binding–deficient mutants, was able to rescue the repopulation activity of null HSCs (Figure 6A-C). Retroviral expression of EGR1 was able to increase Hif-1α level in Sclpfn1 cells (Figure 6D,F), and WT pfn1 expression also up-regulated EGR1 and Hif-1α expression (Figure 6E-F). By contrast, the actin binding–deficient mutant and the poly-proline binding–deficient mutant pfn1 had a lower ability to elevate EGR1 and Hif-1α expression (Figure 6E-F). It is of note that the proline-binding mutants skewed myeloid/B-lymphoid lineage repopulation (Figure 6C). Taken together, these data suggest that the actin binding and poly-proline binding activities of pfn1 are important for its regulation of HSC activity.
In summary, we provide strong evidence that pfn1 plays critical roles in the maintenance of multiple cell fates and metabolism of HSCs. As summarized in supplemental Figure 6, the loss of pfn1 in HSCs leads to increased mobilization and the metabolism switch, and apoptosis of HSCs over time in vivo, all of which contribute to BM failure.
Discussion
It is well established that pfn1 plays critical roles in cell survival, proliferation, and motility in various cell types.2-5,8,27 Its in vivo role in stem cells, however, has not been studied. Here we showed that Pfn1 plays multiple essential roles in HSCs and is required for steady-state hematopoiesis. Different from its migration/proliferation roles in many cell types,3,5-7 pfn1 is key to maintaining the retention and quiescence of HSCs in the BM. It is also critical for the survival but plays no significant role in differentiation or homing of HSCs. Therefore, although pfn1 is ubiquitously expressed, it has specific function in HSCs. This function is partially contributed by its up-regulation of the expression of EGR1, which has been shown to be required for the quiescence and retention of HSCs in the BM.17,18 Although it is known that the forced expression of EGR1 causes changes in differentiation,28 we demonstrated that it can partially rescue the Sclpfn1 HSC phenotype. Importantly, EGR1 is known to be able to bind to and activate the promoter of Hif-1α,22 as confirmed in our system. Therefore, at least partially through the axis of pfn1/Gα13/EGR1, pfn1 regulates BM retention of HSCs that contributes to maintaining their glycolytic metabolism. Treatment of pfn1-deficient mice with NAC reversed the ROS level and loss of quiescence of HSCs, suggesting that pfn1-maintained metabolism is required for the quiescence of HSCs. We further demonstrated that pfn1’s function in HSCs depends on its actin-binding and poly-proline–binding activities.
Recently, we demonstrated that HSCs have a distinct metabolic profile, and Meis1 and Hif-1a are involved in the regulation of this unique metabolism.1,15 However, the relationship between metabolism and cell fates (quiescence, self-renewal, differentiation, apoptosis, and motility) of HSCs is not known. Here, our results suggest that a metabolic quiescence is mechanistically linked to the cell cycle quiescence of stem cells. We showed that pfn1 plays an important role in regulation of the metabolism of HSCs. Deletion of pfn1 in HSCs leads to down-regulation of the metabolism master regulator Hif-1α and a switch from glycolysis to mitochondrial respiration. Importantly, this metabolic change happened earlier than the loss of quiescence of HSCs. It is well known that mitochondrial oxidation is accompanied by significantly increased ROS production,29-33 and the free radical scavenger NAC restored quiescence of pfn1-null HSCs. Therefore the metabolic preference of HSCs regulates HSC cell cycle status. We also found that quiescent HSCs (at G0) have higher levels of glycolysis than proliferating HSCs (at G1). Therefore, the altered metabolic profiling of the pfn1-null HSCs may also partially contribute to the cell cycle change. We speculate that a feedback loop controls the metabolism and cell cycle state of stem cells. Further investigation will be needed to uncover the underlying mechanism for this regulation.
The relationships among different cell fates of HSCs are complicated. For example, whether homing and mobilization are 2 sides of the same coin is not clear, although in many cases homing and mobilization are clearly coupled. Expression of integrin α4/β1 and SDF1/CXCR4 are needed for both homing enhancement and retention of HSCs in BM.34,35 However, Gαs is critical for the homing but not the mobilization of HSCs.36 That there are meaningful molecular distinctions between homing, engraftment, and retention processes is also demonstrated by our finding that pfn1 deficiency uncouples these events. Unlike Gαs, pfn1 is essential for the mobilization but not homing of HSCs. Our results also suggest that quiescence and mobilization/survival of HSCs can be independent events. More detailed studies, including those using high-resolution, real-time imaging tools to elucidate the roles of pfn1 in regulation of the activities of HSCs in and outside of BM, will provide important new insights into the relationships among HSCs, niche, and metabolism.
Here we demonstrated that both the actin-binding and the poly-proline–binding abilities of pfn1 are essential to HSC activity. In future studies, it will be important to dissect which of these 2 activities of pfn1 is responsible for BM retention, survival, differentiation, and metabolic preference of HSCs. The role of the proline-binding function will be particularly interesting. It is known that profilins interact with >12 proline-rich peptide-containing proteins, including those involved in cytoskeleton polymerization (Arp2/3), focal adhesion (Ena-vasodilator stimulated phosphoprotein, Palladin, Mena, and vasodilator-stimulated phosphoprotein), synaptic scaffold synthesis (Delphilin, Aczonlin, Drebrin and Gephyrin), membrane trafficking (valosin containing protein, Clathrin, and Annexin I), ρ GTPase signaling (WAS/WASL interacting protein, WAS protein family, member 1, Wiskott-Aldrich syndrome, formin-related gene in leukocytes and mDiaphanous), and nuclear export (Exportin 6 and survival of motor neuron 1).37,38 Here we added 1 more molecule, Gα13, to this list, and our study suggested that EGR1, as a target of Gα13, is a mediator of pfn1’s function in HSC BM retention and links pfn1 to Hif-1α up-regulation and glycolytic metabolism. It is noteworthy that the Pfn-null HSC phenotype is much more severe than that of the EGR1-null mice. Given the large number of pfn1-interacting proteins, it is likely that the pfn1/Gα13/EGR1 axis represents a branch of the pfn1 downstream effector network. The clarification of the contribution of these and other molecules to pfn1 function in HSCs will lead to a significantly improved understanding of the interplay between stem cells and their niches and the pathology of HSC-related diseases.
Supplementary Material
Acknowledgments
The authors thank Dr Joachim R. Goethert from Universitaetsklinikum Essen and Dr Reinhard Fassler from the Max Planck Institute of Biochemistry for kindly providing SCL-CRE/ER mice and pfn1fl/fl mice, respectively.
This work was supported by National Institutes of Health (National Cancer Institute) grant 1R01CA172268, Department of Defense grant W81XWH-10-1-0429, Cancer Prevention Research Institute of Texas grant RP100402, National Natural Science Foundation of China grant 21328503, the Program for Professor of Special Appointment (Eastern Scholar), Shanghai, China, Robert A. Welch Foundation grant I-1834, and the Gabrielle’s Angel Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.Z., Z.L., and C.C.Z. contributed to design, experimental performance, interpretation, and writing; R.T.B., M.C., R.J.D., and H.S. contributed to experimental performance, interpretation, and writing; and F.K., X.K., X.L. Q.W., and G.-Q.C. contributed to experimental performance.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hesham Sadek, 5323 Harry Hines Blvd, Dallas, TX 75390-8573; e-mail: Hesham.Sadek@UTSouthwestern.edu; and Cheng Cheng Zhang, 5323 Harry Hines Blvd, Dallas, TX 75390-9133; e-mail: Alec.Zhang@UTSouthwestern.edu.
References
- 1.Simsek T, Kocabas F, Zheng J, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7(3):380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA. 2001;98(7):3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttcher RT, Wiesner S, Braun A, et al. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 2009;28(8):1157–1169. doi: 10.1038/emboj.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120(4):717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- 5.Ding Z, Lambrechts A, Parepally M, Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci. 2006;119(Pt 19):4127–4137. doi: 10.1242/jcs.03178. [DOI] [PubMed] [Google Scholar]
- 6.Rust MB, Kullmann JA, Witke W. Role of the actin-binding protein profilin1 in radial migration and glial cell adhesion of granule neurons in the cerebellum. Cell Adhes Migr. 2012;6(1):13–17. doi: 10.4161/cam.19845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun SP, Ryu JM, Jang MW, Han HJ. Interaction of profilin-1 and F-actin via a β-arrestin-1/JNK signaling pathway involved in prostaglandin E(2)-induced human mesenchymal stem cells migration and proliferation. J Cell Physiol. 2011;226(2):559–571. doi: 10.1002/jcp.22366. [DOI] [PubMed] [Google Scholar]
- 8.Bae YH, Ding Z, Das T, Wells A, Gertler F, Roy P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci USA. 2010;107(50):21547–21552. doi: 10.1073/pnas.1002309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoidakis J, Makridakis M, Zerefos PG, et al. Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol Cell Proteomics. 2012;11(4):M111.009449. [DOI] [PMC free article] [PubMed]
- 10.Göthert JR, Gustin SE, Hall MA, Green AR, Göttgens B, Izon DJ, Begley CG. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 2005;105(7):2724–2732. doi: 10.1182/blood-2004-08-3037. [DOI] [PubMed] [Google Scholar]
- 11.Zheng J, Huynh H, Umikawa M, Silvany R, Zhang CC. Angiopoietin-like protein 3 supports the activity of hematopoietic stem cells in the bone marrow niche. Blood. 2011;117(2):470–479. doi: 10.1182/blood-2010-06-291716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J, Umikawa M, Cui C, et al. Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature. 2012;485(7400):656–660. doi: 10.1038/nature11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynh H, Zheng J, Umikawa M, et al. IGF binding protein 2 supports the survival and cycling of hematopoietic stem cells. Blood. 2011;118(12):3236–3243. doi: 10.1182/blood-2011-01-331876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Umikawa M, Zhang S, et al. Ex vivo expanded hematopoietic stem cells overcome the MHC barrier in allogeneic transplantation. Cell Stem Cell. 2011;9(2):119–130. doi: 10.1016/j.stem.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kocabas F, Zheng J, Thet S, et al. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood. 2012;120(25):4963–4972. doi: 10.1182/blood-2012-05-432260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287(5459):1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 17.Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2(4):380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vara Prasad MV, Dhanasekaran N. GTPase deficient mutant of G(alpha13) regulates the expression of Egr-1 through the small GTPase Rho. Oncogene. 1999;18(8):1639–1642. doi: 10.1038/sj.onc.1202461. [DOI] [PubMed] [Google Scholar]
- 20.Miharada K, Karlsson G, Rehn M, Rörby E, Siva K, Cammenga J, Karlsson S. Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell. 2011;9(4):330–344. doi: 10.1016/j.stem.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Takubo K, Goda N, Yamada W, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7(3):391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Sperandio S, Fortin J, Sasik R, Robitaille L, Corbeil J, de Belle I. The transcription factor Egr1 regulates the HIF-1alpha gene during hypoxia. Mol Carcinog. 2009;48(1):38–44. doi: 10.1002/mc.20454. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto K, Araki KY, Naka K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1(1):101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Pervaiz S, Taneja R, Ghaffari S. Oxidative stress regulation of stem and progenitor cells. Antioxid Redox Signal. 2009;11(11):2777–2789. doi: 10.1089/ars.2009.2804. [DOI] [PubMed] [Google Scholar]
- 25.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12(4):1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnaraju K, Hoffman B, Liebermann DA. Early growth response gene 1 stimulates development of hematopoietic progenitor cells along the macrophage lineage at the expense of the granulocyte and erythroid lineages. Blood. 2001;97(5):1298–1305. doi: 10.1182/blood.v97.5.1298. [DOI] [PubMed] [Google Scholar]
- 29.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20(4):145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 30.Nohl H, Hegner D. Do mitochondria produce oxygen radicals in vivo? Eur J Biochem. 1978;82(2):563–567. doi: 10.1111/j.1432-1033.1978.tb12051.x. [DOI] [PubMed] [Google Scholar]
- 31.Miquel J, Economos AC, Fleming J, Johnson JE., Jr Mitochondrial role in cell aging. Exp Gerontol. 1980;15(6):575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- 32.Turrens JF. Superoxide production by the mitochondrial respiratory chain. Biosci Rep. 1997;17(1):3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- 33.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552(Pt 2):335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott LM, Priestley GV, Papayannopoulou T. Deletion of alpha4 integrins from adult hematopoietic cells reveals roles in homeostasis, regeneration, and homing. Mol Cell Biol. 2003;23(24):9349–9360. doi: 10.1128/MCB.23.24.9349-9360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peled A, Petit I, Kollet O, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283(5403):845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 36.Adams GB, Alley IR, Chung UI, et al. Haematopoietic stem cells depend on Galpha(s)-mediated signalling to engraft bone marrow. Nature. 2009;459(7243):103–107. doi: 10.1038/nature07859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14(8):461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Holt MR, Koffer A. Cell motility: proline-rich proteins promote protrusions. Trends Cell Biol. 2001;11(1):38–46. doi: 10.1016/s0962-8924(00)01876-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.