Abstract
Electrophilic DNA adducts produced following oxidative stress can form DNA-protein cross-links (DPCs), dramatically altering genomic maintenance pathways. Complete characterization of DPCs has been hindered, in part, because of a lack of comprehensive techniques for their analysis. We have, therefore, established a proteomics approach to investigate sites of cross-link formation using N6-(3-oxo-1-propenyl)-2′-deoxyadenosine (OPdA) as an electrophilic DNA adduct produced from oxidative stress. OPdA was reacted with albumin, and reduced with NaBH4 to stabilize DPCs. Using LC-MS/MS proteomics techniques, high-resolution peptide sequence data were obtained; however, using a database searching strategy, adducted peptides were only identified in samples subjected to chemical depurination. This strategy revealed multiple oxopropenyl adenine-lysine adducts and oxopropenyl-lysine adducts with the most reactive lysines identified to be Lys256 and Lys548. Manual interrogation of the mass spectral data provided evidence of OPdA deoxynucleoside conjugates to lysines and cross-links that underwent facile collision-induced dissociation to release an unmodified peptide without subsequent fragmentation. These fragmentations precluded adduct detection and peptide sequencing using database searching methods. Thus, comprehensive analysis of DPCs requires chemical depurination of DNA-protein reaction mixtures followed by a combination of database-dependent and manual interrogation of LC-MS/MS data using higher-energy collision-induced dissociation. In the present case, this approach revealed that OPdA selectively modifies surface lysine residues and produces nucleoside-protein cross-links and oxopropenyl lysine.
INTRODUCTION
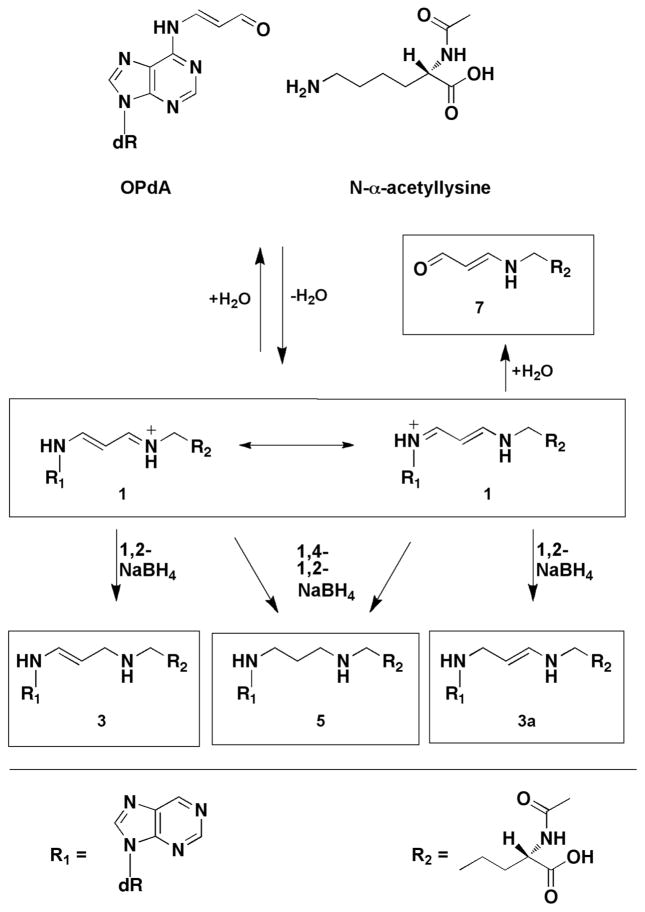
Bi-functional electrophiles can react with DNA-protein complexes to form DNA-protein cross-links (DPCs). Such cross-links represent significant impediments to the dynamic interactions required for DNA replication and transcription inter alia.1 Remarkably little is known about the structure and function of DPCs, especially ones that arise from endogenous metabolic processes. Base propenal and malondialdehyde are products of DNA peroxidation and lipid peroxidation, respectively, and they react with DNA bases to form oxopropenyl adducts to exocyclic amino groups.2–5 N6-Oxopropenyl-deoxyadenosine (OPdA) couples to N-α-acetyllysine to form an enamino-imine crosslink (Figure 1).6 Acrolein, another product of oxidative stress, forms an oxopropyl adduct with 2′-deoxyguanosine that couples to lysine-containing peptides to form cross-links.7 In contrast to the oxopropenyl-derived cross-link from base propenal, the oxopropyl-derived cross-link from acrolein is unstable to hydrolysis and cannot be isolated without chemical reduction.
Figure 1. Formation of covalent cross-links between N-α-acetyllysine and OPdA.
The formation of OPdA and N-α-acetyllysine cross-links is depicted above. R1 represents a portion of OPdA while R2 represents a portion of N-α-acetyllysine, as indicated. Reproduced from Szekely, J., Rizzo, C. J., and Marnett, L. J. (2008) Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J. Am. Chem. Soc. 130, 2195-2201. 6 Copyright 2008 American Chemical Society.
Additional agents that have been shown to lead to DPC formation include methylglyoxal, ionizing radiation, chromium, nitrogen mustards, cisplatin and 1,2,3,4- diepoxybutane (DEB).8–16 This work has provided a wealth of information regarding in vitro and cellular formation of DPCs, with subsets of these studies identifying specific proteins involved in DPC formation using mass spectrometry-based proteomic analyses. Comprehensive structural and chemical analysis of DPCs using mass spectrometry has been lacking, however, N7-guanine adducts have been studied including diepoxybutane (DEB).15, 17, 18 This bi-functional electrophile reacts with the N-7 position of 2′-deoxyguanosine residues in DNA to form an epoxyethyl adduct that couples to nucleophilic sites in proteins.19 Elegant work has comprehensively characterized the sites of modification by DEB on O6-alkylguanine DNA alkyltransferse (AGT) using MS-based proteomics techniques employing neutral thermal hydrolysis to release the adducted guanine and facilitate analysis.15 These methods are effective in the characterization of DPCs containing N-7 purine adducts, which undergo facile depurination; however, they may not be amendable to all classes of DPCs.
We sought to develop methods with which to investigate the DPCs formed from endogenous products of metabolic oxidation. We were particularly interested in methods that could be used with DPCs containing nucleic acid adducts to exocyclic amino groups. We chose to use OPdA for these exploratory investigations because its enamino-imine cross-link to lysine is relatively stable and can be reduced by either NaBH4 or NaCNBH3 to very stable dihydro or tetrahydro derivatives.6 Albumin was chosen as the protein component for method development because it has been used to characterize many protein-electrophile reactions, and a large inventory of known adduction sites is available.
Here we show that standard proteomics techniques using data-dependent analysis reveal a rich assortment of DPCs formed from the reaction of OPdA with albumin. These DPCs all contained deribosylated base adducts rather than nucleoside adducts. Manual interrogation of the data from the samples revealed the presence of nucleoside adducts that were not detected with database searching methods because of the facile loss of 2′-deoxyribose during the collision-induced dissociation used to generate peptide sequence information. We also discovered that a subset of OPdA-derived DPCs contain bonds within the cross-link that are quite labile to collision-induced cleavage. These adducts also require manual interrogation methods to enable their detection and analysis. Our results reveal the importance of chemical lability in the analysis of DPCs and suggest methods that can be used for proteomic analysis to provide information about the nature of the adducting species and the sequence of the peptide to which it is attached.
EXPERIMENTAL PROCEDURES
Materials
BSA was obtained from New England Biolabs (Beverly, MA). OPdA was prepared as previously described and was kindly provided by Dr. Carmelo Rizzo (Vanderbilt University).20 N-α-acetyllysine, trichloroacetic acid (TCA), dithiothreitol, iodoacetamide (Sigma Ultra), and NaBH4 were obtained from Sigma-Aldrich (St. Louis, MO). Trypsin (gold) was obtained from Promega (Madison, WI). 2,2,2-Trifluoroethanol (TFE) was obtained from Acros Organics (Pittsburgh, PA).
Determination of the extent of OPdA and N-α-acetyllysine cross-link formation
N-α-acetyllysine (200 mM) was mixed with 2 mM OPdA in 200 μL reactions containing 50 mM sodium phosphate buffer (pH 7.4), and aliquots of the reaction mixture were injected onto a Waters HPLC system utilizing a 717 Plus autosampler, 1525 Binary HPLC pump, and 2996 photodiode array detector in 1 h intervals for 15 h. Analytes were separated using a Supelco Discovery C18 HPLC column, 25 cm × 4.6 mm with 5 μm particle size at a flow rate of 1 mL/min. The mobile phase solvents were 10 mM ammonium acetate, pH 7.8 (solvent A) and acetonitrile (solvent B) delivered in a 17 min gradient consisting of the following: 0–10.5 min, 5–20% B; 10.5–12.5 min, 20–80% B; 12.5–16 min, 80% B; 16–17 min, 5% B. Analytes were identified by comparison of retention time and absorbance properties to standards. OPdA displays a maximum absorbance at 324 nm and a retention time of 8.5 min while the OPdA-N-α-acetyllysine cross-link absorbs at 348 nm with a retention time of 7.4 min (Supplemental Figure 11).6 Peaks were collected and analyzed using MS to confirm the presence of cross-links.
Reaction of BSA with OPdA
Purified BSA (100 μg; 1.5 nmol) was incubated with increasing concentrations of OPdA (reconstituted in H2O) in 200 μL reaction mixtures containing 50 mM sodium phosphate (pH 7.4) for 6 h at room temperature. OPdA concentrations were as follows: 0.007, 0.15, 0.6, 1.8, and 2.4 mM. These represent a molar ratio of OPdA:BSA of 1:1, 25:1, 100:1, 300:1, and 400:1, respectively. Following incubation, NaBH4 was added to each sample to a final concentration of 50 mM, and reactions were incubated for an additional 30 min. Samples were then processed and analyzed using LC-MS/MS as described.
Liquid chromatography-coupled tandem mass spectrometric (LC-MS/MS) analysis of BSA modification by OPdA
Samples were precipitated with either acetone (ice cold, 6X sample volume) overnight at −20 °C or with 25% TCA on ice for 1 h. Following incubation, samples were centrifuged at 18,000 xg at 4 °C and precipitates were washed with cold acetone, dried, and reconstituted in 50 mM Tris-HCl (pH 8.0), containing 50% TFE. Samples were reduced with dithiothreitol, carbamidomethylated with iodoacetamide, diluted 5-fold with 100 mM Tris-HCl (pH 8.0) (to obtain a final solution containing 10% TFE), and digested with sequencing-grade trypsin overnight. Following digestion, samples were acidified and diluted in 0.1% formic acid. The resulting solutions of trypsin-generated peptides were loaded onto a capillary reverse phase analytical column (360 μm O.D. × 100 μm I.D.) using an Eksigent NanoLC Ultra HPLC and autosampler. The analytical column was packed with 20 cm of C18 reverse phase material (Jupiter, 3 μm beads, 300Å Phenomenex), directly into a laser-pulled emitter tip. Peptides were eluted at a flow rate of 500 nL/min, and the mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). A 90-min gradient was performed, consisting of the following: 0–10 min, 2% B; 10–50 min, 2–40% B; 50–60 min, 40–95% B; 60–65 min, 95% B; 65–70 min 95–2% B; 70–90 min, 2% B. Eluting peptides were mass analyzed on an LTQ Orbitrap XL mass spectrometer or LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Gradient-eluted peptides were introduced into the mass spectrometers via nanoelectrospray ionization, and the instruments were operated using data-dependent methods with dynamic exclusion enabled. Full scan (m/z 400–2000 or m/z 300–2000) spectra were acquired with the Orbitrap (resolution 60,000), and the five most abundant ions (LTQ Orbitrap XL) or sixteen most abundant ions (LTQ Orbitrap Velos) in each MS scan were selected for fragmentation via collision-induced dissociation (CID) in the LTQ ion trap. An isolation width of 2 m/z and normalized collision energy of 35% were used to generate MS2 spectra. Activation times of 30 ms and 10 ms were using for LTQ Orbitrap XL and LTQ Orbitrap Velos LC-MS/MS analyses, respectively. Dynamic exclusion settings allowed for a repeat count of 1 or 2 within a repeat duration of 10 s, and with an exclusion duration time of 15 s. For identification of modified peptides, tandem mass spectra were searched against a Bos taurus subset database created from the UniprotKB protein database (www.uniprot.org). Searches were performed using a custom version of SEQUEST (Thermo Scientific) on the Vanderbilt ACCRE Linux cluster. The searches were performed with trypsin specificity, and were configured to include variable modifications of +57.0214 on Cys (carbamidomethylation), +15.9949 on Met (oxidation), and adducts 1–9 on lysine residues. (Figure 3). Spectra were searched using a 2Da mass tolerance for precursor peptide mass. Search results were assembled using Scaffold 3.0 (Proteome Software), where threshold filtering criteria consisted of 95% peptide probability to achieve a peptide false discovery rate estimate of ≤0.2%. Sites of modification were validated by manual interrogation of tandem mass spectra using Xcalibur 2.1 Qual Browser software (Thermo Scientific). Calculations of mass errors (in ppm) of precursor ions mass analyzed in the Orbitrap were performed using the following equation: ((Theoretical mass – Observed mass)/Theoretical mass) × 106. To obtain theoretical masses, the Protein Prospector MS-Product tool was used (v.5.10.4. UCSF).
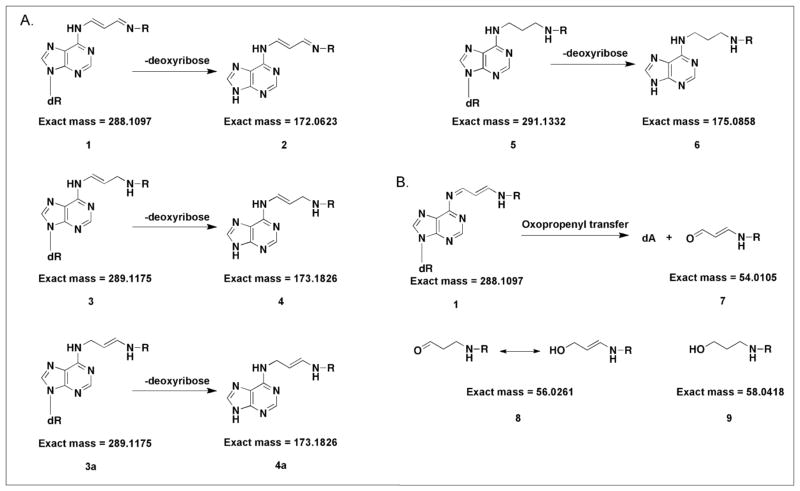
Figure 3. Predicted OPdA adduct structures and masses.
Structures represent theoretical OPdA modifications, including A, unreduced, partially reduced, and fully reduced species, in addition to the loss of 2′-deoxyribose. B, Oxopropenyl transfer of OPdA to lysine residues was also included in the modification search. The exact masses used for the search were calculated and are displayed above.
High-resolution tandem mass spectrometric analyses were performed on OPdA-modified peptides using an LTQ Orbitrap Velos mass spectrometer. All MS and MS/MS spectra were acquired using the Orbitrap as the analyzer, with full scan data collected with resolution 60,000 and MS/MS spectra acquired with resolution 7,500. MS/MS data were generated using either CID or higher-energy collisional dissociation (HCD). An isolation width of 2.5 m/z, 35% normalized collision energy, an MSn automatic gain control target of 4e5, and maximum injection time of 600 ms were used for collection of all high-resolution MS/MS spectra. The reverse phase LC conditions were similar to those described previously. All data were manually interpreted.
RESULTS
Reactivity of OPdA with N-α-acetyllysine
The time course of cross-link formation between OPdA and N-α-acetyllysine was determined by HPLC analysis. Excess N-α-acetyllysine (200 mM) was incubated with OPdA (2 mM) in 50 mM sodium phosphate buffer (pH 7.4) for 15 h during which hourly analyses of the reaction mixture were performed using C18 reversed phase HPLC. The OPdA-N-α-acetyllysine cross-link eluted at 7.4 min and was identified by absorbance at 343 nm (Supplemental Figure 11) as well as by MS analysis.6 OPdA eluted at 8.5 min and exhibited characteristic absorbance at 320 nm (Supplemental Figure 11).6 Peak area was quantified using Empower software, revealing maximum cross-link formation after 6 h of incubation (Supplemental Figure 11). This time course was used to design experiments examining the reaction between OPdA and BSA.
Database search-dependent interrogation of BSA modifications by OPdA
Purified BSA (7.5 μM) was incubated with or without 2.4 mM OPdA for 6 h at room temperature followed by the addition of 50 mM NaBH4 to stabilize imine-containing adducts. Following incubation, samples were prepared for LC-MS/MS analysis by acetone precipitation, followed by resolubilization of the precipitated protein pellet, reduction and carbamidomethylation of cysteine residues, and proteolytic digestion with sequencing-grade trypsin. Following digestion, proteolytic peptides were analyzed via LC-MS/MS using an LTQ-Orbitrap Velos mass spectrometer with collision-induced dissociation (CID) employed to induce peptide fragmentation (Figure 2).
Figure 2. Workflow schematic for identification of OPdA protein modifications.
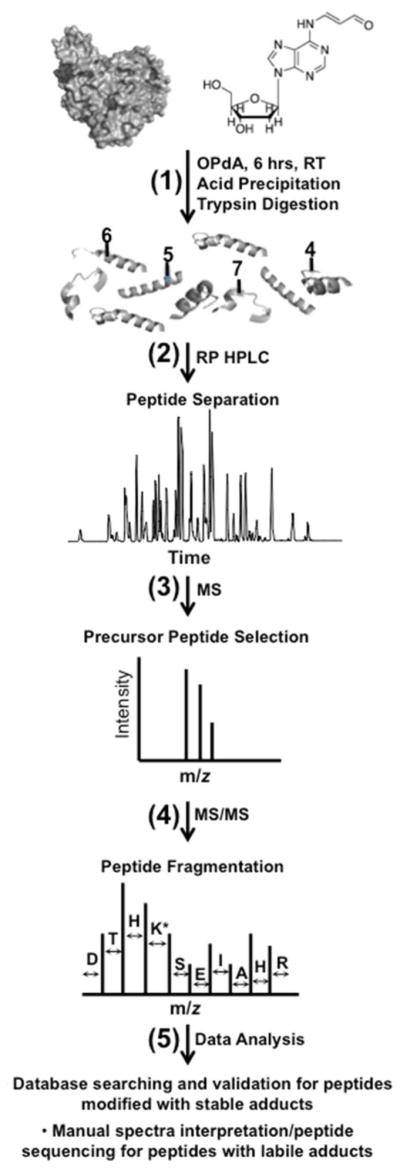
Purified BSA was incubated with OPdA for 6 hr at room temperature and subjected to tryptic digestion as described (1) (Experimental Procedures). Following digestion, precursor peptides were separated and analyzed via LC-MS (2,3). Abundant peptides were selected for MS/MS in which the peptide backbone was fragmented using collisional-induced dissociation (4). Data was then analyzed using database-searching strategies for stable adducts and manual interpretation for peptides modified with labile adducts (5).
Acquired tandem mass spectra were searched against a bovine protein database using the SEQUEST database-searching algorithm.21 Upon tandem mass spectrometry, a peptide is isolated and fragmented along the peptide backbone, and the resulting product ions are measured to produce an MS/MS spectrum. The most widely used fragmentation technique is CID, which induces fragmentation at the peptide bond, producing a series of b- and y-type product ions. The main principle of peptide sequencing is to use the mass difference between two consecutive product ions to calculate the mass of an amino acid residue within the peptide. This process can be continued until all the residues are determined sequentially. SEQUEST functions by correlating the masses of observed CID-derived b- and y-type product ions resulting from backbone fragmentation with theoretical data derived from known protein sequences. In principle, if residues are adducted, b-and y-ions with mass shifts corresponding to OPdA-derived adducts will be observed. To examine OPdA modifications, adduct structures and their corresponding masses were generated based on previous characterization of cross-links formed between OPdA and N-α-acetyllysine (Figures 1 and 3).6 As displayed in Figure 3, adduct 1 represents an intact unreduced OPdA cross-link, whereas adduct 2 represents loss of 2′-deoxyribose from adduct 1. The corresponding singly reduced cross-linked species (3/3a and 4/4a) as well as doubly reduced cross-linked species (5 and 6) were also considered (Figure 3). Following cross-link formation, hydrolytic cleavage resulting in oxopropenylation of the lysine amine and the release of dA has been demonstrated (Figures 1 and 3).6 To account for these potential products, the masses corresponding to unreduced oxopropenyl transfer (7) as well as singly reduced (8) and doubly reduced (9) adducts were interrogated). Theoretical mass shifts provided in Figure 3 were included in the database search parameters, allowing for identification of adducted residues, if present. Following SEQUEST searching, results were compiled using the Scaffold data assembly program (Experimental Procedures).
Database searching and Scaffold data assembly of each sample revealed 89–93% sequence coverage of BSA, which included an average of 87% lysine residue coverage. However, no nucleoside-peptide cross-links were detected. This was surprising, considering the previously demonstrated stability of reduced OPdA-lysine cross-links throughout MS and NMR analysis.6 Previous work analyzing DPCs using proteomic analyses had employed thermal depurination to induce deribosylation, which can also be achieved using acid-based chemical depurination.12, 14, 15 Acetone precipitation was used in our initial analyses as an alternate protocol to avoid depurination and retain intact OPdA modifications. As this initial strategy did not result in the identification of OPdA modifications, and since glycosidic bonds are labile, we hypothesized that the energy applied throughout MS or MS/MS analyses may induce deribosylation and/or adduct decomposition, impeding adduct identification. Adduct decomposition that occurs during LC-MS/MS would preclude detection using database-searching strategies, because an intact adduct initially present on a precursor peptide will vary in mass from a decomposed adduct present on the resulting product ions following MS/MS. This discrepancy will interfere with the ability to identify adducts using a conventional database searching approach. These considerations prompted us to employ chemical depurination by treatment with TCA so that all adducts would be deribosylated prior to LC-MS/MS analysis. Subsequent to TCA precipitation, samples were digested and prepared for MS as described above. Samples were then analyzed using MS and MS/MS (Experimental Procedures), after which SEQUEST database searching and Scaffold data assembly were performed to examine the presence of adducts 1–9. Chemical depurination using TCA resulted in identification of multiple lysine residues modified with the fully reduced OPdA adduct without 2′-deoxyribose (6) and oxopropenyllysine modification (7) (Table 1). These results demonstrate the need for chemical depurination in the identification of OPdA-protein adducts using data-dependent LC-MS/MS analyses and database searching algorithms.
Table 1. Summary of bovine serum albumin residues adducted by OPdA.
BSA was incubated with increasing amounts of OPdA, and proteomic analyses were performed as described in Experimental Procedures. Residues adducted with an additional mass of 175.0858 (6) and/or 54.0105 (7) are indicated. Modified residue numbers are provided, and numbers displayed in parentheses correspond to the equivalent residue in the human serum albumin crystal structure. All peptides displayed mass errors of ≤3 ppm.
| Residue | Peptide | Molar ratio of OPdA:BSA | ||
|---|---|---|---|---|
| 400:1 | 300:1 | 100:1 | ||
|
| ||||
| LYS-28 | DTHK*SEIAHR |
6 7 |
6 7 |
|
|
| ||||
| LYS-36 (12) | FK*DLGEEHFK | 7 | ||
|
| ||||
| LYS-100 | SLHTLFGDELCK*VASLR | 6 | 6 | |
|
| ||||
| LYS-248 (225) | FPK*AEFVEVTK | 6 | 6 | |
|
| ||||
| LYS-256 (233) | AEFVEVTK*LVTDLTK | 6 | 6 | 6 |
|
| ||||
| LYS-299 (276) | LK*ECCDKPLLEK | 6 | ||
|
| ||||
| LYS-437 (414) | K*VPQVSTPTLVEVSR |
6 7 |
6 | |
|
| ||||
| LYS-463 | CCTK*PESER | 6 | 6 | |
|
| ||||
| LYS-489 (466) | LCVLHEK*TPVSEK |
6 7 |
6 | |
|
| ||||
| LYS-498 (475) | VTK*CCTESLVNR | 7 | ||
|
| ||||
| LYS-548 (525) | K*QTALVELLK |
6 7 |
6 7 |
6 |
Characterization of stably formed protein adduction species induced by OPdA
Following identification of 6- and 7-adducted peptides using database searching, manual interrogation of both the MS and MS/MS data was performed to verify adduction sites. First, theoretical precursor peptide masses were used to generate extracted ion chromatograms (XICs) corresponding to the identified precursor ions (Figure 4). Each XIC was evaluated for the presence of a monoisotopic peak displaying a mass error of less than 5 ppm relative to the theoretical precursor mass. In addition to XIC-based analysis and confirmation of monoisotopic peaks, individual tandem mass spectra were manually interrogated for b-and y-type product ions corresponding to the predicted product ions of the adducted peptide. At least 40% of each theoretical b-and y-product ion series was present in the observed product ion spectra of positively identified peptides, and 75% sequence coverage was obtained for each peptide validated. To validate the site of adduction, at least two ions within a product ion series with a mass shift consistent with the modification were required.
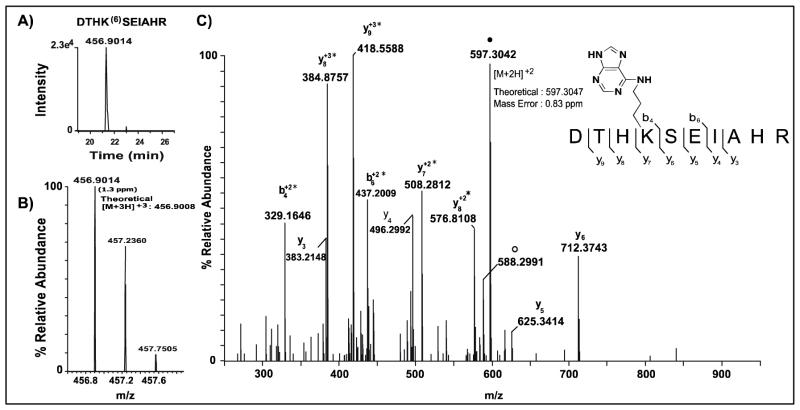
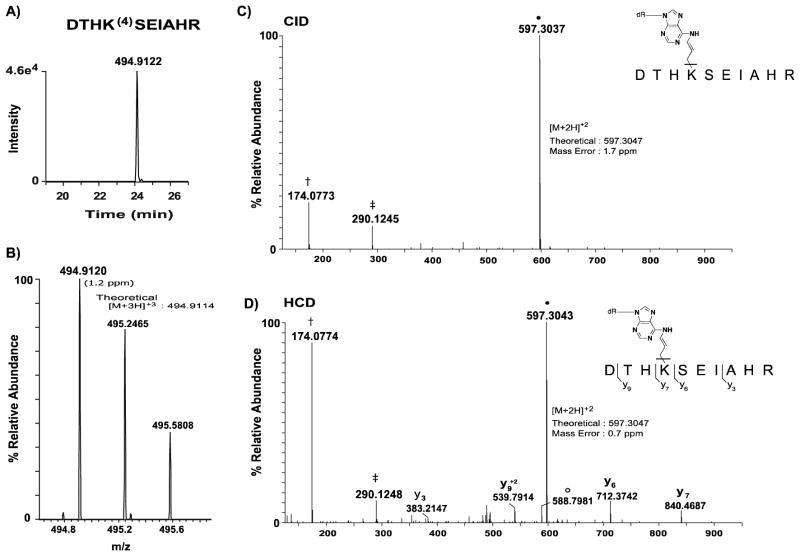
Figure 4. Extracted ion chromatograms and corresponding MS spectra of precursor ions observed for OPdA-modified peptide DTHK(6)SEIAHR.
A, A window of ± 10 ppm around the theoretical m/z values for the observed precursor ion for peptide DHTK(6)SEIAHR was used to generate the extracted ion chromatogram (XIC) shown. B, The mass spectrum for the precursor peptide is annotated with the observed m/z values provided above the corresponding monoisotopic peak. The theoretical value of the precursor ion, adjacent to the observed peak, was used to calculate ppm mass error (shown in parentheses) for the identified peptide form. C, Targeted high-resolution CID spectrum of the 6-adducted DTHK(6)SEIAHR precursor peptide (m/z: 456.90). Brackets adjacent to the peptide sequence indicate sites of amide bond fragmentation at the peptide backbone that occurred with CID to generate b- and y-type product ions. Observed b- and y-ions are annotated above the corresponding product ion peaks in the spectrum. Unmodified DTHKSEIAHR precursor peptide product is represented by ●. Water loss from unmodified DTHK(6)SEIAHR precursor peptide is indicated by ○. Asterisks indicate residues with mass shifts consistent with 6-adduction. The mass error observed for the unmodified peptide product is 0.83 ppm and is shown adjacent to the corresponding ion peak. Mass errors for all ions observed was less than 4 ppm. Unless otherwise indicated, product ions are singly-protonated.
This analytical approach is demonstrated with a representative 6-adducted peptide, DTHK(6)SEIAHR (Lys 28) (Figure 4). The XIC was generated using the modified triplyprotonated ([M+3H]+3) form of the precursor peptide (Figure 4A), with the observed ion displaying a 1.3 ppm mass error relative to theoretical (Figure 4B). The corresponding high-resolution CID MS/MS spectrum displayed a series of b-and y-ions representing 80% of the peptide sequence, as well as several product ions (y7–y9, b4, and b6) that displayed mass shifts consistent with 6-adduction at Lys-28 (Figure 4C). The presence of a monoisotopic peak with accurate mass corresponding to the precursor peptide mass as well as multiple b-and y-product ions with mass additions consistent with 6-adduction confirms the identity of the peptide and the site of OPdA modification at Lys-28. All peptides identified with OPdA-derived modifications were interrogated using this comprehensive analytical method resulting in the confirmation of nine 6-adducted lysine residues including Lys-28, Lys-100, Lys-248, Lys-256, Lys-299, Lys-437, Lys-463, Lys- 489, and Lys-548 (Table 1). Peptides containing adducts 1–5 were not identified using this database searching approach.
In addition to the analysis of intact OPdA adducts, various forms of oxopropenyl transfer products (7–9) were also investigated. Adduct 7 modification was confirmed using the methodologies described above, resulting in the verification of six 7-adducted lysine residues including Lys-28, Lys-36, Lys-437, Lys-489, Lys-498, and Lys-548 (Table 1). Neither singly reduced nor doubly reduced oxopropenyl adducted peptides (adducts 8 and 9) were identified in this analysis.
Mass spectrometric characterization of labile OPdA protein modifications
Data analysis conducted using a database-search strategy in conjunction with chemical depurination demonstrated that OPdA-derived adducts (6 & 7) can be found on lysine residues. As acetone precipitation, which retains intact adducts, did not result in the identification of OPdA-derived adducts, we hypothesized that energy applied throughout LC-MS/MS analysis results in deribosylation and/or adduct decomposition, precluding the detection of a subset of adducts (1–5). Since our goal was to thoroughly describe all adduction species induced by OPdA and implement methods for their detection, we adopted a strategy of manual examination of mass spectra to investigate the formation of adducts 1–5. To begin our analysis, we examined the formation of 5-adducted peptides, as adduct 5 is the 2′-deoxyribose-containing precursor of adduct 6, which was identified using database-searching analyses (Figures 3 & 4). We did not detect any adducts in the acetone precipitated samples, so we focused our attention on the TCA precipitated samples. The rationale for analyzing these data is two-fold, 1) our manual interrogation approach requires previously identified peptide data, which was only available from the TCA precipitated samples, and 2) incomplete depurination in these samples would result in retention of 1-, 3-, and 5-adducted peptides, allowing for their analysis using manual interrogation.
6-adducted peptide sequences were used as a platform for determining precursor peptide masses of theoretical 5-adducted peptides. Theoretical peptide masses containing adduct 5 modifications were calculated using the identified peptides modified by adduct 6. Calculated masses converted to m/z values were then used to generate XICs, which were analyzed for the presence of monoisotopic peaks with the appropriate accurate mass relative to theoretical. For all peptide sequences interrogated, chromatographic and monoisotopic peaks consistent with 5-adducted peptides were identified, providing evidence for their formation. The XIC and monoisotopic peak of a representative 5- adducted peptide, DTHK(5)SEIAHR is presented in Figure 5A. These results indicate that although 5-adducted peptides were not identified in our initial algorithm-based database search, they are present in solution and peptide precursors are readily identifiable using manual interrogation techniques.
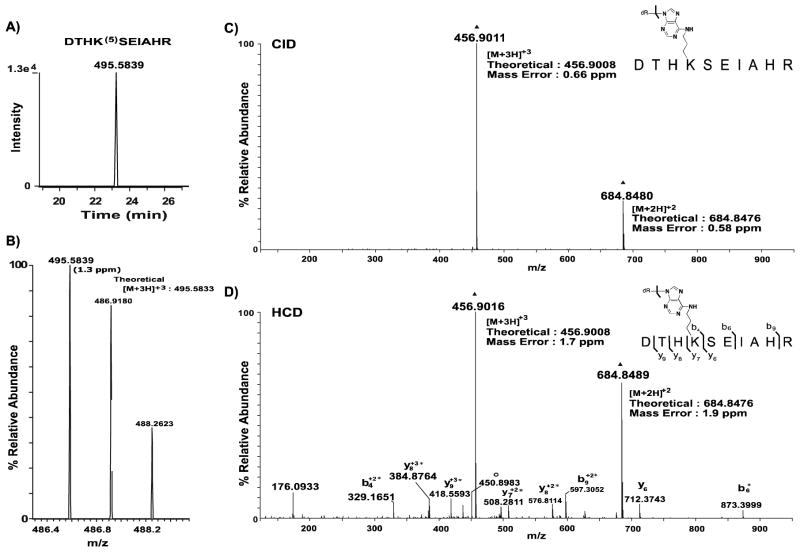
Figure 5. Extracted ion chromatograms and corresponding MS spectra of precursor ions observed for OPdA-modified peptide DTHK(5)SEIAHR.
A, The XIC was generated as described in Figure 4A. B, The mass spectrum for the precursor peptide is annotated with the observed m/z values provided above the corresponding monoisotopic peak. The theoretical value of the precursor ion is adjacent to the observed peak and was used to calculate ppm mass error (shown in parentheses) for the identified peptide form. C, High-resolution targeted CID spectrum of precursor 5-adducted DTHK(5)SEIAHR peptide (m/z: 495.58). Dissociation of the glycosidic bond, as indicated by the bracket, results in the formation of intact peptide products with mass shifts consistent with 6- adduction. This peptide is present as both doubly- and triply-protonated ions, denoted by ▲. D, High-resolution targeted HCD spectrum of precursor 5-adducted DTHK(5)SEIAHR peptide (m/z: 495.58). Glycosidic bond cleavage was observed in addition to peptide backbone fragmentation. Asterisks indicate residues with mass shifts consistent with adduction by 6. Water loss from the 6-adducted DTHK(6)SEIAHR product is denoted with ○, and the intact 6-adducted DTHK(6)SEIAHR peptide product is denoted with ▲. Mass errors observed for the [M+2H]+2 and [M+3H]+3 6-adducted peptide products are provided in both spectra and mass errors for all product ions observed were less than 4.5 ppm. Unless otherwise indicated, product ions are singly-protonated.
The presence of 5-adducted precursor peptides raises the possibility that loss of 2′-deoxyribose from these species may be a source of 6-adducted peptides identified in the database search (Figure 7). To investigate this possibility, extracted chromatograms of 5- and 6-adducted peptides were examined to compare retention times following chromatographic separation. Independent formation will result in differential elution times whereas MS-induced formation of 6-adducted peptides from 5-adducted species will display co-eluting profiles. Comparison of chromatographic peaks corresponding to 5- and 6-adducted peptides reveal retention times of 21 and 23 mins, respectively, indicating that both 5- and 6-adducted peptides are present in solution prior to chromatographic separation and ionization.
Figure 7. Schematic of the fragmentation profile of OPdA adduction species.
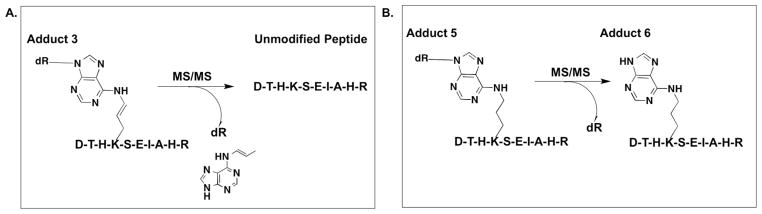
A, 3- adducted DTHK*SEIAHR is formed stably in solution; however, the collision energy employed during MS/MS induces the loss of both OPdA and 2′-deoxyribose, resulting in unmodified peptide. B, 5-adducted DTHK*SEIAHR is also formed stably in solution, but loses the 2′-deoxyribose moiety upon application of collision energy during MS/MS. This results in the formation of a stable 6-adducted peptide.
The presence of 5-adducted peptide precursors indicates stability of these peptides throughout chromatographic separation and initial MS analysis, again presenting the question of why these modified peptides were not detected with database searches. Initial studies involved data-dependent-triggered MS/MS of observed precursor ions, as described in Experimental Procedures. Manual interrogation of acquired data revealed that ions corresponding to 5-adduced precursor peptides were indeed chosen for MS/MS. We therefore manually examined these data, and analysis of the CID spectra of these peptides demonstrated a surprising result. Rather than observing characteristic peptide backbone fragmentation as was seen for the 6-adducted precursor peptide (Figure 5C), two major product ions, m/z 456.9011 and m/z 684.8480, were observed in the tandem mass spectra of the 5-adducted peptide (Figure 5C). Upon further investigation, these ions were found to correlate to the [M+3H]+3 and [M+2H]+2 charge states of a 6-adducted peptide sequence, indicating the loss of 2′-deoxyribose from 5-adducted peptides occurs during MS/MS analysis (Figure 5C). CID is a threshold energy dissociation process in which more labile bonds present within a molecule will preferentially dissociate upon collisional activation.22 CID of 5-adducted peptides leads to the formation of 6-adducted peptide product ions, which, together with the absence of peptide backbone fragmentation, demonstrates the labile nature of the glycosidic bond. CID results in preferential dissociation of the 2′-deoxyribose moiety and little or no fragmentation of the peptide backbone thereby precluding the detection of 5-adducted peptides using standard database searching strategies.
To confirm the identity of 5-adducted peptides, higher-energy collisional dissociation (HCD) was utilized in an attempt to generate amide bond backbone cleavage, which would provide sequence information. While multiple dissociation methods may be employed for peptide fragmentation, HCD often results in a wide range of fragmentation pathways and has the potential to generate a richer product ion spectrum relative to CID.23, 24 This dissociation technique was used in a targeted MS/MS analysis of 5- adducted DTHK(5)SEIAHR, which resulted in the cleavage of the 2′-deoxyribose moiety, as was observed for CID (Figure 5D). HCD, however, also led to fragmentation of the peptide backbone, which produced b-and y-product ions found to correspond to predicted 6-adducted peptide dissociation products (Figure 5D). HCD fragmentation resulted in 60% sequence coverage of the peptide, with both b- and y-ions (y6–y9, b4, b6 and b9) consistent with the 6-adducted peptide identified (Figure 5D). These results demonstrate that 5-adducted peptides are present prior to chromatographic analysis; however, upon CID and HCD fragmentation, 2′-deoxyribose is lost from the molecule, resulting in peptide fragmentation ions corresponding to 6-modification (Figure 7). These data highlight the inherent instability of the 2′-deoxyribose moiety during CID and HCD fragmentation, as well as the stability of adduct 6, as it remains covalently bound to peptides throughout fragmentation. 2′-Deoxyribose instability precludes identification of intact peptides modified with OPdA-derived adducts using the previously applied database-search strategies. However, these data demonstrate that chemical depurination prior to MS analyses creates stable OPdA-derived modifications that can be readily identified.
Confirmation of 5-adducted peptides despite their absence in the initial database search results led to the investigation of adducts 1–4 using the manual precursor-based search strategy described above. Adducts 3 and 3a as well as 4 and 4a could not be discriminated using the analytical methods employed in this analysis. As such, both 3 and 3a will be referred to as adduct 3 and adducts 4 and 4a will be referenced as adduct 4 (Figure 3). The lack of sequence information for peptides containing these adducts required the use of peptide sequences previously identified to be modified by adduct 6, similar to the approach utilized to identify 5-adducted peptides. These theoretical peptide masses were used to generate XICs, which were analyzed for the presence of monoisotopic peaks corresponding to representative peptides. This analysis resulted in no detection of peptides containing adduct masses consistent with 1, 2, and 4; however, several 3-adducted peptides were identified. A chromatographic peak corresponding to the theoretical mass of a representative 3-adducted peptide, DTHK(3)SEIAHR, can be found in Figure 6A. A correlating monoisotopic peak with m/z 494.9120 and 1.2 ppm mass error relative to the theoretical peptide mass is shown in Figure 6B. In order to further confirm the presence of 3-adducted DTHK(3)SEIAHR, MS/MS analysis was performed.
Figure 6. Extracted ion chromatograms and corresponding MS spectra of precursor ions observed for OPdA-modified peptide DTHK(3)SEIAHR.
A, The XIC was generated as described in Figure 4A. B, The mass spectra for the precursor peptide is annotated with the observed m/z values provided above the corresponding monoisotopic peak. The theoretical value of the precursor ion is adjacent to the observed peak, and was used to calculate ppm mass error (shown in parentheses) for the identified peptide form. C, High-resolution targeted CID spectrum of precursor 3-adducted DTHK(3)SEIAHR peptide (m/z: 494.91). Dissociation of the OPdA adduct, as indicated by the bracket, results in the formation of an intact unmodified peptide product (●). Dissociative adductderived products include the partially reduced OPdA adduct with (‡) and without (†) 2′-deoxyribose. D, High-resolution targeted HCD spectrum of precursor 3-adducted DTHK(3)SEIAHR peptide (m/z: 494.91). Glycosidic bond and adduct cleavage was observed in addition to unmodified peptide fragmentation. Intact unmodified precursor peptide (●) was observed as well as a loss of water from this peptide (○). Mass errors observed for the [M+2H]+2 unmodified peptide products are provided in both spectra, and mass errors for all product ions observed were less than 4.5 ppm. Unless otherwise indicated, product ions are singly-protonated.
Collision-induced fragmentation of 3-adducted DTHK(3)SEIAHR demonstrated the formation of three major ions, m/z 597.3037, m/z 290.1245, and m/z 174.0773, representing unmodified precursor peptide and dissociated OPdA adducts with and without 2′-deoxyribose, respectively (Figure 6C). These results are consistent with the fragmentation of OPdA from the peptide, leading to its release and the formation of an unmodified peptide product ion (Figure 7). The instability of singly reduced OPdA adducts is demonstrated by the liberation of this adduct from 3-adducted peptides during MS/MS analysis as well as the lack of detection of 4-adducted peptides in both the MS and MS/MS analysis. The absence of 4-adducted peptides indicates that singly reduced linkages between OPdA and amine groups are more (or equally) labile to fragmentation than the glycosidic bonds described above for 5-adducted peptides. In this instance, the collisional energy applied to induce peptide backbone fragmentation results in the cleavage of both 2′-deoxyribose and partially reduced (3) OPdA adducts from modified peptides.
To further clarify the fragmentation products of DTHK(3)SEIAHR, targeted HCD MS/MS of this peptide was performed. This higher-energy dissociation method resulted in fragmentation comparable to CID, in that OPdA was cleaved from the peptide. The use of HCD, however, also induced peptide backbone fragmentation, producing multiple band y-product ions consistent with the unmodified peptide (Figure 6D). These data demonstrate that 3-adducted peptides are indeed formed by OPdA adduction; however, upon HCD or CID fragmentation, the entire singly reduced OPdA adduct is lost, resulting in an unmodified peptide product (Figure 7).
Site-Selectivity of BSA Modification by OPdA
From 56 total lysine residues within BSA, an average of 49 were represented in the covered sequence of each sample. Of the 49 representative lysines, 11 were found to be modified by adduct 6 and/or adduct 7 (Table 1). Two residues within BSA, Lys-256 and Lys-548, were modified at low OPdA concentrations (0.6 mM – 2.4 mM), identifying these residues as the most susceptible to OPdA modification (Table 1). At higher OPdA concentrations, modification by adducts 6 and 7 was observed at Lys-28, Lys-437, Lys- 548, and Lys-489, whereas Lys-36 and Lys-498 were exclusively modified by 7. Lys-100, Lys-248, Lys-256, Lys-299, and Lys463 were modified by 6 but did not display a mass shift consistent with 7. Representative chromatograms and mass spectra of all confirmed 6-adducted peptides can be found in Supporting Information. Positively identified adduction sites were mapped to the crystal structure of HSA (PDB ID: 1AO6), revealing preferential modification at surface-exposed lysine residues (Figure 8). The absence of peptide information for adducts 4 and 5 prevented comprehensive analyses of these adduction sites. However, examination of modification sites on peptide sequences identified from analysis of 6 adduction revealed several precursor peptides displaying less than 5 ppm mass error compared to theoretical masses (data not shown).
Figure 8. Mapping of OPdA-modified residues on human serum albumin.
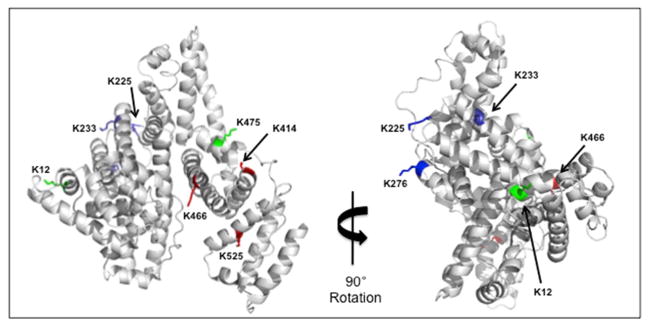
Crystal structure of human serum albumin (PDB: 1AO6) displaying highlighted residues corresponding to lysine residues adducted on BSA by OPdA. Residues displayed in green correspond to oxopropenyl transfer (7) modification. Those in blue correspond to 6 modification and red residues correspond to those that displayed both 6 and 7 modifications.
Amino Acid Specificity of OPdA
HPLC analysis of covalent cross-link formation between OPdA and purified amino acids revealed stable cross-links formed with N-α-acetyllysine but not with N-acetylhistidine or N-α-acetylarginine (Supplemental Figure 11 and data not shown). Examination of the reaction of N-acetylcysteine with OPdA revealed the formation of a very unstable, short-lived species, which is currently under investigation. MS analysis of OPdA adduction at arginine, histidine, and cysteine residues within BSA did not result in positive identification of modification. Therefore, these analyses suggest OPdA to be a lysine-specific electrophile that forms Schiff base products with the ε-amino group in lysine residues.
These results demonstrate the stability of doubly reduced OPdA-amine linkages throughout MS/MS analysis and highlight the inherent instability of singly reduced OPdA-amine linkages to mass spectrometric analyses. In addition, these findings emphasize the utility of manual examination and interpretation of mass spectra when analyzing protein adduction products of deoxyribonucleic acids.
DISCUSSION
Examination of the chemistry of DNA-protein cross-link formation and stability is an underexplored area of genomic research that has important implications for cellular physiology. The reactivity of OPdA with N-α-acetyllysine predicts its potential to form covalent protein-nucleic acid cross-links, which we have explored in this manuscript. Characterization of protein modification sites increases our understanding of the reactive potential of electrophilic DNA adducts and the mechanisms by which OPdA, specifically, can interact with proteins. Proteomic analysis of DPCs has not been adequately characterized, which may be influenced by the labile nature of the glycosidic bond present within all nucleosides, impacting detection using mass spectrometric techniques. The use of collision-induced dissociation for peptide fragmentation followed by automated database searching programs to identify peptides is a typical approach employed for the analysis of protein modifications. Nucleoside adduction products represent a unique category of adducts, and their chemical nature can preclude detection using conventional mass spectrometric data analysis methodologies. Indeed, we found that a manual data interpretation strategy was necessary to fully and accurately characterize DPCs.
Using a database searching approach in which tandem mass spectra were searched against a bovine protein database with parameters included for identification of potential adduct masses, we identified 11 lysine residues within BSA that were modified by OPdA (Table 1). These data revealed amino acid selectivity at lysine residues, specificity for surface-exposed sites, and exclusive modification by adducts 6 and 7 (Figure 8) (Table 1). Manual data analysis methods were required to accurately and thoroughly characterize 3- and 5-adducted peptides (Figure 2). These methods included manual interrogation of LC-MS chromatograms, monoisotopic peaks of peptide precursors, and tandem mass spectra. Fragmentation characteristics of these adducted peptides, including a lack of peptide backbone fragmentation during CID, precluded the identification of these adducted peptides using a standard database searching approach. We found that HCD fragmentation followed by manual data interpretation was required in the characterization of nucleic acid-protein cross-links, as cleavage of the labile glycosidic bond leads to the loss of 2′-deoxyribose and limits peptide backbone cleavage necessary for peptide sequencing and identification.
Applying both automated database searching and manual data interpretation, adducts 3 and 5–7 were identified as a result of the reaction of OPdA with BSA; however, only adducts 6 and 7 were found to be stable throughout MS analysis. Distinct chromatographic peaks corresponding to 5- and 6-adducted peptides indicate these species are present independently prior to chromatographic separation (Figures 4A & 5A). This observation demonstrates the formation of 6-adducted peptides in solution, indicating a loss of 2′-deoxyribose prior to MS analysis as a result of chemical depurination. As the glycosidic bond is labile in acidic conditions, the methodologies implemented in this study for MS-based analyses utilized TCA precipitation to chemically depurinate adducted peptides.25 Had TCA precipitation not been employed, initial analyses would not have identified 6-adducted peptides, highlighting the need for particular consideration of this method when analyzing DNA adducts using standard proteomics techniques. With regard to OPdA adduction species, TCA precipitation resulted in partial cleavage of the glycosidic bond of 5-adducted peptides in solution, allowing detection of 6-adducted peptides using database searching of tandem mass spectra. Without initial identification of 6-adducted peptides, characterization of OPdA-adducted peptides would have been significantly hindered.
Based on the evidence of the formation of both 3- and 5-adducted peptides and their instability to MS/MS analysis, we hypothesized that the lack of adduct detection in acetone-precipitated samples was a result of this instability. We therefore implemented our manual analysis method to examine the formation of 3- and 5-adducted peptides in the acetone-precipitated samples. Interrogation of the data for the presence of DTHK(3)SEIAHR and DTHK(5)SEIAHR revealed XIC, MS, and MS/MS data very similar to that demonstrated in Figures 5 & 6 (data not shown). These results highlight the inability to identify intact adducts using typical database-searching strategies and the importance of implementing HCD fragmentation and manual data interrogation when examining labile DPCs.
Unreduced OPdA adducts 1 and 2 were not observed, presumably due to hydrolysis or oxopropenyl transfer that can occur following their formation or reduction by NaBH4. As described above, OPdA adduction product 4 was not observed due to the inherent instability of the singly reduced OPdA-amine cross-link, 3. Adducts 8 and 9, although explored in this analysis, were not present, consistent with previous reports on the inability of NaBH4 to reduce similar oxopropenyl moieties.26 Understanding the behavior of OPdA adducts during mass spectrometric analysis provides a foundation for the development of automated methods to analyze OPdA and other reactive derivatives of DNA in vitro and in cellular systems. Application of HCD fragmentation along with manual spectral interpretation facilitated identification and characterization of the majority of identified products, many of which were not detected with conventional database searching. Methods described here resulted in comprehensive characterization of OPdA adduction products, providing a framework for expanding this work to the analysis of DPCs. Although we anticipate that sites of modification will vary depending on the oligonucleotide and the protein being examined, the methods described provide a framework for performing these studies.
The examination of covalent cross-links formed between lysine residues and electrophilic DNA adducts by our group and others is shedding light on a relatively underexplored area of DNA damage research. The ability of DNA adducts to form covalent cross-links with protein has wide-reaching cellular implications from blocking normal genomic maintenance processes to triggering aberrant signaling cascades. The chemical properties shared by all DPCs presents a challenge for analytical interrogation, and we describe analytical procedures enabling identification of labile DPCs. The work outlined in this manuscript is the first description of OPdA modification of protein residues and describes a thorough analytical method with which to interrogate protein modification by this and other DNA adducts.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by research grants from the National Institutes of Health [R37 CA087819 to L.J.M.] and [F32 CA159701-01 to S.C.S.].
The authors would like to thank the Vanderbilt University Proteomics Core, in particular Salisha Hill, for assistance in the analysis of protein modification by OPdA. We would also like to thank Dr. Carmelo Rizzo (Vanderbilt University) for providing the OPdA nucleoside and Philip Kingsley (Vanderbilt University) for technical assistance in establishing the HPLC method described. We would also like to thank Dr. James Galligan and Dr. Matthew Windsor for the careful reading and analysis of this manuscript. We are grateful to Dr. Carol Rouzer for assistance with editing the manuscript.
ABBREVIATIONS
- BSA
bovine serum albumin
- CID
collision-induced dissociation
- HCD
higher-energy collisional dissociation
- HPLC
high performance liquid chromatography
- HSA
human serum albumin
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- M1dG
3-(2′-deoxy-β-D-erythro-pentofuranosyl)-pyrimido[1,2-α]-purin-10(3H)-one
- MDA
malondialdehyde
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- OPdA
N6-(3-oxo-1-propenyl)-2′-deoxyadenosine
- ppm
parts per million
- TCA
trichloroacetic acid
- TFE
2,2,2-trifluoroethanol
- XIC
extracted ion chromatogram
Footnotes
SUPPORTING INFORMATION AVAILABLE: Figures displaying chromatograms and mass spectra of 6-modified peptides may be found in the supplemental material. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 3.Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J Clin Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc Natl Acad Sci U S A. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, Taghizadeh K, Dedon PC. Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J Biol Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 6.Szekely J, Rizzo CJ, Marnett LJ. Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J Am Chem Soc. 2008;130:2195–2201. doi: 10.1021/ja074506u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz AJ, Lloyd RS. 1, N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J Biol Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 8.Murata-Kamiya N, Kamiya H. Methylglyoxal, an endogenous aldehyde, crosslinks DNA polymerase and the substrate DNA. Nucleic Acids Res. 2001;29:3433–3438. doi: 10.1093/nar/29.16.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker S, Murray D, Zheng J, Li L, Weinfeld M. A method for the isolation of covalent DNA-protein crosslinks suitable for proteomics analysis. Anal Biochem. 2005;344:204–215. doi: 10.1016/j.ab.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 10.Zhitkovich A, Voitkun V, Kluz T, Costa M. Utilization of DNA-protein cross-links as a biomarker of chromium exposure. Environ Health Perspect. 1998;106(Suppl 4):969–974. doi: 10.1289/ehp.98106s4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker JM, Parish JH, Curtis JP. DNA-DNA and DNA-protein crosslinking and repair in Neurospora crassa following exposure to nitrogen mustard. Mutat Res. 1984;132:171–179. doi: 10.1016/0167-8817(84)90035-x. [DOI] [PubMed] [Google Scholar]
- 12.Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloster M, Kostrhunova H, Zaludova R, Malina J, Kasparkova J, Brabec V, Farrell N. Trifunctional dinuclear platinum complexes as DNA-protein cross-linking agents. Biochemistry. 2004;43:7776–7786. doi: 10.1021/bi030243e. [DOI] [PubMed] [Google Scholar]
- 14.Michaelson-Richie ED, Ming X, Codreanu SG, Loeber RL, Liebler DC, Campbell C, Tretyakova NY. Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2011;10:2785–2796. doi: 10.1021/pr200042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem Res Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J Biol Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 17.Wickramaratne S, Mukherjee S, Villalta PW, Scharer OD, Tretyakova NY. Synthesis of sequence-specific DNA-protein conjugates via a reductive amination strategy. Bioconjug Chem. 2013;24:1496–1506. doi: 10.1021/bc400018u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solivio MJ, Nemera DB, Sallans L, Merino EJ. Biologically relevant oxidants cause bound proteins to readily oxidatively cross-link at Guanine. Chem Res Toxicol. 2012;25:326–336. doi: 10.1021/tx200376e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tretyakova N, Sangaiah R, Yen TY, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N-7-guanine adducts of diepoxybutane. Chem Res Toxicol. 1997;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- 20.Szekely J, Wang H, Peplowski KM, Knutson CG, Marnett LJ, Rizzo CJ. “One-pot” syntheses of malondialdehyde adducts of nucleosides. Nucleosides Nucleotides Nucleic Acids. 2008;27:103–109. doi: 10.1080/15257770701795797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eng JK, Mccormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 22.Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. J Mass Spectrom. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- 23.Michalski A, Neuhauser N, Cox J, Mann M. A Systematic Investigation into the nature of tryptic HCD spectra. J Proteome Res. 2012;11:5479–5491. doi: 10.1021/pr3007045. [DOI] [PubMed] [Google Scholar]
- 24.Frese CK, Altelaar AF, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJ, Mohammed S. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQOrbitrap Velos. J Proteome Res. 2011;10:2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 25.Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchida K, Sakai K, Itakura K, Osawa T, Toyokuni S. Protein modification by lipid peroxidation products: formation of malondialdehyde-derived N(epsilon)-(2-propenol)lysine in proteins. Arch Biochem Biophysa. 1997;346:45–52. doi: 10.1006/abbi.1997.0266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.