Abstract
Pathogens that use multiple host species are an increasing public health issue due to their complex transmission, which makes them difficult to mitigate. Here, we explore the possibility of using networks of ecological interactions among potential host species to identify the particular disease-source species to target to break down transmission of such pathogens. We fit a mathematical model on prevalence data of Mycobacterium ulcerans in western Africa and we show that removing the most abundant taxa for this category of pathogen is not an optimal strategy to decrease the transmission of the mycobacterium within aquatic ecosystems. On the contrary, we reveal that the removal of some taxa, especially Oligochaeta worms, can clearly reduce rates of pathogen transmission and should be considered as a keystone organism for its transmission because it leads to a substantial reduction in pathogen prevalence regardless of the network topology. Besides its potential application for the understanding of M. ulcerans ecology, we discuss about how networks of species interactions can modulate transmission of multi-host pathogens.
1) Introduction
Multi-host pathogens represent an increasing concern for human public health [1]. Among the infectious diseases considered as emerging, more than 60% use at least three different host species [2]. The increasing burden of these generalist infections, i.e., that can infect a wide range of host at each stage of life cycle, is especially threatening considering their sensitivity to current environmental fluctuations [3–5], e.g., biodiversity alteration [6,7].
Consequently, mitigating transmission of these pathogens is a research priority [1]. Nevertheless, such control efforts are known to be especially difficult and complex [8] since each host species can serve as a “refuge” for the pathogen, allowing its re-emergence after the end of a specific host species control effort. During the last several decades, public health consortiums have even claimed that pathogen eradication is not possible in the presence of complex cycles in wildlife, like for sylvatic cycles in Yellow Fever [8]. Today, the only attempt, and success, of elimination of these infections has concerned disease transmitted between domestic animals [9] where it was possible to target, by vaccination and/or by culling, the host species that are the most responsible for pathogen transmission. However, such a strategy does not seem possible for wildlife diseases, especially those embedded in speciose communities or in the environment [10,11].
An interesting alternative to control these infections would be to identify “critical point(s)” in the dispersal of these pathogens, i.e., a species or a group of species that is mandatory in the transmission chain. Such critical point(s) exist for some diseases, especially those that are vector-borne where the insect life cycle requires blood meals. Identifying the critical point(s) in other disease systems offers the opportunity of control for pathogens [12]. It would be then especially useful to identify this kind of Achilles’ heel for multi-host pathogen infections involved within complex and heterogeneous networks of transmission.
The increasing recognition of ecological theory in infectious diseases literature is perfectly timed to address this problem. Indeed, parasitism is one of the possible types of ecological interaction between two species. Today, it is well demonstrated that many networks of ecological interactions contains some “critical points” [13,14], which are required species to sustain the whole connectivity within the community. Applying this concept of “keystone” species to the network of pathogen transmission would be especially relevant in an epidemiological context. A pathogen transmitted by many different host species could go through a given number of hosts that do not amplify the transmission, but are still required to connect the amplifying hosts between them.
Here, we aim to test the hypothesis that such multi-host systems can entertain one or several “keystone species” for pathogen transmission that can represent an Achilles’ heel for controlling disease. To reach this goal, we analyze the case of Mycobacterium ulcerans, a pathogen transmitted in aquatic environments through complex networks of ecological interactions [15], in 27 distinct aquatic communities in Ghana, western Africa [16]. We develop a mathematical model that we fit against the prevalence observed for 68 aquatic invertebrate taxa within each of these communities in order to estimate the networks of pathogen transmission for each of them. Then, we show that removing almost half of host taxa from our model is required to observe a significant decrease in pathogen prevalence in most aquatic communities. On the contrary, we show that removing just one host taxon, i.e. Oligochaeta worms, within and across aquatic localities, can decrease by almost 50% the prevalence predicted by our model. Thus, regardless of the network topology, removing of this taxon leads to a substantial decrease in prevalence. We conclude that this taxon can play a role of “keystone” hosts for M. ulcerans transmission. We then discuss the implications of these results for the control of such environmentally-persistent microbes and conclude that characterizing disease webs using community ecology can help to improve the knowledge of such pathogen transmission, and then contribute to mitigate their undesirable effects and stimulate future epidemiological studies.
2 Materials and Methods
2.1) Biological model
Mycobacterium ulcerans causes the disease Buruli ulcer that leads to serious skin ulcerations in humans. Despite the fact that this mycobacterium was first described 60 years ago [17], its ecology and life-cycle is still a matter of debate [15,18,19]. The path of M. ulcerans transmission to humans remains unclear and two main hypotheses have been proposed [20–23]. The first one implies that the microbe is transmitted through the aquatic environment, and M. ulcerans could infect humans who have frequent contact with contaminated water through swimming or through body injuries that may facilitate the introduction of the microbe into the skin [20,24]. The second hypothesis, which has received particular attention in recent years despite the absence of ecological evidences to support it [21], suggest that M. ulcerans could be transmitted through the bite of aquatic bugs [19,22,25]. The accumulation of many controversies in the characterization of different potential host species for M. ulcerans transmission with time [15] suggest that this mycobacterium could be transmitted among both a large host spectrum, which are embedded within networks of heterospecific interactions within aquatic environments, and a diversity of more or less heterogeneous species communities.
2.2) Data
The study sites, invertebrate field sampling, and M. ulcerans detection methods have been detailed elsewhere [26], and are briefly described hereafter. In June 2004 and August 2005, 27 waterbodies associated with reported Buruli ulcer human communities in southern Ghana were sampled (summary of this dataset is given in supplementary materials S5).
Within each waterbody, two 10 – 20 m transects were measured parallel to the shoreline and positioned through the dominant macrophyte plant community. Along each transect we randomly placed two 1m2 PVC quadrats and collected invertebrates by sweeping within the quadrat with a 500 μm mesh dipnet. Three sweeps of the dipnet were performed from the water surface to the bottom substrate for comprehensive sampling of specimens in the water column. The two quadrats were combined into a single composite sample. All contents were washed through a 500 μm sieve and preserved in 100% ethanol for laboratory identification and PCR assays.
Samples were analyzed in a two-step procedure described in detail by Williamson et al [26]. Small invertebrates were analyzed in pools of 3–15 individuals, whereas larger specimens were tested individually. DNA was extracted using a protocol adapted from Lamour and Finley [27]. Presumptive identification of M. ulcerans in invertebrates was based on PCR detection of the enoyl reduction domain (ER) in mlsA that encodes the lactone core of the mycolactone toxin, the major virulence determinant of M. ulcerans. All samples were screened for the presence of the ER gene, which has been evaluated for M. ulcerans by Williamson et al. (2008).
Hereafter, we use the terminology of host taxon to refer to host “carriage” taxon that may participate in M. ulcerans life cycle. They may, or may not, allow mycobacteria proliferation in their body, or simply house those cells onto or inside the body as a passive material without any duplication.
2.3) Epidemiological model
If the assumption of that transmission of M. ulcerans is well described by a Susceptible, Infectious and Recovered (SIR) within the aquatic community, in which individuals are classified according to their infectious status [28,29], the following multi-host model can be used to analyze its transmission among a network of different hosts:
| (1) |
| (2) |
| (3) |
Where index i represents a given taxon, and n is the number of taxa in the local community. This model follows the tradition of mathematical modeling of micro-parasites infecting different kind of hosts [28,29]. Each (host or carrier) taxon has specific birth and death rates (bi and di) that are quantified through allometric laws (see Supplementary materials and [30]). Since the presence of immune reactions against M. ulcerans in invertebrate aquatic organisms is still enigmatic [31], our model analysis explores the possibility of no immunity (γ=0) or short immunity (γ=7 days.ind−1).
2.4) Network of disease transmission
Our model assumes that pathogen is spreaded through a transmission network, caracterized by the matrix βij, which is different for each local ecological community studied. These networks consider only pathogen transmission, and thus do not involve other direct interaction between each taxa (i.e., abundance of each taxa are assumed to be independent from each other).
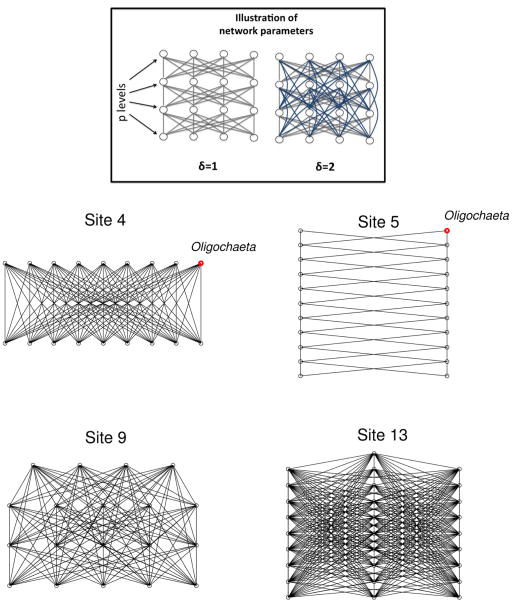
Our networks are designed through two parameters (schematized at the top of figure 1): number of different levels in the network (p) and number of inter-connected levels (δ). First, we assume that the different aquatic taxa are distributed along p different levels according to their abundance (less abundant taxa are at the top of network). The number of present levels, ρ, goes from 1 to n, allows to produce a gradient between the case where each taxon is situated at an unique level to the case where each taxon is situated at different levels. Within a given level, we assume that each taxon is connected to all other taxa of δ lower and upper levels. Then, the number of inter-connecting levels, δ, goes from 1 to ρ to yield a gradient between a situation where each taxon is connected to only the levels closest to it to the situation where all levels are connected together. Finally, for the sake of simplicity, we assume that all inter-taxa transmission rates are equal between them.
Figure 1.
Illustration of pathogen transmission networks. (Framed) Illustration of the network parameters used to construct network models (p levels and δ number of inter-connecting levels). The blue lines represent the difference between δ=1 and δ=2. (Bottom) Examples of pathogen transmission network between species estimated for four different sites selected because they represent a large variety of network topologies encountered in our data. (A) ρ = 2, δ = 2, (B) ρ = 20, δ = 1, (C) ρ = 4, δ = 4, (D) ρ = 12, δ = 11
This network generation can yield a very wide range of networks topologies wherein food web is a very specific case [32]. However, it is worth mentioning here that we consider “disease transmission web” as being completely independent of the “food web”. Indeed, predation is definitely not the only possibility of pathogen transmission between two host species [33], so mycobacterium spillover will not necessarily match with energy fluxes between species from prey to predator.
2.5) Fitting the model to empirical data
Taxa abundances are given by our dataset, allometric laws are used to estimate birth and death rates (Supplementary materials) and two cases of recovery are considered (with and without immunity). Consequently, for each locality present in our study, we have to estimate four parameters relative to the construction of transmission networks: (i) inter-taxa contact rate (βij), (ii) intra-taxon contact rate (βij), (iii) number of levels within the network (ρ), and (iv) number of inter-connected levels (δ). Then, we compare iteratively the maximal pathogen prevalence predicted for each aquatic community by our model with the prevalence observed in the field to estimate the correct network of pathogen transmission for each locality (see Supplementary materials for the estimation algorithm).
3) Results
3.1 Model fitting
Networks of pathogen transmission for each location in Ghana have been estimated through our epidemiological model (Figure 1). The resulting prevalences across aquatic taxa are strongly correlated with the observed ones, whatever the assumption about immunity (with immunity: r=0.9761, p-value<0.0001, and without: r=0.9975, p-value<0.0001). Nevertheless, considering a likelihood error structure in order to derive an AIC (see supplementary materials) does not allow rejecting any hypothesis regarding immunity or not. Consequently, both situations are considered in the present work.
3.2) The influence of removing many host taxa on pathogen transmission within each community
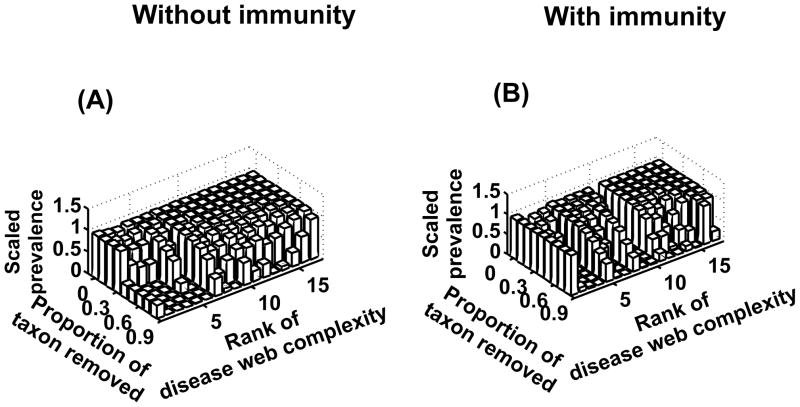
Here, we aim to understand how many host species have to be removed in order to significantly decrease pathogen prevalence within aquatic localities. We use the ratio between disease prevalence with all (host) taxa and disease prevalence without a proportion of the less abundant (host) taxa from each local community. We analyze especially how the complexity of transmission networks, characterized by the ratio between the number of levels ρ and the number of connected levels δ, acts on the number of host taxa that have to be removed.
Figure 2 shows that local aquatic communities with many transmission pathways between host taxa (i.e. that would occur with high species number in localities) are more robust when facing removal of host taxa. Indeed, a higher number of links within this category of network allows maintaining an efficient transmission despite lower community abundance due to the removal of the less abundant taxa. In other words, the higher complexity within this type of network allows mycobacterial transmission through “secondary” pathways even though an important (host) taxon was removed from the system.
Figure 2.
Stability of M. ulcerans transmission across the different aquatic communities in the network transmission models without (A) and with immunity (B). Scaled prevalence represents the decrease of pathogen prevalence within each local community after removing a given proportion of the less abundant host taxa.
3.3) The influence of removing one host taxon on pathogen transmission over all aquatic communities
Here, we wish to understand whether the loss of some specific (host) taxa could significantly decrease M. ulcerans prevalence across the different communities. To quantify this impact, we calculate, from the mathematical model detailed in the main text, the number of infectious individuals in each community with and without the focused host taxon, that we average across all communities. The resulting ratio gives the proportional decrease of prevalence due to the removal of this taxon.
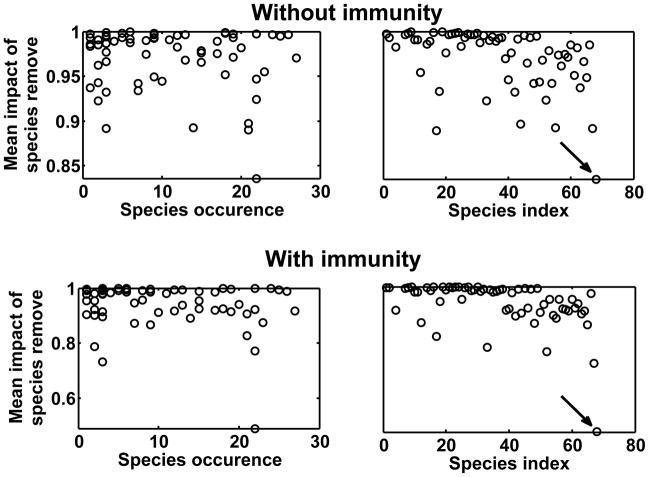
We observe that the most frequent taxa, i.e., present in many aquatic communities across the 27 different sites, are not necessarily those that have strongest influence on pathogen transmission (Figure 3A,C). This lack of relationship is somewhat surprising since removal of taxa that are very frequent can decrease prevalence within all communities while less frequent taxa do not. On the other hand, it shows that some other taxa (Figure 3B,D) are very important for M. ulcerans transmission since their removal within their host community leads to a very sharp decrease in pathogen prevalence.
Figure 3.
Relationships between the “mean impact of taxon removal”, representing proportional decrease of disease prevalence across all communities, and (A,C) occurrence of host taxon (number of times where this taxon has been observed among the 27 aquatic communities) or (B,D) their identity. Arrows indicate the Oligochaeta taxon (see main text for further details).
Figure 3 also shows that one host taxon in particular has a strong impact on pathogen prevalence across the 27 different aquatic sites, whatever we accept or not an immunity against mycobacteria in invertebrates. The removal of the Oligochaeta taxon led to a 15% decrease of disease prevalence across communities in the model without immunity, and to a 55% reduction in the one with immunity.
4) Conclusion and Discussion
In this paper, we have demonstrated that removing one “keystone” taxon, the Oligochaeta, can dramatically decrease prevalence of M. ulcerans across the different aquatic communities. Then, we have fitted a mathematical model against this dataset to estimate the networks of pathogen transmission (Figure 1). These parameterized networks have been used to show that many different host taxa have to be removed to significantly decrease the prevalence of the mycobacterium within each aquatic community (Figure 2). But, we have also shown that removing one single taxon, the Oligochaeta worms, can significantly decrease pathogen prevalence - by 15% to 55% - among all the aquatic communities (Figure 3).
Our study relies on the assumption that M. ulcerans is transmitted through a web of ecological interactions between potential host carriers in the aquatic environment. So, depending on the ecological complexity in space and time and changes in the composition of aquatic communities, transmission of M. ulcerans in nature should take different pathways. This first application of a mathematical model to the study of M. ulcerans transmission in aquatic ecosystems may be considered too simple and other routes of transmission may be important [15], especially direct acquisition from water or soil where the mycobacteria would be environmentally-persistent micro-organisms. In the supplementary materials, we show that considering this route of transmission, i.e., free mycobacterium particles in water, in our model shows significantly lower fidelity to prevalence values observed (r=0.90, p-value<0.05 without immunity, and r=0.30, p-value=0.6851 with immunity). Consequently, we consider the model with transmission within ecological webs, notably through organism-to-organism contacts, as the most likely scenario of transmission according to the field observations.
For the case of M. ulcerans, the keystone property of the worm taxon Oligochaeta makes sense from an ecological point of view. This taxon is one of the most abundant and most common taxon within and across studied localities, but some other taxa could have been intuitively better keystone taxa than the Oligochaeta, especially Baetidae or Chironomidae that are among the most abundant taxonomic groups. This finding underlines once again the high potential of network models in infectious disease ecology. Due to its major place within the network, the removal of Oligochaeta individuals led to a dramatic decrease of M. ulcerans prevalence in aquatic communities. Interestingly, this (host) taxon was the only one found to be infected in field conditions during microbiological trials in previous works [34]. More experimental studies are clearly needed, nevertheless our work suggests that this category of aquatic host organisms, and to a lesser extent some other taxa exhibiting a similar pattern, such as the Nematode roundworms, may play a central role in the transmission pathways of M. ulcerans in Africa.
Thus, ecological niche of Oligochaeta worms, and to a less extent of Nematodes, in disease webs allows these categories of host taxa to act as a major source of infection for other taxa in the web. Aquatic nematodes are generally grazers feeding upon bacterial films and free protozoans at the bottom of the water colum, and aquatic Oligochaeta worms are limivorous organisms that feed upon algae and detritus found on the mud. These feeding styles might contribute to accumulate mycobacterial cells from mud and facilitate its transmission through concentration in disease-webs.
From a multi-host pathogens point of view, we have shown that most efficient target for removal, in terms of disease control, is not always the most common host species, nor the host species with the highest prevalence of the mycobacterium (Figure 3). This finding, despite still needing to be confirmed on other aquatic ecosystems and in both epidemic and endemic sites for Buruli ulcer, is clearly encouraging because it opens new doors for mitigating epidemics of such systems. Even if controlling Oligochaeta in the case of M. ulcerans seems not to be applicable today, the environmental conditions that may favor this taxonomic group to flourish should do. In addition, some other host-disease systems like rodent-borne diseases could benefit from such an approach.
Our results have to be considered within the existing literature on pathogen transmission through networks [35]. Indeed, numerous studies have shown that pathogens are transmitted through networks at very different scales, from small communities [36] to a worldwide scale [37]. At the scale of ecological communities, a lot of work has been done regarding pathogen transmission within food webs [38]. In such a case, it has been demonstrated that some host species are critical to maintain parasite diversity and, consequently, host diversity because of the structuring role of parasites in food webs. In our study, we show that some host species are crucial for pathogen transmission, even without structuring host communities. We believe that this is an important result because it opens perspectives for disease control without, potentially, altering irreparably host diversity locally.
As shown here for the transmission of M. ulcerans, networks of species interactions could be a central component of disease transmission for many pathogens with an indirect life cycle or multiple host solutions. From an ecological perspective, disease transmission through these networks has, to our knowledge, never been demonstrated in previous studies. However, Lafferty et al. [38,39] have shown that parasite taxa are key components in the stability of salt-marsh communities as they improve the connectivity of ”food-webs“ in those ecosystems. Our study, on the other hand, demonstrated how the characteristics of the community of taxa (i.e., community organization and/or presence of keystone taxa) could affect the transmission of a pathogen within a network of species interactions. Another important aspect not studied in the two previous research works is the fluctuation in population dynamics of the different taxonomic groups. Further studies should combine these approaches to give a more global picture of the real interactions between parasites and their host taxa communities.
Supplementary Material
Acknowledgments
BR thanks Institut de Recherche pour le Développement and Université Pierre and Marie Curie. JFG thanks Institut de Recherche pour le Développement, Centre National de la Recherche Scientifique and the French School of Public Health. This work was partially supported by a grant from ANR EREMIBA 05 SEST 008-02, and has benefited from an “Investissement d’Avenir” grant managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-0025). This work was also funded by the World Health Organization and the NIH. The project described was supported by Grant Number R01TW007550 from the Fogarty International Center through the NIH/NSF Ecology of Infectious Diseases Program, and grant number R03AI062719. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Center or the National Institutes of Health. The authors thank Kevin Carolan and Andres Garchitorena for insightful discussion and the members of the WHO Buruli ulcer initiative for their constructive discussions at the 2008 annual meeting held at WHO headquarters in Geneva, Switzerland. We are grateful to Dr. E. Ampadu of the Ghana Ministry of Health for kindly providing the Buruli ulcer case data and Dr. Kingsley Asiedu or the World Health Organization for his continued support of our research. We would also like to thank Todd White and Rebecca Kolar for field assistance and invertebrate collections and identifications.
References
- 1.Woolhouse ME, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 2.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolhouse MEJ. Emerging diseases go global. Nature. 2008;451:898–899. doi: 10.1038/451898a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daszak P, Cunningham AA, Hyatta D. Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 5.Collinge SH, Ray C, editors. Disease ecology: Community structure and pathogen dynamics. Oxford University Press; 2006. [Google Scholar]
- 6.Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roche B, Guégan JF. Ecosystem dynamics, biological diversity and emerging infectious diseases. Comptes Rendus Biologie. 2011;334 :385–392. doi: 10.1016/j.crvi.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson RC. The eradication of infectious diseases. Parasitology today. 1998;14:469. doi: 10.1016/s0169-4758(98)01309-x. [DOI] [PubMed] [Google Scholar]
- 9.Keeling MJ, Woolhouse MEJ, May RM, Davies G, Grenfell BT. Modelling vaccination strategies against foot-and-mouth disease. Nature. 2003;421:136–142. doi: 10.1038/nature01343. [DOI] [PubMed] [Google Scholar]
- 10.Dobson A. Population dynamics of pathogens with multiple host species. Am Nat. 2004;164(Suppl):S64–S78. doi: 10.1086/424681. [DOI] [PubMed] [Google Scholar]
- 11.Roche B, Dobson AP, Guégan J-F, Rohani P. Linking community and disease ecology: The impact of species diversity on pathogen transmission. Phil Trans R Soc Lond B. 2012;367:1604. doi: 10.1098/rstb.2011.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chizyuka HG, Mulilo JB. Methods currently used for the control of multi-host ticks: their validity and proposals for future control strategies. Parassitologia. 1990;32:127–132. [PubMed] [Google Scholar]
- 13.Mills L, Soule MES. Keystone species concept in ecology and conservation. BioScience. 1993;43:219–225. [Google Scholar]
- 14.Jordán F. Keystone species and food webs. Philos Trans R Soc Lond B Biol Sci. 2009;364:1733–1741. doi: 10.1098/rstb.2008.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merritt RW, Walker ED, Small PLC, Wallace JR, Johnson PDR, et al. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS neglected tropical diseases. 2010;4:e911. doi: 10.1371/journal.pntd.0000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asiedu K, Etuaful S. Socioeconomic implications of Buruli ulcer in Ghana: a three-year review. Trans R Soc Trop Med & Hyg. 1998;59:1015–1022. doi: 10.4269/ajtmh.1998.59.1015. [DOI] [PubMed] [Google Scholar]
- 17.MacCallum P, Tolhurst JC, Buckle G, Sissons HA. A new mycobacterial infection in man. J Pathol Bacteriol. 1948;60:93–122. [PubMed] [Google Scholar]
- 18.Johnson PDR, Stinear T, Small PLC, Plushke G, Merritt RW, et al. Buruli Ulcer (M. ulcerans Infection): New Insights, New Hope for Disease Control. PLoS Med. 2005;2(4):e108. doi: 10.1371/journal.pmed.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsollier L, Robert R, Aubry J, Andre JS, HK, et al. Aquatic insects as a vector for Mycobacterium ulcerans. Appl Environ Microbiol. 2002;68:4623–4628. doi: 10.1128/AEM.68.9.4623-4628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duker AA, Portaels F, Hale M. Pathways of Mycobacterium ulcerans infection: A review. Environ Internat. 2006;32:567–573. doi: 10.1016/j.envint.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Benbow ME, Williamson H, Kimbirauskas R, McIntosh MD, Kolar R, et al. A largescale field study on aquatic invertebrates associated with Buruli ulcer disease: Are biting water bugs likely vectors? Emerg Inf Dis. 2008;14:1247–1254. doi: 10.3201/eid1408.071503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portaels F, Elsen P, Guimaraes-Peres A, Fonteyne PA, Meyers WM. Insects in the transmission of Mycobacterium ulcerans infection. Lancet. 1999;353:986. doi: 10.1016/S0140-6736(98)05177-0. [DOI] [PubMed] [Google Scholar]
- 23.Portaels F, Meyers WM, Ablordey A, Castro AG, Chemlal K, et al. First Cultivation and Characterization of Mycobacterium ulcerans from the Environment. PLoS Negl Trop Dis. 2008;2:e178. doi: 10.1371/journal.pntd.0000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aiga H, Amano T, Cairncross S, Adomako J, Domako JA, et al. Assessing water-related risk factors for Buruli ulcer: a case-control study in Ghana. Am J Trop Med Hyg. 2004;71:387–392. [PubMed] [Google Scholar]
- 25.Marsollier L, Aubry J, Coutanceau E, Saint André J-P, Small PL, et al. Colonization of the salivary glands of Naucoris cimicoides by Mycobacterium ulcerans requires host plasmatocytes and a macrolide toxin, mycolactone. Cellular Microbiology. 2005;7:935–943. doi: 10.1111/j.1462-5822.2005.00521.x. [DOI] [PubMed] [Google Scholar]
- 26.Williamson HR, Benbow ME, Nguyen KD, Beachboard DC, Kimbirauskas RK, et al. Distribution of Mycobacterium ulcerans in Buruli Ulcer Endemic and Non-Endemic Aquatic Sites in Ghana. PLoS Negl Trop Dis. 2008;2:e205. doi: 10.1371/journal.pntd.0000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamour K, Finley L. A strategy for recovering high quality genomic DNA from a large number of Phytophthora isolates. Mycologia. 2006;98:514–517. doi: 10.3852/mycologia.98.3.514. [DOI] [PubMed] [Google Scholar]
- 28.Anderson RM, May RM. Infectious diseases of humans: Dynamics and control. Oxford Science Publications; 1991. [Google Scholar]
- 29.Keeling MJ, Rohani P. Modeling Infectious Diseases. Princeton University Press; Princeton: 2008. [Google Scholar]
- 30.De Leo GA, Dobson AP. Allometry and simple epidemic models for microparasites. Nature. 1996;379:720–722. doi: 10.1038/379720a0. [DOI] [PubMed] [Google Scholar]
- 31.Loker ES, Adema CM, Zhang S-M, Kepler TB. Invertebrate immune systems - not homogeneous, not simple, not well understood. Immunol Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pimm SL. Food webs. University Of Chicago Press; 2002. [Google Scholar]
- 33.McCallum H, Barlow N, Hone J. How should pathogen transmission be modelled? Trends Ecol Evol. 2001;16:295–300. doi: 10.1016/s0169-5347(01)02144-9. [DOI] [PubMed] [Google Scholar]
- 34.Fyfe JAM, Lavender CJ, Johnson PDR, Globan M, Sievers A, et al. Development and Application of Two Multiplex Real-Time PCR Assays for the Detection of Mycobacterium ulcerans in Clinical and Environmental Samples. Appl Environ Microbiol. 2007;73(15):4733–4740. doi: 10.1128/AEM.02971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paull SH, Song S, Mcclure KM, Sackett LC, Kilpatrick AM, et al. From superspreaders to disease hotspots3: linking transmission across hosts and space. Front Ecol Environ. 2012;10:75–82. doi: 10.1890/110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrari MJ, Bansal S, Meyers LA, Bjornstad ON, Bjørnstad ON. Network frailty and the geometry of herd immunity. Proc Roy SOC Lond B. 2006;273:2743–2748. doi: 10.1098/rspb.2006.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colizza V, Barrat A, Barthelemy M, Valleron A-JA-J, Vespignani A. Modeling the worldwide spread of pandemic influenza: Baseline case and containment interventions. PLoS Med. 2007;4:e13. doi: 10.1371/journal.pmed.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lafferty KD, Dobson AP, Kuris AM. Parasites dominate food web links. Proc Natl Acad Sci U S A. 2006;103:11211–11216. doi: 10.1073/pnas.0604755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafferty KD, Allesina S, Arim M, Briggs CJ, De Leo G, et al. Parasites in food webs: the ultimate missing links. Ecol Lett. 2008;11:533–546. doi: 10.1111/j.1461-0248.2008.01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.