Abstract
Ins(1,4,5)P3 is a classical intracellular messenger: stimulus-dependent changes in its levels elicits biological effects through its release of intracellular Ca2+ stores. The Ins(1,4,5)P3 response is “switched off” by its metabolism to a range of additional inositol phosphates. These metabolites have themselves come to be collectively described as a signaling “family”. The validity of that latter definition is critically examined in this review. That is, we assess the strength of the hypothesis that Ins(1,4,5)P3 metabolites are themselves “classical” signals. Put another way, what is the evidence that the biological function of a particular inositol phosphate depends upon stimulus dependent changes in its levels? In this assessment, examples of an inositol phosphate acting as a cofactor (i.e. its function is not stimulus-dependent) do not satisfy our signaling criteria. We conclude that Ins(3,4,5,6)P4 is, to date, the only Ins(1,4,5)P3 metabolite that has been validated to act as a second messenger.
Keywords: Adenosine deaminase; AKT; β-cells; Calcium; cAMP; CaMKII; Chloride channel; ClC3; Compartmentalization; DNA repair; Endosomes; ERK; Frizzled receptor; GAP1IP4BP; mRNA export; Ins(1,4,5)P3; Ins(1,4,5)P4 receptor; Ins(1,3,4)P3; Ins(1,3,4,5)P4; Ins(1,3,4,5)P4 receptor; Ins(1,4,5,6)P4; Ins(3,4,5,6)P4; Ins(1,3,4,5,6)P5; InsP6; Insulin; IPMK; IPK2; IP5K; ITP; ITPK1; ITPKB; Lymphocytes; Ku; Neutrophils; Protein phosphatase; PtdIns(4,5)P2; PtdIns(3,4,5)P3; PH domain; PTEN; RASA3; Transcription; Wnt ligand
13.1 Introduction
The receptor-dependent, Ins(1,4,5)P3-mediated mobilization of intracellular Ca2+ stores (Streb et al. 1983) is now a textbook signal transduction pathway (see Taylor’s 2011 chapter). When this signaling activity was discovered, it was certainly a paradigm-altering concept. That novel idea was also highly contagious. After it emerged that Ins(1,4,5)P3 is further phosphorylated to isomers of InsP4, InsP5 and InsP6 (Fig. 13.1), we (Shears 1989) and others (Irvine and Schell 2001; York et al. 2001) proposed that some of these “higher” inositol phosphates might also have important signaling roles. This notion of “orphan signals” (Menniti et al. 1990) certainly captured the imagination (see Ismailov et al. 1996; York et al. 2001). Furthermore, the dramatic digital analogy that paints the inositol ring as a “six-bit signaling scaffold” (York et al. 2001; York 2006) implies that there are signaling roles for many members of the inositol phosphate family. However, the thesis of this review is that, except for one notable exception, we are still a long way from confirming that these “orphan” inositol phosphates truly function as classic second messengers.
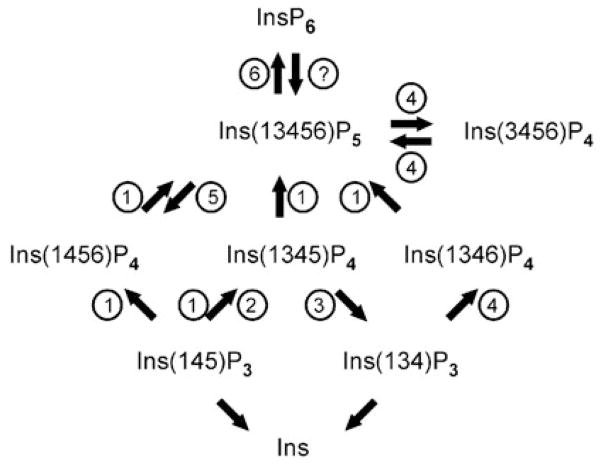
Fig. 13.1.
Inositol phosphate metabolism. The figure shows the pathway of Ins(1,4,5)P3 metabolism. The numbers in circles indicate the different enzymes that are involved: 1, IPK2/IPMK; 2, Ins(1,4,5)P3 3-kinases; 3, Ins(1,4,5)P3/ Ins(1,3,4,5)P4 5-phopshatase; 4, ITPK1; 5, PTEN; 6, IP5K. There is a candidate for the question mark—MIPP—but it is uncertain how that enzyme can access its substrate (see text for details). The inositol pyrophosphates are not shown in this figure (they are the subject of a separate chapter (Saiardi 2011)). The enzymes that dephosphorylate Ins(1,4,5)P3 and Ins(1,3,4)P3 to inositol are not shown, as this review is only concerned with metabolites that have received attention as being cellular signals. Note that the positional specificity of IPK2/IPMK shows phylogenetic variation. In yeasts, Ins(1,4,5)P3 is phosphorylated primarily to Ins(1,4,5,6)P4, in mammals the product is predominantly Ins(1,3,4,5)P4, and the enzyme in flies produces roughly equal quantities of both InsP4 isomers
In anticipation that our colleagues might raise their eyebrows in reaction to that last sentence, we will quickly provide some clarification. The second messenger concept (Robison et al. 1968) owes its existence to the discovery of cyclic AMP (Rall and Sutherland 1958; Sutherland and Rall 1958), a diffusible molecule that, in response to an extracellular stimulus, is generated at (or released from) a particular subcellular site. In this example, a change in the concentration of cAMP leads to the modification of a protein kinase activity. If a soluble inositol phosphate is to act as an intracellular signal it must also exhibit a stimulus-dependent change in its concentration. Additionally, the “information” encoded by the inositol phosphate should be converted—transduced—from one chemical form to another. These are significant criteria in the current context.
Take InsP6 for example. This polyphosphate is an essential co-factor for adenosine deaminase, an mRNA editing enzyme; without InsP6 at its core, the protein does not fold correctly and is devoid of catalytic activity (Macbeth et al. 2005). This is unarguably an important function, but there is no evidence that InsP6 serves as a stimulus-dependent regulator of this enzyme. In fact, in add-back experiments, a maximal effect of InsP6 upon deaminase activity was attained at a polyphosphate concentration of 1 μM (Macbeth et al. 2005), which is 10–50-fold less than the concentration that is always present in cells (Pittet et al. 1989; Oliver et al. 1992; Barker et al. 2004; Bunce et al. 1993). That is, the deaminase should always be fully InsP6-activated by default. A similar argument can be made of the requirement that S. cerevisiae has for InsP6, in order that mRNA can be exported out of the nucleus (Alcázar-Román et al. 2006; York et al. 1999). Here, InsP6 acts by stimulating the ATPase activity of Dbp5, which is envisaged to be a molecular ratchet that pulls mRNA out of the nucleus (Yu et al. 2004). This function is fulfilled by only 0.1 μM InsP6, a small fraction of the total cellular concentration. Again, the role of InsP6 in this process is almost certainly as a cofactor rather than as the “regulator” or “signal” that it was originally proposed to be (York et al. 1999). Once more, let’s acknowledge that these are important discoveries. They are just not examples of cell signaling activities.
Thus, in the current review it is our intention to assess the evidence that a biological response can be attributed to a stimulus-dependent alteration in the levels of a particular inositol phosphate. While Ins(1,4,5)P3 clearly fulfils that criterion, it will not be discussed here as it is the dedicated subject of another chapter (Taylor 2011). The inositol “pyrophosphates” are also reserved for the attention of a separate chapter (Saiardi 2011). Finally, the emphasis in this review is on the inositol phosphates themselves.
13.2 Ins(1,3,4,5)P4
Receptor dependent Ca2+ mobilization arises not just by Ca2+ release from intracellular stores, but also through Ca2+ entry across the plasma membrane; the fact that the two processes are tightly linked lies at the heart of the “capacitative calcium entry” hypothesis (Putney 1986), which was subsequently refined and repackaged as “store-operated calcium entry” (Hoth and Penner 1992; Parekh and Penner 1997). In the immediate aftermath of Ins(1,4,5)P3 being discovered to release Ca2+ from intracellular stores (Streb et al. 1983), a 3-kinase was discovered that phosphorylated Ins(1,4,5)P3 (Irvine et al. 1986). At that time, it was not known how Ca2+ release was coupled to Ca2+ entry. Thus, Ins(1,3,4,5)P4 became a prime suspect for signaling Ca2+ entry (Irvine 1986). The initial evidence seemed highly incriminating: stimulus-dependent increases in cellular levels of Ins(1,4,5)P3 are followed shortly afterwards by several-fold increases in levels of Ins(1,3,4,5)P4 (Batty et al. 1985). That is a valuable credential if Ins(1,3,4,5)P4 is to be a cellular signal. Furthermore, Irvine’s group (Irvine and Moor 1986) reported that micro-injection of Ins(1,3,4,5)P4 into sea urchin eggs raised the fertilization envelope in a manner that was dependent upon extracellular [Ca2+]. To explain how Ins(1,3,4,5)P4 might mediate a physiological activity that was dependent upon the Ca2+ outside the cell, it was proposed that this inositol phosphate somehow stimulates Ca2+ entry (Irvine and Moor 1986).
However, as (Schell 2010) noted in a recent review, we are still searching to identify an Ins(1,3,4,5)P4-based signaling cascade, now more than 20 years after Irvine’s initial experiments. Not in the least because Ins(1,3,4,5)P4 has been a discordant beast, producing conflicting data in the hands of different groups. For example, the early results obtained from the sea urchin eggs proved impossible to reproduce in subsequent studies (Irvine and Moor 1987; Crossley et al. 1988). Irvine and colleagues were unable to account for this difficulty (Irvine and Moor 1987), and so they turned to other model systems in which they could test their hypothesis (Changya et al. 1989; Morris et al. 1987; Loomis-Husselbee et al. 1996). For example, they used Ca2+-activated K+ channels in mouse lacrimal acinar cells as a readout of the degree of Ca2+ mobilization, and they reported that Ins(1,3,4,5)P4 augmented the Ins(1,4,5)P3-induced Ca2+ response (Morris et al. 1987). Initially, a stimulation of Ca2+ entry by Ins(1,3,4,5)P4 was put forward as the explanation (Morris et al. 1987), although subsequently (Changya et al. 1989) it was suggested to be more likely that Ins(1,3,4,5)P4 somehow augmented intracellular Ca2+ mobilization. That idea was also consistent with a separate series of experiments performed with permeabilized L-1210 cells (Loomis-Husselbee et al. 1996). In contrast, Bird et al. (Bird et al. 1991; Bird and Putney 1996), who also worked with mouse lacrimal cells, reported that Ins(1,4,5)P3 by itself maximally activates Ca2+-activated K+ channels; Ins(1,3,4,5)P4 was not required. After some debate of this topic in the literature (see (Putney 1992; Irvine 1992)), Irvine and colleagues (Smith et al. 2000) later described how the two group’s use of slightly different cell preparations might largely explain the contrary data. (Bird et al. 1991) had studied cells that had been in primary culture for up to 24 h. In contrast, Irvine’s group had used freshly isolated cells. So, which laboratory used the most appropriate cell preparation? Irvine and colleagues (Smith et al. 2000) accepted that the continuous morphology of the endoplasmic reticulum that is observed in cultured cells reflects a more physiologically-relevant model; this organelle is somewhat fragmented in freshly isolated lacrimal cells. Thus, Irvine et al. (Smith et al. 2000) accept that their model might be less biologically relevant. Nevertheless, they (Smith et al. 2000) argued, the effects of Ins(1,3,4,5)P4 that they observed had to be an exaggeration of a physiological event, rather than an artifact. Perhaps, they suggested, Ins(1,3,4,5)P4 acted by influencing the continuity of endoplasmic reticulum. However, later studies (Brough et al. 2005) did not support that hypothesis.
In their later studies Irvine and colleagues (Changya et al. 1989; Smith et al. 2000) somewhat de-emphasized a possible role for Ins(1,3,4,5)P4 in regulating Ca2+ entry into cells. Others (Hermosura et al. 2000) have also shown that store-operated Ca2+ entry is not activated by Ins(1,3,4,5)P4. Yet, there continue to be occasional electrophysiological demonstrations of plasma membrane Ca2+ currents being stimulated by Ins(1,3,4,5)P4, although such data have not been placed in a physiologically-adequate context. In one example of this genre, (Luckhoff and Clapham 1992), 30 μM Ins(1,3,4,5)P4 was reported to enhance Ca2+-activated Ca2+ current in excised inside-out patches. However, such high levels of Ins(1,3,4,5)P4 are not biologically relevant. In most cases, cellular Ins(1,3,4,5)P4 levels after receptor activation would not be expected to exceed 3–4 μM (Guse et al. 1993; Barker et al. 1992; Huang et al. 2007), and initial work with optical sensors do not reveal any compartmentalization of Ins(1,3,4,5)P4 synthesis (Sakaguchi et al. 2010). (Luckhoff and Clapham 1992) were able to get lower concentrations of Ins(1,3,4,5)P4 to activate a plasma membrane current when Mn2+ was used as a Ca2+ surrogate, but neither the ion selectivity of this current, nor its biophysical characteristics, match those of store-operated calcium entry (Parekh and Penner 1997).
During their work with lacrimal cells, Irvine and colleagues (Changya et al. 1989; Smith et al. 2000) went to great lengths to prove that their effects of Ins(1,3,4,5)P4 upon Ca2+ mobilization could not be explained by inhibition of Ins(1,4,5)P3 metabolism (Changya et al. 1989; Smith et al. 2000). It is therefore somewhat ironic that Penner and colleagues (Hermosura et al. 2000) concluded that in certain circumstances inhibition of Ins(1,4,5)P3 metabolism by Ins(1,3,4,5)P4 was indeed a genuine mechanism by which Ca2+ mobilization can be enhanced. More precisely, it was argued that Ins(1,3,4,5)P4 set “a discriminatory time window for coincidence detection that enables selective facilitation of Ca2+ influx by appropriately timed low-level receptor stimulation”. However, to achieve these effects in vitro, 5–20 μM concentrations of Ins(1,3,4,5)P4 were required (Hermosura et al. 2000), which, as mentioned above, are well above those that prevail inside cells.
Ins(1,3,4,5)P4 has also been reported to inhibit Ca2+ signaling. For example, in vitro Ins(1,3,4,5)P4 can inhibit Ins(1,4,5)P3 from binding to its receptor (Hermosura et al. 2000); Putney’s group (Bird and Putney 1996) had discovered this phenomenon some years earlier, but they had expressed concern that the levels of Ins(1,3,4,5)P4 that were required were too high to be physiologically relevant. In yet another twist in this tale, Ins(1,3,4,5)P4 was reported to promote Ca2+ re-uptake into the endoplasmic reticulum of permeabilized T51B liver cells (Hill et al. 1988; Boynton et al. 1990). That particular observation has never been reproduced in another cell type, and neither has a mechanistic rationale been developed.
Genetic perturbation of Ins(1,3,4,5)P4 production has also failed to yield a consistent picture of the polyphosphate’s putative role in Ca2+ mobilization. For example, Ca2+ signals in thymocytes were unaffected when Ins(1,3,4,5)P4 synthesis was compromised upon knock-out of the type B Ins(1,4,5)P3 3-kinase gene (ITPKB) (Pouillon et al. 2003). Yet, in B-lymphocytes from Itpkb−/− mice it was reported (Marechal et al. 2007) that there is a reduction in receptor-mediated Ca2+ signaling. Adding further to the confusion, Miller and colleagues (2007, 2009) observed the opposite effect; B-lymphocytes from their knock-out mice showed enhanced Ca2+ mobilization. The latter phenotype does have a quite facile explanation. Ca2+ mobilization might well be expected to be enhanced when the half-life of Ins(1,4,5)P3 is prolonged following loss of a significant route of Ins(1,4,5)P3 metabolism, i.e., an Ins(1,4,5)P3 kinase. Moreover, cells that have more robust Ins(1,4,5)P3 signals might be expected to have more depleted Ca2+ stores, and hence higher rates of store-dependent Ca2+ entry (Jia et al. 2008). However, Miller et al. (2007, 2009) interpreted their data quite differently. They argued that Itpkb−/− lymphocytes have lost an inhibitor of store-operated Ca2+ entry—Ins(1,3,4,5)P4—and that, they concluded, is why Ca2+ mobilization is enhanced in those cells.
To more directly pursue their hypothesis, Miller et al. (2007, 2009) used a cell-permeant analogue of Ins(1,3,4,5)P4. The addition of this analogue attenuated both receptor-dependent (anti-IgM) and receptor-independent (thapsigargin-mediated) increases in cytosolic [Ca2+] (Miller et al. 2007, 2009). In many of their experiments, Miller et al. reported that cellular Ca2+ levels were reduced immediately upon the addition of cell-permeant Ins(1,3,4,5)P4. The speed of those responses is, perhaps, unexpected, in view of a report (Li et al. 1997) that a delay of at least a minute should be expected, which, apparently, cannot be eliminated by increasing the concentration of the cell permeant analogue. One of the reasons for this lag is the time it takes time for the analogue to diffuse across the membrane. Additionally, each phosphate group is “protected” by two butyryloxymethyl groups (Li et al. 1997), all eight of which must be removed by intracellular esterases before the Ins(1,3,4,5)P4 can be liberated. These technical considerations raise a concern that the immediate effect of the cell permeant Ins(1,3,4,5)P4 might be non-physiological. Yet, it does seems that the response is specific: Miller et al. (2007) added cell-permeant Ins(1,4,5,6)P4 in control experiments. That had no effect upon the cell’s Ca2+ responses. Nevertheless, a more direct demonstration of Ins(1,3,4,5)P4 inhibiting a physiologically-relevant pathway for Ca2+ entry, for example in an electrophysiological assay, would be helpful.
Is it possible to rationalize all of these different reported effects of Ins(1,3,4,5)P4 upon Ca2+ mobilization? And what are we to make of experimental observations where even the order of addition of Ins(1,4,5)P3 and Ins(1,3,4,5)P4 determines whether or not an effect upon Ca2+ fluxes is observed (Loomis-Husselbee et al. 1996)? One group (Hermosura et al. 2000) has argued that the somewhat confused and inconsistent literature reflects the actions of Ins(1,3,4,5)P4 being complex and multifaceted. Perhaps, they say, the precise molecular actions of Ins(1,3,4,5)P4 are cell-specific, varying with the strength of receptor activation, or subcellular localization of Ins(1,4,5)P3-metabolic enzymes, or heterogeneity of intracellular Ca2+ stores, or differences in Ins(1,4,5)P3 receptor subtypes, or other regulatory factors. However, we put it to the jury that the case for Ins(1,3,4,5)P4 being a cellular signal for Ca2+ mobilization is unproven. Judgement should be reserved until a specific signaling activity can be reproducibly demonstrated using physiologically-relevant concentrations of Ins(1,3,4,5)P4 and, moreover, an Ins(1,3,4,5)P4-sensitive signaling entity with a defined role in Ca2+ signaling would need to be identified.
Of course, Ins(1,3,4,5)P4 could have other signaling roles that do not involve Ca2+. Indeed, Itpkb−/− mice are immunologically compromised (Pouillon et al. 2003). For example, thymocytes in Itpkb−/− mice synthesize almost no Ins(1,3,4,5)P4 and their developmental program fails (Pouillon et al. 2003). Itpkb−/− mice also exhibit defective B-lymphocyte development (Miller et al. 2007; Marechal et al. 2007) and neutrophil migration is compromised (Jia et al. 2007). We will briefly discuss three recent developments that suggest there may be Ins(1,3,4,5)P4 “receptors” that can help explain the nature of these Itpkb−/− phenotypes.
One intriguing protein to which Ins(1,3,4,5)P4 binds tightly and specifically is the Ras-Gap that was originally named GAP1IP4BP (Cullen et al. 1995). An exhaustive study in which the levels of GAP1IP4BP were genetically manipulated (Walker et al. 2002) led to the conclusion that this protein did not exert any influence upon Ca2+ mobilization. Nevertheless, there has recently been renewed interest in GAP1IP4BP, which has been re-christened as RASA3. The Ins(1,3,4,5)P4-binding region of RASA3 is now known to be a PH domain, which also binds to PtdIns(4,5)P2 (Cozier et al. 2000). Could competition between Ins(1,3,4,5)P4 and PtdIns(4,5)P2 have some signaling significance? In CHO cells, receptor-dependent PLC activation (and hence Ins(1,3,4,5)P4 accumulation) did not affect the membrane-association of GAP1IP4BP/RASA3 (Cozier et al. 2000). However, in a subsequent study with COS cells (Marechal et al. 2007), in which ITPKB was over-expressed, RASA3 was dislodged from the plasma membrane upon PLC activation. Furthermore, 30 min pre-incubation of cells with cell-permeant Ins(1,3,4,5)P4 also caused RASA3 to translocate from the plasma membrane to the cytosol (Marechal et al. 2007). This may represent a new signaling function for Ins(1,3,4,5)P4 in B-lymphocytes (Marechal et al. 2007). It was proposed that Ins(1,3,4,5)P4 regulates the intracellular location and hence the activity of a RASA3-ERK signaling pathway that controls pro-apoptotic BIM gene expression (Marechal et al. 2007). As noted elsewhere (Sauer and Cooke 2010), the further development of this hypothesis would be helped by a demonstration that Ins(1,3,4,5)P4 acts in this manner in lymphocytes.
Ins(1,3,4,5)P4 has also been reported to be an inhibitory signal for neutrophil function, by competing with PtdIns(3,4,5)P3 for binding to the PH domain of AKT (Jia et al. 2007). A cell-permeant Ins(1,3,4,5)P4 analogue was shown to diminish receptor-dependent recruitment of AKT-PH domain to the plasma membrane (Jia et al. 2007). Moreover, neutrophils from Itpkb−/− mice exhibit up-regulated AKT activity because, it was argued, more of that kinase can translocate to the plasma membrane in the absence of Ins(1,3,4,5)P4 (Jia et al. 2007).
Another proposed “receptor” for Ins(1,3,4,5)P4 is the interleukin-2 tyrosine kinase ITK, which phosphorylates and activates PLC-γ. For ITK to regulate PLC-γ, the kinase must first translocate to the plasma membrane, courtesy of the affinity for PtdIns(3,4,5)P3 of the protein’s pleckstrin homology domain. Interestingly, this receptor-dependent translocation process is inhibited in thymocytes prepared from Itpkb−/− mice (Huang et al. 2007). In that latter study it was further reported that, in vitro, Ins(1,3,4,5)P4 promotes the association of the PH-domain of ITK with an immobilized, short acyl-chain analogue of PtdIns(3,4,5)P3. An attractive feature of this phenomenon is that it was observed when using concentrations of Ins(1,3,4,5)P4 (1 μM) that are physiologically relevant. However, it is not clear exactly how Ins(1,3,4,5)P4 has this effect. In fact, as discussed above, it would normally be expected that Ins(1,3,4,5)P4 would compete with PtdIns(3,4,5)P3 for the same ligand-binding domain. To resolve this apparent paradox, Huang et al. (2007) have proposed that, if ITK were to oligomerize, then the binding of Ins(1,3,4,5)P4 to one subunit might allosterically enhance the affinity of another subunit for PtdIns(3,4,5)P3. However, that idea is not consistent with FRET analysis that has revealed cytoplasmic ITK to be monomeric in vivo (Qi et al. 2006; Qi and August 2009). Oligomerization of ITK only occurs after the kinase has already translocated to the plasma membrane (Qi et al. 2006). The folded state of monomeric ITK (Qi and August 2009) suggests that it alters its conformation upon its transfer to the plasma membrane. Thus, the alternative “induced-fit” hypothesis that Huang et al. (2007) have proposed is arguably a more likely explanation for their in vitro data. Here, initial binding of Ins(1,3,4,5)P4 is suggested to alter the conformation of ITK so that it gains an increased ligand affinity, but particularly for PtdIns(3,4,5)P3. The viability of the latter proposal is critically dependent upon accurate determinations of the relative affinities of PtdIns(3,4,5)P3 and Ins(1,3,4,5)P4. It would therefore be useful to determine these binding parameters using an in vitro technique that is closer to a physiological context than is the use of a soluble PtdIns(3,4,5)P3 analogue immobilized to beads. The preferred method (Narayan and Lemmon 2006) is to incorporate “natural” PtdIns(3,4,5)P3 into phospholipid vesicles, and then determine the affinities of the lipid (and the competing headgroup) by using surface plasmon resonance. Additionally, rather than characterizing just the PH domain fragment of ITK (Huang et al. 2007), it would be more physiological to use full-length protein.
In all three of the examples described above—RASA3, AKT and ITK—it will be important to address the concern (Irvine et al. 2006) that an Itpkb−/− phenotype could be an unpredictable consequence of a loss of non-catalytic (scaffolding?) activities of the type B Ins(1,4,5)P3 3-kinase, rather than a reduction in Ins(1,3,4,5)P4 levels. This question could be pursued by studying if the phenotype of the Itpkb−/− cells can be rescued by expression of ITPKB, and not by a kinase-dead Itpkb mutant. That being said, it also needs to be established that the biological effects that have been attributed to Ins(1,3,4,5)P4 are not actually performed by one or more of its metabolites (Fig. 13.1). For example, the elimination from cells of Ins(1,4,5)P3 3-kinase activity can lead to a reduction in levels of Ins(1,3,4,5,6)P5 and InsP6 (Leyman et al. 2007). Ins(1,3,4)P3 is another important metabolite of Ins(1,3,4,5)P4. Loss of the latter in Itpkb−/− cells (Pouillon et al. 2003) will uncouple the link between PLC activity and the Ins(3,4,5,6)P4-signaling cascade (see below). That is, cells that have reduced ITPK activity will also be encumbered by an inability to synthesize the Ins(3,4,5,6)P4 signal. That could modify cell function in a number of ways (see below). To take one pertinent example, the ClC3 Cl− channel that Ins(3,4,5,6)P4 regulates (Mitchell et al. 2008) plays an important role in neutrophil migration (Volk et al. 2008) which, as mentioned above, is defective in Itpkb−/− cells.
13.3 Ins(1,4,5,6)P4
In yeast (Odom et al. 2000; Saiardi et al. 2000) and in flies (Seeds et al. 2004), Ins(1,4,5,6)P4 is formed from Ins(1,4,5)P3 by the kinase activity of Ipk2. In a 2000 study (Odom et al. 2000) evidence was presented that this synthesis of Ins(1,4,5,6)P4 was necessary for the function of an ArgR-Mcm1 transcriptional complex that regulated the expression of ornithine transaminase. The authors of that study assayed transcriptional control in wild-type and ipk2Δ cells using “growth on ornithine as a sole nitrogen source”. However, this interpretation of the data has been disputed by others (Dubois et al. 2000), who reported that the slower growth of ipk2Δ cells was a general phenotype rather than being specific to the nutrient source. This is an important point; if slowed cell growth were to be a general phenotype, then it would no longer provide a specific readout of the expression of ornithine transaminase. Unfortunately, it has never been resolved why these two groups came to different conclusions. Yet, despite the controversy, the idea that the catalytic activity of Ipk2 might regulate transcription was taken up in subsequent studies: O’Shea and colleagues found that this kinase activity regulated Pho5 transcription (Steger et al. 2003). Additional genetic experiments indicated it was chromatin remodeling that was regulated by either Ins(1,4,5,6)P4 and/or Ins(1,3,4,5,6)P5 (another product of Ipk2 activity). These authors reported an increased accessibility of a Cla I restriction site in the Pho5 promoter in nuclei that were isolated from cells shifted to Pho5 inducing conditions (Steger et al. 2003). This increased Cla I accessibility was impaired in the ipk2Δ cells (Steger et al. 2003).
But are these phenotypes the direct consequence of altering the expression of the kinase activity of Ipk2? There are far-reaching effects upon many mRNA species in yeast strains in which the catalytic activity of Ipk2 is compromised (El Alami et al. 2003), and so it is important to separate primary regulatory events from secondary consequences. Additionally, metabolic homeostasis utilizes regulatory processes that control the expression of genes encoding metabolic enzymes. Thus, there are many links between gene regulation and metabolic status (McKnight 2003). It might be considered inevitable that control over phosphate supply to yeast, and the regulation of Pho5 expression, must be intertwined with regulation of inositol phosphate synthesis, which of course is a phosphate-consuming process. The big question, therefore, is whether this apparent effect of Ipk2 upon chromatin remodeling reflects a global metabolic control process, or instead is this really a more specific utilization of inositol phosphate turnover to control gene expression? This query could be resolved if we had a molecular justification for the intriguing genetic effects that were described by O’Shea and colleagues (Steger et al. 2003).
A molecular mechanism by which Ipk2 might control Pho5 expression has been put forward; it was proposed that Ins(1,4,5,6)P4 and Ins(1,3,4,5,6)P5 directly stimulate “nucleosome sliding” (Shen et al. 2003; Steger et al. 2003). Nucleosomes are the basic repetitive unit of chromatin: histone octomers around which are wrapped about 150 bp of DNA (Becker and Hörz 2002). Regulatory elements can be exposed when nucleosomes are nudged along the DNA helix by ATP-consuming, nucleosome remodeling factors. Wu and colleagues (Shen et al. 2003) studied nucleosome movement along a Drosophila hsp70 DNA fragment driven by the yeast SWI/SNF chromatin remodeling complex. It was reported that 500 μM of either Ins(1,4,5,6)P4 or Ins(1,3,4,5,6)P5 stimulated this nucleosome sliding (Shen et al. 2003). Unfortunately, as discussed elsewhere (Shears 2004), Wu and colleagues (Shen et al. 2003) studied the effects of inositol phosphates at concentrations that are 500–1700 times higher than estimated cellular levels. In such circumstances, it is difficult for us to accept that those effects are physiologically relevant.
It is even more difficult to imagine Ins(1,4,5,6)P4 being a signal that controls transcription in higher organisms. The mammalian homologue of Ipk2—often called IPMK—phosphorylates Ins(1,4,5)P3 to Ins(1,3,4,5)P4 (Fig. 13.1) rather than to Ins(1,4,5,6)P4 (Saiardi et al. 2001; Nalaskowski et al. 2002). So mammalian cells do not use IPK2/IPMK to regulate levels of Ins(1,4,5,6)P4. It is possible for Ins(1,4,5,6)P4 to be synthesized by dephosphorylation of Ins(1,3,4,5,6)P5, probably by the cytoplasmic and nuclear pools of PTEN that, in these particular locations, cannot access the alternative and better-known substrate, PtdIns(3,4,5)P3 (Caffrey et al. 2001; Otto et al. 2007). In any case, there is no evidence that cellular levels of Ins(1,4,5,6)P4 are receptor regulated (Menniti et al. 1990). (It has been published that Ins(1,4,5,6)P4 levels are elevated in src-transformed fibroblasts (Mattingly et al. 1991), but that result has not been studied further).
There is an environmental pathogen that can perturb Ins(1,4,5,6)P4 metabolism: Some years ago it was noted that the invasion of epithelial cells by Salmonella strongly activated the dephosphorylation of Ins(1,3,4,5,6)P5 to Ins(1,4,5,6)P4 (Eckmann et al. 1997). The evidence indicated that Ins(1,4,5,6)P4 might augment the secretion of salt and fluid that accompanies Salmonella infection (Eckmann et al. 1997). Whether or not this phenomenon might have physiological rather than just pathological relevance has not been established. Subsequently, it was demonstrated that one of the proteins that is required for the pathogen’s virulence, SopB, was responsible for dephosphorylating Ins(1,3,4,5,6)P5 (Norris et al. 1998). A later study (Zhou et al. 2001) established that the main product was Ins(1,4,5,6)P4. However, there was no evidence that virulence itself depends upon Ins(1,3,4,5,6)P5 dephosphorylation (Zhou et al. 2001). Instead, cell invasion by Salmonella appeared to require inositol lipid dephosphorylation by SopB (Hernandez et al. 2004). Indeed, it now seems possible that Ins(1,3,4,5,6)P5 is little more than an off-target substrate for SopB. In any case, the groups that work with SopB now focus on its role in metabolizing inositol lipids rather than the inositol phosphates (Hernandez et al. 2004). Taking all these data into account, we conclude that Ins(1,4,5,6)P4 is not qualified to be described as a cellular signal.
13.4 Ins(1,3,4,5,6)P5 and InsP6
Most nucleated cells synthesize 15–50 μM of both Ins(1,3,4,5,6)P5 and InsP6 (Pittet et al. 1989; Oliver et al. 1992; Barker et al. 2004; Bunce et al. 1993). Undoubtably, the initial proposals that Ins(1,3,4,5,6)P5 and InsP6 might be cellular signals (Heslop et al. 1985; Vallejo et al. 1987; Michell et al. 1988) were strongly influenced by the manner in which these polyphosphates were first discovered in animal cells, that is, as a consequence of studying metabolism of Ins(1,4,5)P3. However, intracellular signals typically are expected to exhibit significant stimulus-dependent changes in their concentrations. In contrast, cellular levels of Ins(1,3,4,5,6)P5 and InsP6 do not respond acutely to most extracellular stimuli, and even when they do, 25–35% changes in their concentrations seem to be an upper limit (Larsson et al. 1997; Pittet et al. 1989).
One dramatic exception emerged in a study (Gao and Wang 2007) of certain signaling events that lie downstream of the so-called Frizzled receptors. Activation of Frizzleds by the Wnt ligands regulates many aspects of embryonic development and adult tissue homeostasis. Wnt ligands can activate PLC and stimulate inositol phosphate accumulation (Slusarski et al. 1997), but Ins(1,3,4,5,6)P5 would normally be expected to be well-insulated from that response (Menniti et al. 1990). It was therefore unexpected when Gao and Wang (Gao and Wang 2007) demonstrated that Ins(1,3,4,5,6)P5 levels increased up to 2.5-fold in the few minutes following activation of the (over-expressed) rat Fz1 receptor by Wnt3a. In vitro data indicated that the biological consequence of this increase in Ins(1,3,4,5,6)P5 was activation of casein kinase II (CK2) and inhibition of GSK3β. The maximally effective concentration of Ins(1,3,4,5,6)P5 in each case was approximately 50 μM, which is approximately what is normally estimated to be present inside mammalian cells (Oliver et al. 1992; Pittet et al. 1989). Both of those effects of Ins(1,3,4,5,6)P5, if they occurred in vivo, would be expected to stabilize β-catenin, enhancing its transcriptional response to Wnt signaling (Gao and Wang 2007).
Let’s first discuss the proposed regulation of CK2. It has been known for some years that Ins(1,3,4,5,6)P5 can activate CK2 in vitro, although it was originally reported by Solyakov et al. (2004) that InsP6 is more efficacious. However, neither Ins(1,3,4,5,6)P5 nor InsP6 affects the activity of purified, native CK2 (Solyakov et al. 2004; Gao and Wang 2007). Instead, it was reported that the inositol phosphates act by reversing the effect of an uncharacterized, heat-stable inhibitor of CK2 that is present in cell lysates (Solyakov et al. 2004). The lack of insight into either the nature of the inhibitor, or the mechanism of action of the polyphosphates, has prevented this hypothesis from developing further. Moreover, it is a popular viewpoint in the CK2 field that this kinase is normally constitutively active and therefore has no requirement to be stimulated (Ruzzene and Pinna 2010). Even if that prevailing opinion were to be incorrect, it is hard to see how a stimulus-dependent increase in Ins(1,3,4,5,6)P5 levels would have any effect upon CK2 that should already be constitutively activated by endogenous InsP6.
The inhibitory effect of Ins(1,3,4,5,6)P5 upon GSK3β is more encouraging because of its specificity: neither Ins(1,4,5)P3, Ins(1,3,4,5)P4 nor InsP6 had any effect (Gao and Wang 2007). The treatment of intact cells with inhibitors of Ins(1,3,4,5,6)P5 synthesis also prevented Wnt3a from inhibiting GSK3β (Gao and Wang 2007). Since Ins(1,3,4,5,6)P5 levels do not typically change in response to short-term receptor activation, it is possible that the response that Gao and Wang (Gao and Wang 2007) observed is specific to signaling through Frizzled receptors. As for possible mechanisms, Ins(1,3,4,5,6)P5 did not inhibit purified GSK3β, so an intermediary seems to be required (Gao and Wang 2007). Further work on this topic would seem to be appropriate.
Aside from that isolated response of Ins(1,3,4,5,6)P5 to activation of the Fz1 receptor, its steady-state levels—and also those of InsP6—do not respond acutely to receptor activation (see above). Thus, when there are reports that Ins(1,3,4,5,6)P5 and InsP6 have biological activity, it has been difficult to place these data in a signaling context. One illustrative example is a report that both Ins(1,3,4,5,6)P5 and InsP6 inhibit protein phosphatases (Larsson et al. 1997). How can this be of regulatory significance if the levels of these polyphosphates do not change acutely? (In this particular case, one might also ask how any signaling specificity could result from two inositol polyphosphates both being broad spectrum inhibitors of PP1, PP2A and PP5). Similarly, it is also difficult to place in a signaling context a report that Ins(1,3,4,5,6)P5 and InsP6 inhibit L-type Ca2+ channels (Quignard et al. 2003); a similar criticism can be made of proposals that InsP6 is a “regulatory factor” in mRNA export and gene translation (Monserrate and York 2010; York et al. 1999). To be fair, we do note that others interpret these data rather differently. For example, it has been proposed that one role for InsP6 is to “set” (Berggren and Barker 2008) the basal state of a number of signaling entities. In particular, there are a number of studies that argue InsP6 establishes the default activities of various beta-cell signaling complexes (Berggren and Barker 2008). Barker and colleagues (2004) have also proposed that this putative global effector role for InsP6 may be of regulatory significance as cells transit through the cell cycle, during which time they estimate that the level of InsP6 may fluctuate by as much as threefold. Nevertheless, those cell-cycle dependent metabolic changes have not been tied to a specific signaling event.
Of course, the situation would be different if, as has been suggested (Larsson et al. 1997; Barker et al. 2002; Otto et al. 2007), the high total cellular levels of Ins(1,3,4,5,6)P5 and InsP6 mask stimulus-dependent alterations in smaller, discrete “signaling” pools of these compounds. That is, significant changes in “local” concentrations of Ins(1,3,4,5,6)P5 and InsP6 could be missed during the analysis of inositol phosphates in entire cell populations. Not so long ago, such a concept would have been labeled as heretical: how could a small and apparently freely-diffusible molecule not be uniformly distributed throughout the cell? However, it is now recognized that a concentration gradient of cAMP across a cell can be maintained by the spatial separation of the adenylyl cyclases from cAMP phosphodiesterases (Zaccolo et al. 2006). Is there any evidence for spatial heterogeneity of enzymes of inositol phosphate metabolism? Indeed there are. Arguably the most dramatic example is the receptor-dependent relocalization of the Ins(1,4,5)P3 3-kinase in hippocampal neurones (Schell and Irvine 2006). In stimulated cells, the kinase moves away from the post-synaptic region of the neuronal spines and into the dendritic shaft. This translocation undoubtably influences Ins(1,4,5)P3-dependent Ca2+ mobilization (Schell and Irvine 2006). But what about the enzymes that metabolize higher inositol phosphates? IPK2/IPMK (Nalaskowski et al. 2002; Odom et al. 2000) and IP5K (York et al. 1999; Brehm et al. 2007) are both concentrated in the nucleus. Moreover, the only known mammalian InsP6 phosphatase—MIPP—is restricted to the lumen of the endoplasmic reticulum (Craxton et al. 1997; Ali et al. 1993). So it is also of interest that plants at least can utilize an ABC-transporter like protein to move InsP6 across membranes (Nagy et al. 2009; Shi et al. 2007). It would be a significant breakthrough in this field if a mammalian homologue could be identified that transported InsP6 across the endoplasmic reticulum so that it could be metabolized by MIPP. However, embryonic fibroblasts made from Mipp−/− mice showed only 30% higher levels of InsP6 than wild-type animals; more discouragingly, the animals exhibited no obvious phenotype (Chi et al. 2000). Is it possible that mammals express another InsP6 phosphatase that we’ve all missed? Certainly we are missing something: we (Yang et al. 2008) have reported that a 20–25% decrease in cell volume following hyperosmotic stress is accompanied by a proportionate decrease in the amount of cellular InsP6, so that its concentration is not altered. This observation indicates that, when the cell deems it necessary, the metabolism of InsP6 can be quite rapid. We really ought to find out how, and why.
Some time ago, Michell’s group (Stuart et al. 1994) also considered this question of whether or not some inositol phosphates might be present inside cellular organelles. They ascertained that 80–90% of the cells’s inositol phosphates, including Ins(1,3,4,5,6)P5 and InsP6, were immediately released into the surrounding medium when the plasma membrane was permeabilized. That observation argues strongly against inositol phosphates being inside membrane-delimited cellular organelles. It can also be argued that no more than about half of the InsP6 pool in intact cells can be “hidden” from the cytoplasm, since the other half is readily accessible to soluble kinases that synthesize the inositol pyrophosphates (Menniti et al. 1993).
On the other hand, InsP6 can bind to membranes, at least in vitro (Cooke et al. 1991). InsP6 is also a structural component of certain cellular proteins (Macbeth et al. 2005); that particular pool of InsP6 would not be expected to be freely exchangeable with the bulk phase. There are other cellular proteins that can bind InsP6, which could also reduce its free concentration in the cytosol (Barker et al. 2002). The punctate intracellular distribution of endogenous IP5K, particularly in nucleoli and so-called stress granules (Brehm et al. 2007), also suggests that, to a degree at least, the synthesis of InsP6 might be compartmentalized. Uncertainty over compartmentalization is unlikely to be resolved until appropriate sensors of the intracellular location of these polyphosphates can be developed. It is our opinion that the status of Ins(1,3,4,5,6)P5 and InsP6 as cellular signals depends upon this question being answered.
There is no doubt that Ins(1,3,4,5,6)P5 and InsP6 still fulfill important biological functions that do not depend upon dynamic changes in their concentrations. For example, both Ins(1,3,4,5,6)P5 and InsP6 are precursors for the inositol pyrophosphates, which are currently the recipients of considerable interest from the signaling community (Saiardi 2011). Another possible function for Ins(1,3,4,5,6)P5 and InsP6 (in vitro at least) is to compete with inositol lipids for binding to certain proteins (Komander et al. 2004; Kavran et al. 1998). It has been speculated that this phenomenon increases the signal-to-noise ratio for PtdIns(3,4,5)P3-dependent functions (Irvine and Schell 2001; Komander et al. 2004). The idea is that binding of soluble inositol phosphates to a protein target helps keep it away from membranes until PtdIns 3-kinase activity is elevated by an appropriate stimulus (this concept is arguably analogous to the proposal (Berggren and Barker 2008) that Ins(1,3,4,5,6)P5 and InsP6 “set” the basal activities of certain signaling entities). Likewise, the binding of Ins(1,3,4,5,6)P5 to PTEN may inhibit that enzyme’s low protein phosphatase activity and possibly contribute to PTEN’s cytoplasmic and nuclear localization in the absence of PtdIns 3-kinase signaling (Caffrey et al. 2001).
Also, as noted in the introduction, InsP6 is an essential cofactor for adenosine deaminase (Macbeth et al. 2005), and, in yeast at least, InsP6 stimulates mRNA export from the nucleus (Alcázar-Román et al. 2006; York et al. 1999). Additionally, by enhancing the interaction of Ku with other proteins, InsP6 stimulates DNA repair through non-homologous end-joining (Cheung et al. 2008). However, since all of these functions for InsP6 can be satisfied by just a small percentage of total cellular InsP6 levels, it is our contention that in these cases this inositol polyphosphate more likely functions as a cofactor rather than as a dynamic “regulator”.
In view of all of these activities of Ins(1,3,4,5,6)P5 and InsP6, it is not surprising that, in mammals, embryonic lethality results from the knock-out of IPK2/IPMK (Frederick et al. 2005) or IP5K (Verbsky et al. 2005a). The knock-down of Ip5K in zebrafish embryos is also phenotypically dramatic: there is disturbance of asymmetric Ca2+ signaling that is important for embryonic patterning (Sarmah et al. 2005). However, those genetic experiments in themselves do not speak to any specific signaling role of Ins(1,3,4,5,6)P5 or InsP6. It is possible that these phenotypes are, in part, consequences of the loss of non-catalytic activities of inositolphosphate kinases (Odom et al. 2000) and/or the absence of more highly phosphorylated metabolites, such as the inositol pyrophosphates, which also function in development (Sarmah and Wente 2010).
13.5 Ins(3,4,5,6)P4
Cellular levels of Ins(3,4,5,6)P4 are around 1 μM in resting cells, and they increase to the 5–10 μM range whenever PLC is activated (Ho and Shears 2002). Ins(3,4,5,6)P4 is a concentration-dependent inhibitor of a CaMKII-activated Cl− conductance that is located in the plasma membrane (Xie et al. 1996, 1998; Ho et al. 2001; Mitchell et al. 2008). At least in mammalian cells, the inhibition of Cl− channel conductance by Ins(3,4,5,6)P4 is an exquisitely specific regulatory process; it is not imitated by any of the many other inositol phosphates that exist inside cells (Ho and Shears 2002; Ho et al. 2000; Xie et al. 1996). In other words, it has been demonstrated that Ins(3,4,5,6)P4 is a receptor-regulated signal, its biological target is known, and Ins(3,4,5,6)P4 acts specifically. This inositol phosphate is undoubtedly an intracellular signal.
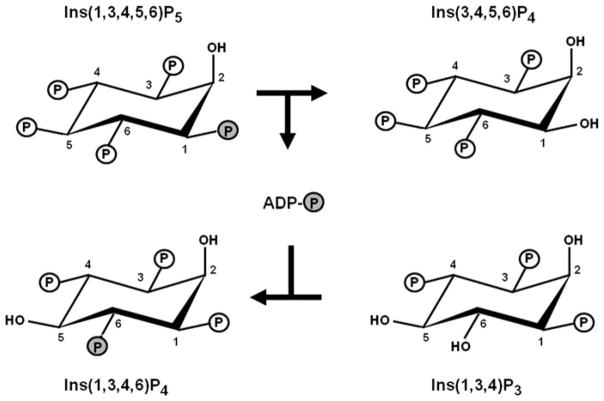
Ins(3,4,5,6)P4 can only be formed in animal cells by receptor-dependent dephosphorylation of Ins(1,3,4,5,6)P5 by an enzyme that is—unfortunately—known as ITPK1 (for Inositol Trisphosphate Kinase). This baptism by the Human Genome Nomenclature Committee seems to have been prompted by the fact that the protein was initially characterized as a 6-kinase activity that phosphorylates Ins(1,3,4)P3 to Ins(1,3,4,6)P4 (Shears et al. 1987; Balla et al. 1987). It is only recently that it has been determined that the trisphosphate kinase activity reflects a more complex, ADP-dependent phosphotransferase activity (Chamberlain et al. 2007; Ho et al. 2002). This is a unique phenomenon in the inositol phosphate field, that explains the molecular mechanism by which Ins(3,4,5,6)P4 levels are coupled to receptor-regulated PLC activity (Fig. 13.2). In its ADP-bound form, ITPK1 dephosphorylates Ins(1,3,4,5,6)P5 to Ins(3,4,5,6)P4. The Ins(3,4,5,6)P4 is released to the bulk phase in exchange for Ins(1,3,4)P3, but the nucleotide—now ATP—remains bound. The tenacity of this binding of adenine nucleotide has been verified by crystallographic data showing that less than 10% of the nucleotide is solvent exposed (Miller et al. 2005; Chamberlain et al. 2007). Thus, the inorganic phosphate that is removed from Ins(1,3,4,5,6)P5 is not released. Instead, it is passed on to the newly-bound Ins(1,3,4)P3, thereby phosphorylating it to Ins(1,3,4,6)P4, which the active-site then exchanges for a new molecule of Ins(1,3,4,5,6)P5, and the entire phosphotransferase cycle is repeated (Fig. 13.2). Importantly, the rate at which Ins(1,3,4,5,6)P5 is dephosphorylated to Ins(3,4,5,6)P4 is stimulated as the rate-limiting concentration of phosphate acceptor—Ins(1,3,4)P3—is increased (Ho et al. 2002). In turn, the cellular levels of Ins(1,3,4)P3—a metabolite of Ins(1,4,5)P3 (Fig. 13.1)—mirrors both the intensity and the duration of receptor-activated PLC activity (Batty et al. 1998; Batty and Downes 1994). In other words, the degree of PLC activity sets Ins(1,3,4)P3 levels, which controls Ins(3,4,5,6)P4 synthesis. This is the molecular basis for the integration of inositol phosphate signaling pathways via human ITPK1.
Fig. 13.2.
The phosphotransferase activity of ITPK1. The figure shows the phosphotransferase activity that is catalyzed by ITPK1. The phosphate group that is transferred between from Ins(1,3,4,5,6)P5 to Ins(1,3,4)P3 is highlighted in by the grey circle
The Ins(1,3,4)P3 6-kinase activity of ITPK1 also plays a metabolic role (Fig. 13.1) in maintaining the size of the cell’s Ins(1,3,4,5,6)P5 pool (Shears et al. 1987; Balla et al. 1987; Verbsky et al. 2005b). It is unclear if it is this metabolic function, or the signaling activities of ITPK1, which explain why mice which are hypomorphic for the Itpk1 allele are susceptible to neural tube defects (Wilson et al. 2009). A complete knock-down of ITPK1 expression is apparently lethal (Verbsky et al. 2005b).
The best characterized biological end-point for Ins(3,4,5,6)P4 action upon Cl− transport is to regulate epithelial salt and fluid secretion (Vajanaphanich et al. 1994; Carew et al. 2000). However, the recent identification of ClC3 as the target of Ins(3,4,5,6)P4, at least in mammalian cells (Mitchell et al. 2008), has greatly expanded the biological repertoire of this inositol phosphate. For example, ClC3 is responsible for the Ins(3,4,5,6)P4-regulated Cl− conductance in hippocampal neurones (Mitchell et al. 2008), which is thought to contribute to the overall regulation of the synaptic efficacy in generating action potentials (Wang et al. 2006). Long-term changes in synaptic efficacy comprise a cellular basis for information storage and memory formation (Bliss and Collingridge 1993). Thus, Ins(3,4,5,6)P4 is a molecule that has the potential to affect neuronal development. It therefore seems pertinent that Ins(3,4,5,6)P4 has also previously been suggested to have the characteristics of a “memory molecule”, because its relatively slow rate of metabolism permit its physiological effects to long outlast the duration of the stimulus that initially prompts intracellular Ins(3,4,5,6)P4 to accumulate (Ho and Shears 2002).
ClC3 that is in the plasma membrane may have other roles, such as tumor cell migration (Mao et al. 2008; Cuddapah and Sontheimer 2010) and the regulation of apoptosis (Claud et al. 2008). We can therefore anticipate that Ins(3,4,5,6)P4 might also regulate these processes. It should be noted, however, that some of the workers in this field (Jentsch 2008; Jentsch et al. 2002) propose that ClC3 is not a plasmalemmal Cl− channel per se, but instead a regulator of other Cl− channels. That argument, if correct, does not devalue the role of Ins(3,4,5,6)P4 in regulating ClC3 function. For example, in one cell type in which our own data are consistent with the ClC3 regulating other Cl− channels, we have shown that ClC3 still mediates the effect of Ins(3,4,5,6)P4 upon plasma membrane Cl− fluxes (unpublished data).
While there is disagreement as to whether or not ClC3 is an independent plasma membrane Cl− channel, it is well recognized that ClC3 also resides in intracellular vesicles such as insulin granules (Barg et al. 2001) and the early endosomal compartment (Zhao et al. 2007; Gentzsch et al. 2003; Stobrawa et al. 2001; Hara-Chikuma et al. 2005; Mitchell et al. 2008). Here, ClC3 contributes to endosomal acidification (Jentsch 2008), although there is uncertainty concerning the exact mechanism. Nevertheless, when a cell-permeant analogue of Ins(3,4,5,6)P4 was added to cells so as to inhibit ClC3, the pH of certain vesicular sub-compartments became more alkaline (Renström et al. 2002; Mitchell et al. 2008). What is the biological significance of this regulation of intravesicular pH? With regard to insulin granules, it has been proposed that their intraluminal acidification is a priming process, without which they become less competent to fuse with the plasma membrane and release their cargo (Barg et al. 2001). In support of this idea, we have shown that alkalinization of insulin granules by Ins(3,4,5,6)P4 has the effect of reducing insulin secretion from pancreatic β-cells (Renström et al. 2002). In many other cell types, the acidification of endosomes and secretory vesicles serves other important functions, including modulation of certain ligand-protein interactions during endocytosis, enzyme targeting, and H+-coupled uptake of small molecules (such as neurotransmitters) (Nishi and Forgac 2002; von and Sorkin 2007). It appears that we have only scratched the surface of our understanding of the biological importance of Ins(3,4,5,6)P4.
It is unfortunate that we do not yet understand the mechanism by which Ins(3,4,5,6)P4 prevents CaMKII from activating ClC3. Data obtained to date indicate that Ins(3,4,5,6)P4 does not inhibit CaMKII activity per se (Xie et al. 1996; Ho et al. 2001; Ho and Shears 2002). Furthermore, in single channel analysis of CaMKII-activated Cl− channels, Ins(3,4,5,6)P4 was not inhibitory, so it is unlikely to act as a direct channel blocker (Ho et al. 2001). Presumably an intermediary protein mediates the action of Ins(3,4,5,6)P4. However, our efforts to identify an Ins(3,4,5,6)P4 “receptor” have so far been disappointingly fruitless (unpublished data). Our work on this important problem is ongoing.
13.6 Conclusions
The metabolic intermediates that accumulate during the dephosphorylation of Ins(1,4,5)P3 and Ins(1,3,4)P3 to inositol are not generally considered to be signaling molecules. The same could be true of at least some of the intermediates in the pathways of phosphorylation of Ins(1,4,5)P3 to InsP6. The concept that inositol is a combinatorial signaling scaffold (York 2006) is intellectually appealing, but it is still not obligatory that all of these inositol phosphates be cellular signals; only at least one of the end products might act in a signaling pathway.
We have argued here that there is no strong evidence that Ins(1,4,5,6)P4 is a cellular signal. As for Ins(1,3,4,5)P4, we have highlighted the confusing and often conflicting observations in the literature concerning proposed actions of this inositol phosphate. Under such circumstances, it is difficult to formulate a general signaling role. Ins(1,3,4,5,6)P5 and InsP6—particularly the latter—clearly have important roles as cofactors, but our conclusion is that we need more concrete evidence before we can claim that these molecules are truly cellular signals. As we have discussed, it may be that further information of cellular compartmentalization may be the savior of these molecule’s signaling credentials. So, other than Ins(1,4,5)P3, that leaves, in our opinion, Ins(3,4,5,6)P4 as the only validated “classical” cellular signal from within this group of molecules.
References
- Alcázar-Román AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:645–647. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Ali N, Craxton A, Shears SB. Hepatic Ins(1,3,4,5)P4 3-phosphatase is compartmentalized inside endoplasmic reticulum. J Biol Chem. 1993;268:6161–6167. [PubMed] [Google Scholar]
- Balla T, Guillemette G, Baukal AJ, Catt KJ. Formation of inositol 1,3,4,6-tetrakisphosphate during angiotensin II action in bovine adrenal glomerulosa cells. Biochem Biophys Res Commun. 1987;146:199–205. doi: 10.1016/0006-291x(87)91095-3. [DOI] [PubMed] [Google Scholar]
- Barg S, Huang P, Eliasson L, Nelson DJ, Obermüller S, Rorsman P, Thévenod F, Renström E. Priming of insulin granules for exocytosis by granular chloride uptake and acidification. J Cell Sci. 2001;114:2145–2154. doi: 10.1242/jcs.114.11.2145. [DOI] [PubMed] [Google Scholar]
- Barker CJ, Wong NS, Maccallum SM, Hunt PA, Michell RH, Kirk CJ. The interrelationships of the inositol phosphates formed in WRK-1 stimulated rat mammary tumour cells. Biochem J. 1992;286:469–474. doi: 10.1042/bj2860469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CJ, Leibiger IB, Leibiger B, Berggren PO. Phosphorylated inositol compounds in beta -cell stimulus-response coupling. Am J Physiol Endocrinol Metab. 2002;283:E1113–E1122. doi: 10.1152/ajpendo.00088.2002. [DOI] [PubMed] [Google Scholar]
- Barker CJ, Wright J, Hughes PJ, Kirk CJ, Michell RH. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty IH, Downes CP. The inhibition of phosphoinositide synthesis and muscarinic-receptor-mediated phospholipase C activity by Li+ as secondary selective consequences of inositol depletion in 1321N1 cells. Biochem J. 1994;297:529–537. doi: 10.1042/bj2970529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty IH, Nahorski SR, Irvine RF. Rapid formation of inositol 1,3,4-tetrakisphosphate following muscarinic receptor stimulation of rat cerebral cortical slices. Biochem J. 1985;232:211–215. doi: 10.1042/bj2320211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty IH, Currie RA, Downes CP. Evidence for a model of integrated inositol phospholipid pools implies an essential role for lipid transport in the maintenance of receptor-mediated phospholipase C activity in 1321N1 cells. Biochem J. 1998;330:1069–1077. doi: 10.1042/bj3301069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Hörz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Berggren PO, Barker CJ. A key role for phosphorylated inositol compounds in pancreatic beta-cell stimulus-secretion coupling. Adv Enzyme Regul. 2008;48:276–294. doi: 10.1016/j.advenzreg.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Bird GStJ, Putney JW., Jr Effect of inositol 1,3,4,5-tetrakisphosphate on inositol trisphosphate-activated Ca2+ signaling in mouse lacrimal acinar cells. J Biol Chem. 1996;271:6766–6770. doi: 10.1074/jbc.271.12.6766. [DOI] [PubMed] [Google Scholar]
- St Bird GJ, Rossier MF, Hughes AR, Shears SB, Armstrong DL, Putney JW., Jr Activation of calcium entry into acinar cells by a non-phosphorylatable inositol trisphosphate. Nature. 1991;352:162–165. doi: 10.1038/352162a0. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Boynton AL, Dean NM, Hill TD. Inositol 1,3,4,5-tetrakisphosphate and regulation of intracellular calcium. Biochem Pharmacol. 1990;40:1933–1939. doi: 10.1016/0006-2952(90)90221-6. [DOI] [PubMed] [Google Scholar]
- Brehm MA, Schenk TM, Zhou X, Fanick W, Lin H, Windhorst S, Nalaskowski MM, Kobras M, Shears SB, Mayr GW. Intracellular localization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. Biochem J. 2007;408:335–345. doi: 10.1042/BJ20070382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough D, Bhatti F, Irvine RF. Mobility of proteins associated with the plasma membrane by interaction with inositol lipids. J Cell Sci. 2005;118:3019–3025. doi: 10.1242/jcs.02426. [DOI] [PubMed] [Google Scholar]
- Bunce CM, French PJ, Allen P, Mountford JC, Moor B, Greaves MF, Michell RH, Brown G. Comparison of the levels of inositol metabolites in transformed haemopoietic cells and their normal counterparts. Biochem J. 1993;289:667–673. doi: 10.1042/bj2890667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey JJ, Darden T, Wenk MR, Shears SB. Expanding Coincident Signaling by PTEN through its Inositol 1,3,4,5,6-Pentakisphosphate 3-phosphatase Activity. FEBS Lett. 2001;499:6–10. doi: 10.1016/s0014-5793(01)02500-5. [DOI] [PubMed] [Google Scholar]
- Carew MA, Yang X, Schultz C, Shears SB. Ins(3,4,5,6)P4 inhibits an apical calcium-activated chloride conductance in polarized monolayers of a cystic fibrosis cell-line. J Biol Chem. 2000;275:26906–26913. doi: 10.1074/jbc.M002316200. [DOI] [PubMed] [Google Scholar]
- Chamberlain PP, Qian X, Stiles AR, Cho J, Jones DH, Lesley SA, Grabau EA, Shears SB, Spraggon G. Integration of inositol phosphate signaling pathways via human ITPK1. J Biol Chem. 2007;282:28117–28125. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changya L, Gallacher DV, Irvine RF, Potter BVL, Petersen OH. Inositol 1,3,4,5-trisphosphate is essential for sustained activation of the Ca2+-dependent K+ current in single internally perfused lacrimal cells. J Membr Biol. 1989;109:85–93. doi: 10.1007/BF01870793. [DOI] [PubMed] [Google Scholar]
- Cheung JC, Salerno B, Hanakahi LA. Evidence for an inositol hexakisphosphate-dependent role for Ku in mammalian nonhomologous end joining that is independent of its role in the DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:5713–5726. doi: 10.1093/nar/gkn572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H, Yang X, Kingsley PD, O’Keefe RJ, Puzas JE, Rosier RN, Shears SB, Reynolds PR. Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol Cell Biol. 2000;20:6496–6507. doi: 10.1128/mcb.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claud EC, Lu J, Wang XQ, Abe M, Petrof EO, Sun J, Nelson DJ, Marks JD, Jilling T. Platelet-activating Factor induced chloride channel activation is associated with intracellular acidosis and apoptosis of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1191–G1200. doi: 10.1152/ajpgi.00318.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke F, Poyner DR, Hawkins P, Erlebach C, Hanley MR. Inositol hexakisphosphate-membranes interactions: the role of metal ions. Biochem Soc Trans. 1991;19:152s. doi: 10.1042/bst019152s. [DOI] [PubMed] [Google Scholar]
- Cozier GE, Lockyer PJ, Reynolds JS, Kupzig S, Bottomley JR, Millard TH, Banting G, Cullen PJ. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J Biol Chem. 2000;275:28261–28628. doi: 10.1074/jbc.M000469200. [DOI] [PubMed] [Google Scholar]
- Craxton A, Caffrey JJ, Burkhart W, Safrany ST, Shears SB. Cloning and expression of rat hepatic multiple inositol polyphosphate phosphatase. Biochem J. 1997;328:75–81. doi: 10.1042/bj3280075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley I, Swann K, Chambers E, Whitaker M. Activation of sea urchin eggs by inositol phosphates is independent of external calcium. Biochem J. 1988;252:257–262. doi: 10.1042/bj2520257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah VA, Sontheimer H. Molecular interaction and functional regulation of ClC-3 by Ca2+/calmodulin-dependent protein kinase II (CaMKII) in human malignant glioma. J Biol Chem. 2010;285:11188–11196. doi: 10.1074/jbc.M109.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Hsuan JJ, Truong O, Letcher AJ, Jackson TR, Dawson AP, Irvine RF. Identification of a specific Ins(1,3,4,5)P4-binding protein as a member of the GAP1 family. Nature. 1995;376:527–530. doi: 10.1038/376527a0. [DOI] [PubMed] [Google Scholar]
- Dubois E, Dewaste V, Erneux C, Messenguy F. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 2000;486:300–304. doi: 10.1016/s0014-5793(00)02318-8. [DOI] [PubMed] [Google Scholar]
- Eckmann L, Rudolf MT, Ptasznik A, Schultz C, Jiang T, Wolfson N, Tsien R, Fierer J, Shears SB, Kagnoff MF, Traynor-Kaplan A. D-myoinositol 1,4,5,6-tetrakisphosphate produced in human intestinal epithelial cells in response to Salmonella invasion inhibits phosphoinositide 3-kinase signaling pathways. Proc Natl Acad Sci U S A. 1997;94:14456–14460. doi: 10.1073/pnas.94.26.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49:457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci U S A. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang HY. Inositol Pentakisphosphate Mediates Wnt/beta-Catenin Signaling. J Biol Chem. 2007;282:26490–26502. doi: 10.1074/jbc.M702106200. [DOI] [PubMed] [Google Scholar]
- Gentzsch M, Cui L, Mengos A, Chang XB, Chen JH, Riordan JR. The PDZ-binding chloride channel ClC-3B localizes to the Golgi and associates with cystic fibrosis transmembrane conductance regulator-interacting PDZ proteins. J Biol Chem. 2003;278:6440–6449. doi: 10.1074/jbc.M211050200. [DOI] [PubMed] [Google Scholar]
- Guse AH, Greiner E, Emmrich F, Brand K. Mass changes of inositol 1,3,4,5,6-pentakisphosphate and inositol hexakisphosphate during cell cycle progression in rat thymocytes. J Biol Chem. 1993;268:7129–7133. [PubMed] [Google Scholar]
- Hara-Chikuma M, Yang B, Sonawane ND, Sasaki S, Uchida S, Verkman AS. ClC-3 chloride channels facilitate endosomal acidification and chloride accumulation. J Biol Chem. 2005;280:1241–1247. doi: 10.1074/jbc.M407030200. [DOI] [PubMed] [Google Scholar]
- Hermosura MC, Takeuchi H, Fleig A, Riley AQM, Potter BVL, Hirata M, Penner R. InsP4 facilitates store-operated calcium influx by inhibition of InsP3 5-phosphatase. Nature. 2000;408:735–740. doi: 10.1038/35047115. [DOI] [PubMed] [Google Scholar]
- Hernandez LD, Hueffer K, Wenk MR, Galan JE. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- Heslop JP, Irvine RF, Tashjian AH, Berridge MJ. Inositol tetrakis- and pentakisphosphates in GH4 cells. J Exp Biol. 1985;119:395–401. doi: 10.1242/jeb.119.1.395. [DOI] [PubMed] [Google Scholar]
- Hill TD, Dean NM, Boynton AL. Inositol 1,3,4,5-tetrakisphosphate induces Ca2+ sequestration in rat liver cells. Science. 1988;242:1176–1178. doi: 10.1126/science.2847317. [DOI] [PubMed] [Google Scholar]
- Ho MWY, Shears SB. Regulation of calcium-activated chloride channels by inositol 3,4,5,6-tetrakisphosphate. In: Fuller CM, editor. Current topics in membranes. Vol. 53. Academic Press; London: 2002. pp. 345–363. [Google Scholar]
- Ho MWY, Carew MA, Yang X, Shears SB. Regulation of chloride channel conductance by Ins(3,4,5,6)P4; a phosphoinositide-initiated signalling pathway that acts downstream of Ins(1,4,5)P3. In: Cockroft S, editor. Frontiers in molecular biology: biology of phosphoinositides. Oxford University Press; Oxford: 2000. pp. 298–319. [Google Scholar]
- Ho MWY, Kaetzel MA, Armstrong DL, Shears SB. Regulation of a human chloride channel: a paradigm for integrating input from calcium, CaMKII and Ins(3,4,5,6)P4. J Biol Chem. 2001;276:18673–18680. doi: 10.1074/jbc.M101128200. [DOI] [PubMed] [Google Scholar]
- Ho MWY, Yang X, Carew MA, Zhang T, Hua L, Kwon Y-U, Chung S-K, Adelt S, Vogel G, Riley AM, Potter BVL, Shears SB. Regulation of Ins(3456)P4 signaling by a reversible kinase/phosphatase. Curr Biol. 2002;12:477–482. doi: 10.1016/s0960-9822(02)00713-3. [DOI] [PubMed] [Google Scholar]
- Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, Sauer K. Positive regulation of Itk PH domain function by soluble IP4. Science. 2007;316:886–889. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Calcium transients: mobilization of intracellular Ca2+ Br Med Bull. 1986;42:369–374. doi: 10.1093/oxfordjournals.bmb.a072154. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Is inositol tetrakisphosphate the second messenger that controls Ca2+ entry into cells? In: Putney JW Jr, editor. Advances in second messenger and phosphoprotein research. Raven Press; New York: 1992. pp. 161–185. [PubMed] [Google Scholar]
- Irvine RF, Moor RM. Micro-injection of inositol 1,3,4,5-tetrakisphosphate activates sea urchin eggs by a mechanism dependent on external Ca2+ Biochem J. 1986;240:917–920. doi: 10.1042/bj2400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF, Moor RM. Inositol(1,3,4,5)tetrakisphosphate-induced activation of sea urchin eggs requires the presence of inositol trisphosphate. Biochem Biophys Res Commun. 1987;146:284–290. doi: 10.1016/0006-291x(87)90723-6. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell M. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Letcher AJ, Heslop JP, Berridge MJ. The inositol tris/tetrakisphosphate pathway—demonstration of Ins(1,4,5)P3 3-kinase activity in animal tissues. Nature. 1986;320:631–634. doi: 10.1038/320631a0. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Lloyd-Burton SM, Yu JC, Letcher AJ, Schell MJ. The regulation and function of inositol 1,4,5-trisphosphate 3-kinases. Adv Enzyme Regul. 2006;46:314–323. doi: 10.1016/j.advenzreg.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismailov II, Fuller CM, Berdiev BK, Shlyonsky VG, Benos DJ, Barrett KE. A biologic function for an “orphan” messenger: D-myo-Inositol 3,4,5,6-tetrakisphosphate selectively blocks epithelial calcium-activated chloride current. Proc Nat Acad Sci U S A. 1996;93:10505–10509. doi: 10.1073/pnas.93.19.10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch TJ. CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit Rev Biochem Mol Biol. 2008;43:3–36. doi: 10.1080/10409230701829110. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ, Stein V, Weinreich F, Zdebik AA. Molecular structure and physiological function of chloride channels. Physiol Rev. 2002;82:503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- Jia Y, Subramanian KK, Erneux C, Pouillon V, Hattori H, Jo H, You J, Zhu D, Schurmans S, Luo HR. Inositol 1,3,4,5-tetrakisphosphate negatively regulates phosphatidylinositol-3,4,5-trisphosphate signaling in neutrophils. Immunity. 2007;27:453–467. doi: 10.1016/j.immuni.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Loison F, Hattori H, Li Y, Erneux C, Park SY, Gao C, Chai L, Silberstein LE, Schurmans S, Luo HR. Inositol trisphosphate 3-kinase B (InsP3KB) as a physiological modulator of myelopoiesis. Proc Natl Acad Sci U S A. 2008;105:4739–4744. doi: 10.1073/pnas.0800218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and Promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter DC, Safrany ST, Alessi DR, van Aalten DM. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Barker CJ, Sjoholm A, Carlqvist H, Michell RH, Bertorello A, Nilsson T, Honkanen RE, Mayr GW, Zwiller J, Berggren PO. Inhibition of phosphatases and increased Ca2+ channel activity by inositol hexakisphosphate. Science. 1997;278:471–474. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- Leyman A, Pouillon V, Bostan A, Schurmans S, Erneux C, Pesesse X. The absence of expression of the three isoenzymes of the inositol 1,4,5-trisphosphate 3-kinase does not prevent the formation of inositol pentakisphosphate and hexakisphosphate in mouse embryonic fibroblasts. Cell Signal. 2007;19:1497–1504. doi: 10.1016/j.cellsig.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Li W, Schultz C, Llopis J, Tsien RY. Membrane-permeant esters of inositol polyphosphates, chemical synthesis and biological applications. Tetrahedron. 1997;53:12017–12040. [Google Scholar]
- Loomis-Husselbee JW, Cullen PJ, Dreikausen UE, Irvine RF, Dawson AP. Synergistic effects of inositol 1,3,4,5-tetrakisphosphate on inositol 2,4,5-trisphosphate-stimulated Ca2+ release do not involve direct interaction of inositol 1,3,4,5-tetrakisphosphate with inositol trisphosphate binding sites. Biochem J. 1996;314:811–816. doi: 10.1042/bj3140811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckhoff A, Clapham DE. Inositol 1,3,4,5-tetrakisphosphate activates an endothelial Ca2+-permeable channel. Nature. 1992;355:356–358. doi: 10.1038/355356a0. [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Chen L, Xu B, Wang L, Li H, Guo J, Li W, Nie S, Jacob TJ, Wang L. Suppression of ClC-3 channel expression reduces migration of nasopharyngeal carcinoma cells. Biochem Pharmacol. 2008;75:1706–1716. doi: 10.1016/j.bcp.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Marechal Y, Pesesse X, Jia Y, Pouillon V, Perez-Morga D, Daniel J, Izui S, Cullen PJ, Leo O, Luo HR, Erneux C, Schurmans S. Inositol 1,3,4,5-tetrakisphosphate controls proapoptotic Bim gene expression and survival in B cells. Proc Natl Acad Sci U S A. 2007;104:13978–13983. doi: 10.1073/pnas.0704312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly RR, Stephens LR, Irvine RF, Garrison JC. Effects of transformation with the v-src oncogene on inositol phosphate metabolism in rat-1 fibroblasts: D-myoinositol 1,4,5,6-tetrakisphosphate is increased in v-src transformed rat-1 fibroblasts and can be synthesised from d-myoinositol 1,3,4-trisphosphate in cytosolic extracts. J Biol Chem. 1991;266:15144–15153. [PubMed] [Google Scholar]
- McKnight S. Gene switching by metabolic enzymes—how did you get on the invitation list? Cell. 2003;174:150–152. doi: 10.1016/s0092-8674(03)00563-4. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Oliver KG, Nogimori K, Obie JF, Shears SB, Putney JW., Jr Origins of myoinositol tetrakisphosphates in agonist-stimulated rat pancreatoma cells. Stimulation by bombesin of myo-inositol 1,3,4,5,6-pentakisphosphate breakdown to myo-inositol 3,4,5,6-tetrakisphosphate. J Biol Chem. 1990;265:11167–11176. [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Michell RH, King CE, Piper CJ, Stephens LR, Bunce CM, Guy GR, Brown G. Inositol lipids and phosphates in erythrocytes and HL60 cells. J Gen Physiol. 1988;43:345–355. [PubMed] [Google Scholar]
- Miller GJ, Wilson MP, Majerus PW, Hurley JH. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell. 2005;18:201–212. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Miller AT, Sandberg M, Huang YH, Young M, Sutton S, Sauer K, Cooke MP. Production of Ins(1,3,4,5)P4 mediated by the kinase Itpkb inhibits store-operated calcium channels and regulates B cell selection and activation. Nat Immunol. 2007;8:514–521. doi: 10.1038/ni1458. [DOI] [PubMed] [Google Scholar]
- Miller AT, Beisner DR, Liu D, Cooke MP. Inositol 1,4,5-trisphosphate 3-kinase B is a negative regulator of BCR signaling that controls B cell selection and tolerance induction. J Immunol. 2009;182:4696–4704. doi: 10.4049/jimmunol.0802850. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Wang X, Zhang G, Gentzsch M, Nelson DJ, Shears SB. An Expanded Biological Repertoire for Ins(3,4,5,6)P(4) through its Modulation of ClC-3 Function. Curr Biol. 2008;18:1600–1605. doi: 10.1016/j.cub.2008.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Morris AP, Gallacher DV, Irvine RF, Petersen OH. Synergism of inositol trisphosphate and tetrakisphosphate in activating Ca2+-dependent K+ channels. Nature. 1987;330:653–655. doi: 10.1038/330653a0. [DOI] [PubMed] [Google Scholar]
- Nagy R, Grob H, Weder B, Green P, Klein M, Frelet A, Schjoerring JK, Brearley CA, Martinoia E. The Arabidopsis ATP-binding cassette protein ATMRP5/ATABCC5 is a high-affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J Biol Chem. 2009 doi: 10.1074/jbc.M109.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T, Forgac M. The vacuolar (H+)-ATPases—nature’s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci U S A. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Oliver KG, Putney JW, Jr, Obie JF, Shears SB. The interconversion of inositol 1,3,4,5,6-pentakisphosphate and inositol tetrakisphosphates in AR4–2J cells. J Biol Chem. 1992;267:21528–21534. [PubMed] [Google Scholar]
- Otto JC, Kelly P, Chiou ST, York JD. Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc Natl Acad Sci U S A. 2007;104:15653–15658. doi: 10.1073/pnas.0705729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh AB, Penner R. Store depletion and calcium influx. Physiol Rev. 1997;77:901–930. doi: 10.1152/physrev.1997.77.4.901. [DOI] [PubMed] [Google Scholar]
- Pittet D, Schlegel W, Lew DP, Monod A, Mayr GW. Mass changes in inositol tetrakis- and pentakisphosphate isomers induced by chemotactic peptide stimulation in HL-60 cells. J Biol Chem. 1989;264:18489–18493. [PubMed] [Google Scholar]
- Pouillon V, Hascakova-Bartova R, Pajak B, Adam E, Bex F, Dewaste V, Van Lint C, Leo O, Erneux C, Schurmans S. Inositol 1,3,4,5-tetrakisphophate is essential for T lymphocyte development. Nat Immunol. 2003;4:1136–1143. doi: 10.1038/ni980. [DOI] [PubMed] [Google Scholar]
- Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- Putney JW., Jr . Inositol phosphates and calcium entry. In: Putney JW, editor. Advances in second messenger and phosphoprotein research. Raven Press; New York: 1992. pp. 143–160. [PubMed] [Google Scholar]
- Qi Q, August A. The Tec family kinase Itk exists as a folded monomer in vivo. J Biol Chem. 2009;284:29882–29892. doi: 10.1074/jbc.M109.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Sahu N, August A. Tec kinase Itk forms membrane clusters specifically in the vicinity of recruiting receptors. J Biol Chem. 2006;281:38529–38534. doi: 10.1074/jbc.M609180200. [DOI] [PubMed] [Google Scholar]
- Quignard JF, Rakotoarisoa L, Mironneau J, Mironneau C. Stimulation of L-type Ca2+ channels by inositol pentakis- and hexakisphosphates in rat vascular smooth muscle cells. J Physiol. 2003;549:729–737. doi: 10.1113/jphysiol.2002.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem. 1958;232:1065–1076. [PubMed] [Google Scholar]
- Renström E, Ivarsson R, Shears SB. Ins(3,4,5,6)P4 inhibits insulin granule acidification and fusogenic potential. J Biol Chem. 2002;277:26717–26720. doi: 10.1074/jbc.C200314200. [DOI] [PubMed] [Google Scholar]
- Robison GA, Butcher RW, Sutherland EW. Cyclic AMP. Annu Rev Biochem. 1968;37:149–174. doi: 10.1146/annurev.bi.37.070168.001053. [DOI] [PubMed] [Google Scholar]
- Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Saiardi A. Phosphoinositides and inositol phosphates. 2011. [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci U S A. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi R, Tainaka K, Shimada N, Nakano S, Inoue M, Kiyonaka S, Mori Y, Morii T. An in vivo fluorescent sensor reveals intracellular ins(1,3,4,5)P4 dynamics in single cells. Angew Chem Int Ed Engl. 2010;49:2150–2153. doi: 10.1002/anie.200903951. [DOI] [PubMed] [Google Scholar]
- Sarmah B, Wente SR. Inositol hexakisphosphate kinase-2 acts as an effector of the vertebrate Hedgehog pathway. Proc Natl Acad Sci U S A. 2010;107:19921–19926. doi: 10.1073/pnas.1007256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Sauer K, Cooke MP. Regulation of immune cell development through soluble inositol-1,3,4,5-tetrakisphosphate. Nat Rev Immunol. 2010;10:257–271. doi: 10.1038/nri2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MJ. Inositol trisphosphate 3-kinases: focus on immune and neuronal signaling. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-009-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MJ, Irvine RF. Calcium-triggered exit of F-actin and IP(3) 3-kinase A from dendritic spines is rapid and reversible. Eur J Neurosci. 2006;24:2491–2503. doi: 10.1111/j.1460-9568.2006.05125.x. [DOI] [PubMed] [Google Scholar]
- Seeds AM, Sandquist JC, Spana EP, York JD. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J Biol Chem. 2004 doi: 10.1074/jbc.M408295200. [DOI] [PubMed] [Google Scholar]
- Shears SB. Metabolism of the inositol phosphates produced by receptor activation. Biochem J. 1989;260:313–324. doi: 10.1042/bj2600313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. How versatile are inositol phosphate kinases? Biochem J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Parry JB, Tang EKY, Irvine RF, Michell RH, Kirk CJ. Metabolism of D-myo-inositol 1,3,4,5-tetrakisphosphate by rat liver, including the synthesis of a novel isomer of myo-inositol tetrakisphosphate. Biochem J. 1987;246:139–147. doi: 10.1042/bj2460139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu W-H, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Schellin K, Li B, Faller M, Stoop JM, Meeley RB, Ertl DS, Ranch JP, Glassman K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat Biotechnol. 2007;25:930–937. doi: 10.1038/nbt1322. [DOI] [PubMed] [Google Scholar]
- Slusarski DC, Corces VG, Moon RT. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997;390:410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- Smith PM, Harmer AR, Letcher AJ, Irvine RF. The effect of inositol 1,3,4,5-tetrakisphosphate on inositol trisphosphate-induced Ca2+ mobilization in freshly isolated and cultured mouse lacrimal cells. Biochem J. 2000;347:77–82. [PMC free article] [PubMed] [Google Scholar]
- Solyakov L, Cain K, Tracey BM, Jukes R, Riley AM, Potter BV, Tobin AB. Regulation of casein kinase-2 (CK2) activity by inositol phosphates. J Biol Chem. 2004 doi: 10.1074/jbc.M403239200. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobrawa SM, Breiderhoff T, Takamori S, Engel D, Schweizer M, Zdebik AA, Bösl MR, Ruether K, Jahn H, Draguhn A, Jahn R, Jentsch TJ. Disruption of ClC-3, a chloride channel expressed on synaptic vesicles, leads to a loss of hippocampus. Neuron. 2001;29:185–196. doi: 10.1016/s0896-6273(01)00189-1. [DOI] [PubMed] [Google Scholar]
- Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial store in pancreatic cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–68. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Anderson KL, French PJ, Kirk CJ, Michell RH. The intracellular distribution of inositol polyphosphates in HL60 promyeloid cells. Biochem J. 1994;303:517–525. doi: 10.1042/bj3030517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- Taylor C. Phosphoinositides and inositol phosphates. 2011. [Google Scholar]
- Vajanaphanich M, Schultz C, Rudolf MT, Wasserman M, Enyedi P, Craxton A, Shears SB, Tsien RY, Barrett KE, Traynor-Kaplan AE. Long-term uncoupling of chloride secretion from intracellular calcium levels by Ins(3,4,5,6)P4. Nature. 1994;371:711–714. doi: 10.1038/371711a0. [DOI] [PubMed] [Google Scholar]
- Vallejo M, Jackson T, Lightman S, Hanley MR. Occurrence and extracellular actions of inositol pentakis- and hexakisphosphate in mammalian brain. Nature. 1987;330:656–658. doi: 10.1038/330656a0. [DOI] [PubMed] [Google Scholar]
- Verbsky J, Lavine K, Majerus PW. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc Natl Acad Sci U S A. 2005a;102:8448–8453. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW. The pathway for the production of inositol hexakisphosphate in human cells. J Biol Chem. 2005b;280:1911–1920. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- Volk AP, Heise CK, Hougen JL, Artman CM, Volk KA, Wessels D, Soll DR, Nauseef WM, Lamb FS, Moreland JG. ClC-3 and IClswell are required for normal neutrophil chemotaxis and shape change. J Biol Chem. 2008;283:34315–34326. doi: 10.1074/jbc.M803141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von ZM, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19:436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Kupzig S, Lockyer PJ, Bilu S, Zharhary D, Cullen PJ. Analysing the role of the putative inositol 1,3,4,5-tetrakisphosphate receptor GAP1IP4BP in intracellular Ca2+ homeostasis. J Biol Chem. 2002;277:48779–48785. doi: 10.1074/jbc.M204839200. [DOI] [PubMed] [Google Scholar]
- Wang XQ, Deriy LV, Foss S, Huang P, Lamb FS, Kaetzel MA, Bindokas V, Marks JD, Nelson DJ. CLC-3 channels modulate excitatory synaptic transmission in hippocampal neurons. Neuron. 2006;52:321–333. doi: 10.1016/j.neuron.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Hugge C, Bielinska M, Nicholas P, Majerus PW, Wilson DB. Neural tube defects in mice with reduced levels of inositol 1,3,4-trisphosphate 5/6-kinase. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0904172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Kaetzel MA, Bruzik KS, Dedman JR, Shears SB, Nelson DJ. Inositol 3,4,5,6-tetrakisphosphate inhibits the calmodulin-dependent protein kinase II-activated chloride conductance inT84 colonic epithelial cells. J Biol Chem. 1996;271:14092–14097. doi: 10.1074/jbc.271.24.14092. [DOI] [PubMed] [Google Scholar]
- Xie W, Solomons KRH, Freeman S, Kaetzel MA, Bruzik KS, Nelson DJ, Shears SB. Regulation of Ca2+-dependent Cl- conductance in T84 cells: cross-talk between Ins(3,4,5,6)P4 and protein phosphatases. J Physiol (Lond) 1998;510:661–673. doi: 10.1111/j.1469-7793.1998.661bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Reece JM, Cho J, Bortner CD, Shears SB. The nucleolus exhibits an osmotically regulated gatekeeping activity that controls the spatial dynamics and functions of nucleolin. J Biol Chem. 2008;283:11823–11831. doi: 10.1074/jbc.M800308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- York JD. Phosphoinositides and inositol phosphates. 2011. [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- York JD, Guo S, Odom AR, Spiegelberg BD, Stolz LE. An expanded view of inositol signaling. Adv Enzyme Regul. 2001;41:57–71. doi: 10.1016/s0065-2571(00)00025-x. [DOI] [PubMed] [Google Scholar]
- Yu X, Duan KL, Shang CF, Yu HG, Zhou Z. Calcium influx through hyperpolarization-activated cation channels (I(h) channels) contributes to activity-evoked neuronal secretion. Proc Natl Acad Sci U S A. 2004;101:1051–1056. doi: 10.1073/pnas.0305167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M, Di BG, Lissandron V, Mancuso L, Terrin A, Zamparo I. Restricted diffusion of a freely diffusible second messenger: mechanisms underlying compartmentalized cAMP signalling. Biochem Soc Trans. 2006;34:495–497. doi: 10.1042/BST0340495. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Li X, Hao J, Winston JH, Weinman SA. The ClC-3 chloride transport protein traffics through the plasma membrane via interaction of an N-terminal dileucine cluster with clathrin. J Biol Chem. 2007;282:29022–29031. doi: 10.1074/jbc.M703506200. [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen L-M, Hernandez L, Shears SB, Galán JE. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host-cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol. 2001;39:248–259. doi: 10.1046/j.1365-2958.2001.02230.x. [DOI] [PubMed] [Google Scholar]