Abstract
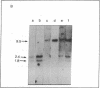
The sequence organization of the structural ovalbumin gene and flanking sequences in native chicken DNA was studied by restriction mapping and filter hybridization using a nick-translated probe generated from pOV230, a recombinant plasmid that contains a full-length ovalbumin DNA synthesized from ovalbumin mRNA. The structural sequences of the ovalbumin gene in native chicken DNA were found to be noncontiguous because at least two restriction endonucleases that do not cut the structural sequence do cleave the natural gene into multiple fragments by cleaving within nonstructural sequences interspersed between the structural sequences. The observation that all ovalbumin DNA-containing sequences were contained within a single DNA fragment generated by BamHI digestion of total chicken DNA has allowed us to construct an inclusive restriction map of the natural ovalbumin gene which contains at least two "insert regions." These regions may be further subdivided into alternating structural and insert sequences. Both insert regions were located within the peptide-coding regions of the gene and the sizes of these insert regions were estimated to be approximately 1.0 and 1.5 kilobase pairs, respectively.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Brack C., Tonegawa S. Variable and constant parts of the immunoglobulin light chain gene of a mouse myeloma cell are 1250 nontranslated bases apart. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5652–5656. doi: 10.1073/pnas.74.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Mandel J. L., Chambon P. Ovalbumin gene is split in chicken DNA. Nature. 1977 Nov 24;270(5635):314–319. doi: 10.1038/270314a0. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977 Sep;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Harris S. E., Means A. R., Mitchell W. M., O'Malley B. W. Synthesis of (3H)DNA complementary to ovalbumin messenger RNA: evidence for limited copies of the ovalbumin gene in chick oviduct. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3776–3780. doi: 10.1073/pnas.70.12.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. The rabbit beta-globin gene contains a large large insert in the coding sequence. Cell. 1977 Dec;12(4):1097–1108. doi: 10.1016/0092-8674(77)90172-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. S., Schimke R. T. Ovalbumin messenger RNA: evidence that the initial product of transcription is the same size as polysomal ovalbumin messenger. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4327–4331. doi: 10.1073/pnas.71.11.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan J. J., Harris S. E., Woo S. L., Robberson D. L., O'Malley B. W. The synthesis and properties of the complete complementary DNA transcript of ovalbumin mRNA. Biochemistry. 1976 Jan 13;15(1):223–233. doi: 10.1021/bi00646a034. [DOI] [PubMed] [Google Scholar]
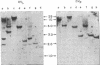
- Monahan J. J., Woo S. L., Liarakos C. D., O'Malley B. W. Ovalbumin gene. Action of restriction endonucleases upon DNA coding sequence. J Biol Chem. 1977 Jul 10;252(13):4722–4728. [PubMed] [Google Scholar]
- O'Malley B. W., Means A. R. Female steroid hormones and target cell nuclei. Science. 1974 Feb 15;183(4125):610–620. doi: 10.1126/science.183.4125.610. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975 Mar;4(3):189–189. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- Schwartz R. J., Tsai M-J, Tsai S. Y., O'Malley B. W. Effect of estrogen on gene expression in the chick oviduct. V. Changes in the number of RNA polymerase binding and initiation sites in chromatin. J Biol Chem. 1975 Jul 10;250(13):5175–5182. [PubMed] [Google Scholar]
- Sharp P. A., Gallimore P. H., Flint S. J. Mapping of adenovirus 2 RNA sequences in lytically infected cells and transformed cell lines. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):457–474. doi: 10.1101/sqb.1974.039.01.058. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sullivan D., Palacios R., Stavnezer J., Taylor J. M., Faras A. J., Kiely M. L., Summers N. M., Bishop J. M., Schimke R. T. Synthesis of a deoxyribonucleic acid sequence complementary to ovalbumin messenger ribonucleic acid and quantification of ovalbumin genes. J Biol Chem. 1973 Nov 10;248(21):7530–7539. [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. M., Tiemeier D. C., Seidman J. G., Peterlin B. M., Sullivan M., Maizel J. V., Leder P. Intervening sequence of DNA identified in the structural portion of a mouse beta-globin gene. Proc Natl Acad Sci U S A. 1978 Feb;75(2):725–729. doi: 10.1073/pnas.75.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., Schwartz R., Kalimi M., Clark J. H., O'Malley B. W. Effects of estrogen on gene expression in chick oviduct: nuclear receptor levels and initiation of transcription. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4228–4232. doi: 10.1073/pnas.72.11.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock R., Sweet R., Weiss M., Cedar H., Axel R. Intragenic DNA spacers interrupt the ovalbumin gene. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1299–1303. doi: 10.1073/pnas.75.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Woo S. L., Chandra T., Means A. R., O'Malley B. W. Ovalbumin gene: purification of the coding strand. Biochemistry. 1977 Dec 27;16(26):5670–5676. doi: 10.1021/bi00645a003. [DOI] [PubMed] [Google Scholar]
- Woo S. L., O'Malley B. W. Hormone inducible messenger RNA. Life Sci. 1975 Oct 10;17(7):1039–1047. doi: 10.1016/0024-3205(75)90322-7. [DOI] [PubMed] [Google Scholar]
- Woo S. L., Rosen J. M., Liarakos C. D., Choi Y. C., Busch H., Means A. R., O'Malley Physical and chemical characterization of purified ovalbumin messenger RNA. J Biol Chem. 1975 Sep 10;250(17):7027–7039. [PubMed] [Google Scholar]