Abstract
Proteins secreted by Helicobacter pylori (H. pylori), an important human pathogen responsible for severe gastric diseases, are reviewed from the point of view of their biochemical characterization, both functional and structural. Despite the vast amount of experimental data available on the proteins secreted by this bacterium, the precise size of the secretome remains unknown. In this review, we consider as secreted both proteins that contain a secretion signal for the periplasm and proteins that have been detected in the external medium in in vitro experiments. In this way, H. pylori’s secretome appears to be composed of slightly more than 160 proteins, but this number must be considered very cautiously, not only because the definition of secretome itself is ambiguous but also because the included proteins were observed as secreted in in vitro experiments that were not representative of the environmental situation in vivo. The proteins that appear to be secreted can be grouped into different classes: enzymes (48 proteins), outer membrane proteins (43), components of flagella (11), members of the cytotoxic-associated genes pathogenicity island or other toxins (8 and 5, respectively), binding and transport proteins (9), and others (11). A final group, which includes 28 members, is represented by hypothetical uncharacterized proteins. Despite the large amount of data accumulated on the H. pylori secretome, a considerable amount of work remains to reach a full comprehension of the system at the molecular level.
Keywords: Helicobacter pylori, Secreted proteins, Periplasmic space, Secretion signal, Secretome
Core tip: This paper summarizes what is known, from the molecular point of view, about the proteins that are secreted by the bacterium and that are generally grouped under the name “secretome”. These proteins play a very relevant role in pathogenesis, as secreted proteins or those that are present on the external surface of the bacterium are responsible of all the interactions with the host.
INTRODUCTION
Helicobacter pylori (H. pylori)[1], the bacterium that affects approximately half of the human population and is responsible for severe diseases, from peptic ulcers to gastric cancer[2-4], can be considered a paradigm for the study of host-pathogen interactions. In fact, as the only bacterium that permanently inhabits the human stomach, its effects on the host can be easily dissected; the bacterium has adapted itself to living in a peculiar and unique environment. For example, H. pylori has reduced its metabolic machinery to a minimum, taking advantage of all of the nutrients available in the stomach. Consequently, H. pylori possesses a relatively small genome of less than 1600 genes[5,6] compared to other Gram-negative prokaryotes, such as Escherichia coli, whose genome includes more than 4000 genes[7]. Despite its small size, a relevant fraction of its proteins, possibly from 30% to 40%, are annotated as “hypothetical proteins”. The function of some of the latter can be hypothesized based on a weak homology with proteins of other bacteria, while that of others is completely unknown.
Among H. pylori proteins, those that are secreted are particularly relevant in the context of interactions with the host. The bacterium uses a set of secreted and translocated proteins (outer membrane adhesins, secreted enzymes, effector proteins, etc.) to adapt itself to the mucosal environment[8]. In addition, most secreted proteins induce some effect on the host immune system. For example, while the H. pylori-derived inflammatory reaction is triggered mainly by the binding of bacterial cells to surface class II major histocompatibility complex molecules, the bacterium is able to evade the host immune response mainly by taking advantage of dendritic cells; its effects on the host immune system are largely due to the secretion of an unidentified heat-stable factor from the bacterium that specifically inhibits interleukin (IL)-12 release from dendritic cells[9]. In addition, the proliferation of lymphocytes was abolished in vitro by a secreted protein with an apparent molecular weight of between 30 and 60 kDa, distinct from that of vacuolating toxin (VacA)[10]. Most of the possible effects on the host of secreted or released proteins have yet to be discovered. Furthermore, secreted factors represent eligible proteins for studies aimed at identifying the most promising targets for the development of new antimicrobial drugs.
In this review, we will attempt to summarize the state of the H. pylori secretome from a molecular point of view, specifically what is known about the structure and function of the secreted proteins.
H. PYLORI SECRETOME
The word “secretome” is intrinsically ambiguous. In fact, the expression “secreted proteins” strictly refers to polypeptides that are transported outside the outer cell membrane through some secretion mechanism. This definition would exclude, for example, integral membrane proteins present in the outer membrane and, as such, potentially in contact with the host cells. On the other hand, there are H. pylori proteins that are highly immunogenic, but they do not possess, at least apparently, a secretion signal; this is the case, for example, of the neutrophil-activating protein (HP-NAP). HP-NAP is a large multimeric protein consisting of 12 identical subunits arranged with 32 symmetry[11] and belonging to the class of Dps or mini-ferritins[12]. The most relevant activity of HP-NAP, even if perhaps secondary for the bacterium itself, is its ability to induce neutrophils to adhere to endothelial cells[13] and its role in immunity, promoting the T helper 1 immune response[14,15]. Once present in the outer space, HP-NAP mediates the interaction of the bacterium with the external surface of the outer membrane of the host cell[16]. HP-NAP is associated with the outer membrane fraction in patients affected by duodenal ulcers or gastric cancer[17], but this presence could be due to the release of the protein, normally present in the cytoplasm, upon autolysis. A similar situation could apply for urease[17,18], the large protein complex responsible for the hydrolysis of urea and the production of ammonia that allows for the survival of H. pylori in the acidic environment of the stomach[19]. These proteins, which have been the subject of several reviews, will not be discussed here. We will mostly concentrate on proteins that bear a secretion signal and that are consequently secreted from the bacterium, either to the periplasm or to the external space to perform a specific function. In addition, proteins that have been experimentally detected as present in the outer space will be discussed in relation to their function.
Five major studies have attempted to analyze H. pylori’s secretome as a whole. Bumann et al[20] have analyzed, using two-dimensional gel electrophoresis followed by mass spectrometry fingerprinting, the extracellular proteins present in a culture of H. pylori. More than thirty secreted proteins were detected, and 26 were chemically identified. A similar proteomic approach, applied by Kim et al[21], identified eighteen secreted proteins. Notably, five proteins were identified as secreted in both studies. Two other proteomic studies were performed using isolates from asymptomatic patients or from those affected by duodenal ulcers or gastric cancer[17] or by two different H. pylori strains grown in a defined serum-free medium[22]. More than 60 unique proteins were identified in the first study, and more than 165 were identified in the second study. Most of the identified targets are common between these four studies, but others are not; in some cases, the identified targets are unique. It is evident that some drawbacks are present in these approaches: (1) because proteins present in the medium are detected, it is difficult to distinguish between molecules that have actually been secreted from those present due to bacterial lysis; (2) protein secretion can depend upon very different environmental conditions, and targets can be missed simply because the conditions for the secretion of a specific target were not fulfilled; (3) in in vitro experiments, the type of cells used and the growth conditions can drastically change the secretion profile (in vivo, the tissue inflammation of the host can also influence the expression of the proteins of the bacterium); and finally, (4) some proteins are strain-specific, and in this review, we have neglected proteins that are not present in the genome of strain 26695.
Very recently, Müller et al[23] used a proteogenomics approach to identify N-terminal export signal peptides. Although these signal peptides identify not only proteins that are secreted in the medium but also those directed to the inner or outer membrane or to the periplasmic space, this approach does not suffer from the aforementioned problem. A total of 72 polypeptide chains bearing an export signal peptide were identified.
In addition to the previous approaches, several papers have reported single secreted H. pylori proteins. These data are listed in Table 1, where the proteins have been tentatively grouped according to their function or to some other rational criteria. In Table 2, we report the proteins involved in the biogenesis of the outer membrane. Some of these proteins are also secreted, and they should also be included in Table 1; however, due to their specialized function, we prefer to group them together. Proteins involved in outer membrane biogenesis represent a complex system, which deserves a separate study and will not be covered further in this review.
Table 1.
Proteins that have been experimentally identified as secreted in one of the reported papers or predicted based on a secretion signal
| Label | Name and/or function | Ref. | PDB code | UniProtKB |
| Outer membrane proteins (Omps) | ||||
| HP0009 | Outer membrane protein (Omp1), HopZ | y | Q7X2J7 | |
| HP0025 | Outer membrane protein (Omp2) | b’, y | O24870 | |
| HP0078/79 | Outer membrane protein (Omp3) | y | O24907/O24908 | |
| HP0127 | Outer membrane protein (omp4), HorB | b’, y, z | O24941 | |
| HP0227 | Outer membrane protein (Omp5) | b’, y | O34523 | |
| HP0229 | Outer membrane protein (Omp6) | b’, y, z | O25015 | |
| HP0232 | UPF0323 lipoprotein HP_0232 | b | O25018 | |
| HP0252 | Outer membrane protein (Omp7), HopF | b’, y | O25034 | |
| HP0253 | Outer membrane protein (Omp8) | y | O25035 | |
| HP0317 | Outer membrane protein (Omp9) | b’, y | O25086 | |
| HP0324 | Outer membrane protein (Omp10) | y | O25091 | |
| HP0410 | Putative neuraminyllactose-binding hemagglutinin | z | O25166 | |
| HP0472 | Outer membrane protein (Omp11) | b’, y, z | O25218 | |
| HP0477 | Outer membrane protein (Omp12) | y | O25222 | |
| HP0486 | Putative outer membrane protein, HofC | b’, y | O25230 | |
| HP0487 | Putative outer membrane protein, HofD | b’ | O25231 | |
| HP0638 | Outer membrane protein (Omp13) | y | O25355 | |
| HP0671 | Outer membrane protein (Omp14) | b’, y | O25382 | |
| HP0694 | Putative outer membrane protein | b’, y | O25401 | |
| HP0706 | Outer membrane protein (Omp15) | b’, z | O25410 | |
| HP0710 | Outer membrane protein (HomA) | z | O25414 | |
| HP0722 | Outer membrane protein (Omp16) | y | ||
| HP0725 | Outer membrane protein (Omp17) | y | Q7X2J9 | |
| HP0797 | Neuraminyllactose-binding hemagglutinin, HpaA | b, y | 3BGH | P55969 |
| HP0896 | Outer membrane protein (Omp19). BabB | b’, y | O25556 | |
| HP0912 | Outer membrane protein (omp20), AlpA | q, u, b’, y, z | O25570 | |
| HP0913 | Outer membrane protein (omp21) | q, u, b’, y | O25571 | |
| HP0923 | Outer membrane protein (Omp22) | y | O25580 | |
| HP1107 | Outer membrane protein (Omp23) | b’, y | O25735 | |
| HP1125 | Outer membrane protein (Omp18), peptidoglycan-associated lipoprotein, PAL | s, y | O25625 | |
| HP1156 | Outer membrane protein (Omp25) | y | O25771 | |
| HP1157 | Outer membrane protein (Omp26) | y | O25772 | |
| HP1177 | Outer membrane protein (Omp27), HopO | b’, y | O25791 | |
| HP1243 | Outer membrane protein (Omp28) BabA | b’, y | O25840 | |
| HP1342 | Outer membrane protein (Omp29) | b’, y | O34523 | |
| HP1395 | Outer membrane protein (Omp30) | y, z | O25945 | |
| HP1456 | Lpp20 lipoprotein | b, y, z | P0A0V0 | |
| HP1469 | Outer membrane protein (Omp31) | b’, y | O26005 | |
| HP1501 | Outer membrane protein, HopW | y, z | O26031 | |
| HP1512 | Iron regulated outer membrane protein | y | O26042 | |
| HP1564 | Outer membrane protein, lipoprotein | u, y | O26084 | |
| HP1571 | RlpA-like lipoprotein | b, y | O26091 | |
| HP0284 | Mechanosensitive ion channel membrane protein | b’ | O25059 | |
| Cag proteins | ||||
| HP0528 | CagX, Cag8 | b’ | O25263 | |
| HP0529 | CagW, Cag9 | b’ | O25264 | |
| HP0532 | CagT, Cag12 | b | P97245 | |
| HP0537 | CagM, Cag16 | b’ | O25270 | |
| HP0540 | CagI, Cag19 | b’ | O25273 | |
| HP0545 | CagD | z | 3CWX, 3CWY | O25277 |
| HP0546 | Cag25 | b’ | O25278 | |
| HP0547 | CagA, Cag26 | b’, y | 4G0H, 4DVY, 4DVZ, 3IEC | P55980 |
| Flagellar components | ||||
| HP0115 | Flagellin B, FlaB | y | Q07911 | |
| HP0126 | Flagellar P-ring protein, FlgL | b | O25028 | |
| HP0295 | Flagellar hook-associated protein 3, HAP3 | y | O25068 | |
| HP0325 | Flagellar L-ring protein | b | O25092 | |
| HP0601 | Flagellin A, FlaA | v, y | P0A0S1 | |
| HP0752 | Flagellar hook-associated protein 2, HAP2 | z | P96786 | |
| HP0870 | Flagellar hook protein FlgE | a, y | P50610 | |
| HP0907 | Hook flagella assembly protein FlgD | a, q | O25565 | |
| HP1119 | Flagellar hook-associated protein 1, HAP1 | y | O25744 | |
| HP1462 | Secreted protein involved in flagellar motility | y | O25998 | |
| HP1557 | Flagellar hook-basal body protein FliE | a | P67708 | |
| Binding or transport proteins | ||||
| HP0243 | Neutophil-activating protein, HP-NAP | y, z | 1JI4, 4EVB, 4EVC, 4EVD, 4EVE, 3T9J, 3TA8 | P43313 |
| HP0298 | Periplasmic dipeptide-binding protein, DppA | z | O25069 | |
| HP0508 | Plasminogen-binding protein pgbA | b’ | O25249 | |
| HP0888/ | Iron (III) dicitrate ABC transporter, ATP-binding protein | v | O05732 | |
| HP0889 | ||||
| HP0970 | Nickel-cobalt-cadmium import protein (NccB) | b’ | O25623 | |
| HP1073 | Copper-binding protein, CopP | z | 1YG0 | Q48271 |
| HP1252 | Oligopeptide ABC transporter periplasmic oligopeptide-binding protein (OppA) | b’ | O25845 | |
| HP1286 | YceI-like acidic stress response, lipocalin | a, e, l, u, b’, z | 3HPE | O25873 |
| HP1561/ | ABC transporter periplasmic binding protein (CeuE) | b’ | O26083/ | |
| HP1562 | O26082 | |||
| VacA and other toxins | ||||
| HP0289 | VacA-like protein, ImaA | d | O25063 | |
| HP0609/HP0610 | VacA-like protein, FaaA | d | O25330/ | |
| O25531 | ||||
| HP0887 | Vacuolating cytotoxin VacA | a, b, g, u, z | 2QV3 | P55981 |
| HP0922 | VacA-like protein, VlpC | d | O25579 | |
| HP0596 | Tumor necrosis factor α-inducing protein, Tip-α | f, m, n | 3VNC, 3GUQ, 3GIO, 2WCQ, 2WCR | O25318 |
| Enzymes | ||||
| Redox systems and the electron transport chain | ||||
| HP0224 | Peptide methionine sulfoxide reductase MsrA/MsrB | b’ | O25011 | |
| HP0231 | Disulphide interchange protein (DsbG, or DscB or HpDsbA) | a, q, u | 3TDG | O25017 |
| HP0330 | Ketol-acid reductoisomerase, IlvC | z | O25097 | |
| HP0377 | Thiol-disulfide interchange protein DsbC | a, b’ | 4FYB, 4FYC | O25140 |
| HP0389 | Superoxide dismutase, SOD | x | 3CEI | P43312 |
| HP0390 | Adhesin-thiol peroxidase, TagD | y | O25151 | |
| HP0485 | Catalase-like protein | b’ | O25229 | |
| HP0824 | Thioredoxin TrxA | a, w | P66928 | |
| HP0825 | Thioredoxin reductase, trxB | b’ | 3ISH, 2Q0K, 2Q0L | P56431 |
| HP0875 | Catalase, katA | x, b’,z | 2IQF, 2A9E, 1QWL, 1QWM | P77872 |
| HP1136 | ATP synthase subunit b | b’ | P56086 | |
| HP1161 | Flavodoxin FldA | a | 2W5U, 2BMV, 1FUE | O25776 |
| HP1212 | ATP synthase subunit c | b’ | P56087 | |
| HP1227 | Cytochrome c-553 | b | O25825 | |
| HP1266 | NADH-ubiquinone oxidoreductase, NQO3 subunit | z | O25856 | |
| HP1458 | Thioredoxin TrxC | a, u, z | O25996 | |
| HP1461 | Cytochrome c551 peroxidase | b’ | O25997 | |
| Putative solenoid proteins | ||||
| HP0160 | Beta-lactamase HcpD | b, o, p | O24968 | |
| HP0211 | Beta-lactamase HcpA, Cysteine-rich 28 kDa protein | b, o, p, b’ | O25001 | |
| HP0235 | Beta-lactamase HcpE | b, o, p, b’ | O25021 | |
| HP0336 | Beta-lactamase HcpB | o | 1KLX | O25103 |
| HP1098 | Beta-lactamase, Cysteine-rich protein C HcpC | a, b, o, q, t, b’, z | 1OUV | O25728 |
| HP0519 | Hypothetical Sel1-like protein | p | Q7X5D6 | |
| HP0628 | Hypothetical Sel1-like protein | p | O25345 | |
| HP1117 | Hypothetical Sel1-like protein, Cys-rich protein X | p | O25742 | |
| HP1124 | Tetratricopeptide-like repeat protein | b’ | O25749 | |
| Proteases | ||||
| HP0570 | Aminopeptidase, PepA | z | O25294 | |
| HP0657 | Processing protease (YmxG) | b’, z | O25371 | |
| HP1012 | Putative Zinc protease | z | O25656 | |
| HP1018/HP1019 | Serine protease (HtrA) | a, f, q, r, z | O25663 | |
| HP1037 | Amino peptidase | b’ | O25681 | |
| HP1350 | Carboxyl-terminal protease | z | O25905 | |
| Other enzymes | ||||
| HP0026 | Citrate synthase | z | P56062 | |
| HP0072 | Urease B (UreB) | a, q, v, y | 1E9Y, 1E9Z | P69996 |
| HP0073 | Urease A (UreA) | y | 1E9Y, 1E9Z | P14916 |
| HP0154 | Enolase, phosphopyruvate hydratase, eno | z | P48285 | |
| HP0194 | Triosephosphate isomerase, tpiA | b’ | 2JGQ | P56076 |
| HP0275 | ATP-dependent nuclease (addB) | b’, z | O25025 | |
| HP0294 | Aliphatic amidase, AimE | z | O25067 | |
| HP0310 | Polysaccharide deacetylase | z | 3QBU, 4LY4 | O25080 |
| HP0323 | Membrane-bound endonuclease (nuc) | b’, y | O25090 | |
| HP0380 | Glutamate dehydrogenase | z | P55990 | |
| HP0392 | Histidine kinase, CheA | b’ | O25153 | |
| HP0672 | Member of the PLP-dependent aminotransferase superfamily clan, AspB | z | 3EZS | O25383 |
| HP1118 | γ-Glutamyltranspetptidase | a, q, b’, z | 3FNM, 2QM6, 2QMC, 2NQO | O25743 |
| HP1178 | Purine nucleoside phosphorylase DeoD-type | b’ | P56463 | |
| HP1186 | Carbonic anhydrase | a, q, z | O25798 | |
| HP1375 | UDP-N-acetylglucosamine acyltransferase, LpxA | z | 1J2Z | O25927 |
| Others | ||||
| HP0010 | GroEL, HSP60 | c | P42383 | |
| HP0011 | GroES | z | P0A0R3 | |
| HP0166 | Response regulator, ArsS | z | O24973 | |
| HP0305 | Putative human regulator of G protein signaling 12 | u, b | O25076 | |
| HP0743 | Rod shape-determining protein | y | P56098 | |
| HP1000 | PARA protein | b’ | O25646 | |
| HP1126 | Protein TolB | b | O25751 | |
| HP0827 | ssDNA-binding 12RNP2 precursor | u | 2KI2 | O25501 |
| HP0835 | Histone-like DNA-binding protein HU | u | O25506 | |
| HP1v1 | Ribosomal protein L1 | u | P56029 | |
| HP1202 | Ribosomal protein L11 | u | P66052 | |
| Hypothetical proteins | ||||
| HP0122 | Hypothetical uncharacterized protein | b | P64653, O24940 | |
| HP0129 | Hypothetical uncharacterized protein | u | O24943 | |
| HP0130 | Hypothetical uncharacterized protein | b’ | O24944 | |
| HP0135 | Hypothetical uncharacterized protein | b | P64655, O24948 | |
| HP0149 | Hypothetical uncharacterized protein | b’ | O24960 | |
| HP0169 | Hypothetical secreted colagenase | b | P56113 | |
| HP0204 | Hypothetical uncharacterized protein | b’ | O24996 | |
| HP0367 | Hypothetical uncharacterized protein | a | O25131 | |
| HP0555 | Hypothetical uncharacterized protein | b’ | O25382 | |
| HP0583 | Hypothetical uncharacterized protein | b’ | O25305 | |
| HP0659 | SurA N-terminal domain protein, SurA chaperone of outer membrane proteins | b’ | O25373 | |
| HP0719 | Hypothetical uncharacterized protein | b’ | O25421 | |
| HP0720 | Hypothetical uncharacterized protein | u | K4NCD4 | |
| HP0721 | Sialic acid-specific adhesion, metal homeostasis | i, u | 2XRH | O25423 |
| HP0781 | Hypothetical uncharacterized protein | b’ | O25470 | |
| HP0783 | Hypothetical uncharacterized protein | b’ | O25742 | |
| HP0902 | Hypothetical uncharacterized protein | u | O25562 | |
| HP0953 | Hypothetical uncharacterized protein | b’, z | O25607 | |
| HP0973 | Hypothetical uncharacterized protein | u, b’ | O26525 | |
| HP1023 | Hypothetical uncharacterized protein | b’ | O25270 | |
| HP1055 | Hypothetical uncharacterized protein | b’ | O25695 | |
| HP1056 | Hypothetical uncharacterized protein | b’ | O25696 | |
| HP1057 | Hypothetical uncharacterized protein | b’ | O25697 | |
| HP1173 | Hypothetical uncharacterized protein | a, b, z | O25787 | |
| HP1285 | lipoprotein, e(P4) family | b’, z | O25872 | |
| HP1454 | Hypothetical uncharacterized protein | a, z | O25993 | |
| HP1527 | Hypothetical uncharacterized protein | b’ | O26055 | |
| HP1580 | Hypothetical uncharacterized protein | b’ | O26100 | |
The first column refers to the symbol for strain ACT 700392/26695, the third to the reference, the forth to the PDB ID code if the X-ray structure has been determined, and the fifth to the UniProtKB code. The proteins are grouped according to their function, as explained in the text. a[20], b’[23]; (b refers to proteins having a signal peptide as reported in Supplementary Table mmc4 of reference[23]), c[142], d[84], e[72], f[151], g[152], h[153], i[148], l[74], m[154], n[88], o[112], p[116], q[123], r[119], s[155], t[156], u[21], v[75], w[91], x[102], y[17], z[22].
Table 2.
Outer membrane biogenesis complex components[153]
| Label | Protein symbol | Name and/or function | Ref. |
| HP0785 | lolA | Periplasmic chaperone | b', b |
| HP0787 | lolC (lolE) | Integral membrane protein | |
| HP1568 | lptA | Periplasmic chaperone | |
| HP0715 | lptB | Inner membrane (IM) ATP-binding-cassette transporter domain | |
| HP1569 | lptC | IM associated lipoprotein | |
| HP1216 | lptD | Outer membranes (OM) lipopolysaccharide (LPS) transport protein | |
| HP1546 | lptE | OM-associated lipoprotein | |
| HP0362 | lptF | Integral membrane protein | |
| HP1498 | lptG | Integral membrane protein | |
| HP1082 | msbA | IM LPS flippase | |
| HP0655 | bamA | OM β-barrel assembly component | b', b |
| HP1378 | bamD | OM-associated lipoprotein | b' |
| HP0786 | secA | Preprotein translocase subunit | |
| HP1300 | secY | Preprotein translocase subunit | |
| HP1203 | secE | Preprotein translocase subunit | |
| HP1255 | secG | Preprotein translocase subunit | |
| HP1550 | secD | Preprotein translocase subunit | |
| HP1549 | secF | Preprotein translocase subunit | |
| HP1551 | yajC | Preprotein translocase subunit | |
| HP1540 | yidC | IM protein translocase component | |
| HP1152 | ffh | Signal recognition particle (SRP) | |
| HP0763 | tfsY | SRP receptor | |
| HP0320 | tatA | Sec-independent translocase | |
| HP1060 | tatB | Sec-independent translocase | |
| HP1061 | tatC | Sec-independent translocase | |
| HP0175 | surA | Peptidyl-prolyl cis-trans isomerase | a, b', q, u, z |
| HP1019 | degP | Serine protease |
OUTER MEMBRANE PROTEINS
The marked H. pylori genetic variability, both in terms of micro- and genome-wide macro-diversity[24], represents a great advantage for gastric environment adaptation and infection transmission. A paradigm of such heterogeneity is represented by proteins that belong to the outer membrane subgroup (Omps). These proteins localize on the bacterial surface and are directly involved in the bacterial-host interaction. Indeed, high variability rates in the expression profile of a representative subset of Omps have been observed by comparing different clinical isolates, demonstrating their importance in H. pylori adaptation to its host[25]. Moreover, in a recent study of H. pylori genomic evolution during human infection by mixed strains, the Omps family was affected by a very high frequency of import and recombination events, suggesting a positive and diversifying selection acting on this protein’s subgroup[26].
More than 60 genes coding for Omps have been predicted by sequencing and comparative genomic approaches[5,27,28], possibly grouped in five paralogous gene families. The largest family (33 members) is composed of the Hop (Helicobacter outer membrane porins) and Hor (Hop-related) proteins, 30 of which have been identified by mass-spectrometry based studies and other techniques, as summarized in this paper (Table 1). Within Hop and Hor, most of the proteins are involved in bacterial adhesion to the stomach epithelium or form channels that allow for the passive transport of nutrients and small hydrophobic molecules. The structures of the members of this family have not yet been determined. However, a topology study concerning the HopE porin, one of the smallest H. pylori Omps, support the hypothesis that, despite the low sequence identity between these family members, a peculiar C-terminus with alternating hydrophobic and hydrophilic residues and a core of approximately 100 residues that are conserved in all of the Hop and Hor proteins define a common scaffold of amphipathic β-strands forming a β-barrel[27].
While the majority of the bacteria reside in the mucous layer, a minor fraction reaches the epithelium, adheres to the surface, guarantees higher colonization density and efficiency, interacts with gastric cells, contributes to immune response evasion and delivers virulence determinants such as CagA toxin and peptidoglycan. Within the Hop/Hor family, BabA (HP1243, Omp28) and SabA (HP0725, Omp17) cover a fundamental role by binding sialylated carbohydrate structures that are enriched on gastric epithelial cell surfaces and substitute the naturally occurring Lewis antigens as a consequence of the strong and chronic inflammation of such tissue elicited by H. pylori[29].
A key role in adhesion has also been identified for HopZ (HP0009, Omp1), HorB (HP0127, Omp4), OipA (Hp0638, Omp13) and finally AlpA and AlpB (HP0912/Omp20 and HP0913/Omp21), which bind laminin immobilized in the host extracellular matrix[30,31].
Hof (H. pylori outer membrane protein family) is another Omps subfamily composed of 8 members, two of which (HofC/Hp0486 and HofD/Hp0487) are exported[23] via an N-terminal signal peptide. Analogously, HomA (HP0710) is the only member of the Hom protein subgroup that is enriched in the extracellular milieu[22], and HP1512 is the only one of the iron-regulated Omps - elsewhere labeled as FecA-/FrpB-like members - that has been identified in the secreted proteome.
HpaA (H. pylori adhesin A), a surface-localized, highly conserved and quite unique protein (HP0797), has been initially proposed to play a role in bacterial adhesion as a sialic acid binding protein and, for this reason, has been identified as a promising candidate for vaccine development. Nevertheless, contradicting results have accumulated in the literature concerning the localization and function of HpaA. HpaA was later demonstrated to be a lipoprotein that plays an essential role in stomach colonization in an in vivo mouse model, whereas no significant differences were observed in the HpaA isogenic mutant compared to the wild-type strain under laboratory conditions[17]. HP0492 and HP0410 are putative paralogous proteins with a high degree of similarity to HpaA, the latter being identified in the secretome by at least one of the studies considered here (Table 1). Interestingly, the crystal structures of both HP0410 and HP0492 have been determined (PDB codes: 3bgh and 2i9i, New York SGX Research Center for Structural Genomics). HP0410, HP0492 and HP0797 share a marked degree of sequence homology, greater than 45%, and the crystal structures of the first two are characterized by a root mean square deviation between equivalent Cα atoms of 2.4 Å. Both HP0410 and HP0492 monomers show an elongated α/β fold and assemble into dimers in a two-layer sandwich arrangement: each monomer contributes a twisted β sheet composed of four extended β strands facing each other and defining the protein core (interface area > 930 Å2), with each monomer being surrounded by four α-helices and a long loop and packed on one face of the β-sheet at the opposite sides of the dimeric assembly (Figure 1).
Figure 1.
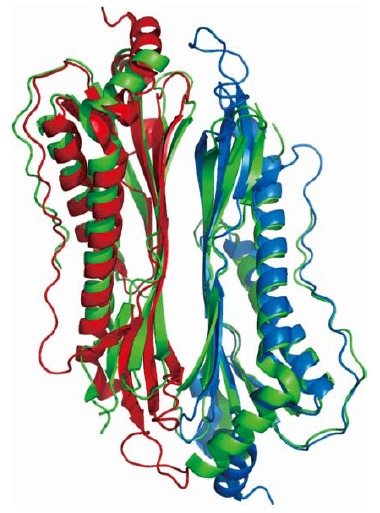
Cartoon view of the superimposed Cα traces of HpaA paralogs. HP0410 dimer (3bgh, green) and HP0492 dimer (2i9i, red and light blue). Cα root mean square deviation between equivalent atoms corresponds to 2.4 Å.
Finally, our classification includes some lipoproteins from the Omps subgroup, including a homolog of the peptidoglycan-associated lipoprotein PAL (HP1125), an RlpA-like lipoprotein (HP1571), and other lipoproteins, such as HP0232 and HP1564. The HpaA protein and its paralogs should also be included in this subgroup. For more comprehensive reviews[28,32].
CAG-PAI PROTEINS
Among the secreted proteins, it is difficult to underestimate the role of the cytotoxic-associated genes pathogenicity island (cag-PAI) in H. pylori virulence[33-35]. The cag-PAI is a genomic insert of 27-28 genes that are present in all type I strains of H. pylori and are absent in type II strains[36-38]. The cag-PAI encodes an effector protein, CagA [Proteins coded by cag-PAI are labeled by the suffix Cag followed either by a Latin or Greek letter, such as CagA, CagB … CagZ, Cagα, etc., or by a number from 1 to 28. Proteins of the type IV secretion system (T4SS) of Agrobacterium tumefaciens, the best characterized among them, are labeled VirB1, VirB2, etc. The correspondence between Cag proteins belonging to type IV secretion system and VirB proteins is, in most cases, well defined] for a T4SS and for other accessory proteins (for a recent review, see[39,40]). Intensive studies have been carried out in the last decades regarding the role of cag-PAI proteins, specifically the effector protein CagA and T4SS components, and their effects on host cells. The latter is a needle-like structure (also called pilus) that crosses the inner and outer membrane of the bacterium and protrudes from the bacterial surface. In general, T4SS systems are used by Gram-negative bacteria to exchange material (proteins or protein-DNA complexes) to and from other organisms[41]. In the case of H. pylori, its major function is the translocation of the CagA effector, and possibly peptidoglycan, into the host cell. In addition, cag-PAI T4SS induces the production of IL-8 from the host, an event independent of CagA translocation[42,43]. CagA is a protein that varies in size from 125 to 145 kDa and that does not bear any significant sequence similarity with other proteins. Once injected into the host cell, CagA is tyrosine-phosphorylated by endogenous kinases at several EPIYA motifs, triggering the binding of CagA to multiple signaling proteins of the host cell. The ability of CagA to interfere with several signaling cascades (including the induction of membrane dynamics, actin-cytoskeletal rearrangements, the disruption of cell-cell junctions, and the proliferative and pro-inflammatory responses) supports the theory of the “master key”, i.e., that CagA contains several interaction domains, each able to bind to different SH2 domains and, in doing so, hijacking different pathways[44]. Very recently, the crystal structures of two large N-terminal portions (residues 1-876 and 1-884) of CagA were obtained[45,46]. This protein comprises three domains and, despite the absence in the crystal structure of some flexible segments, its structure provides a structural basis for understanding the interaction with the targets (Figure 2).
Figure 2.
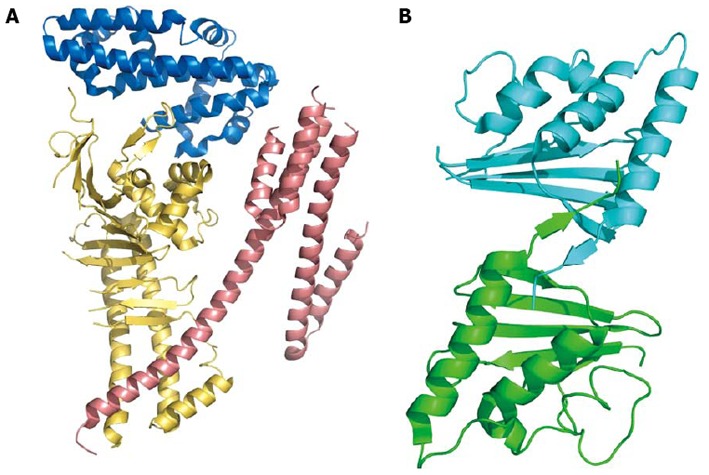
Cartoon model of Cag proteins. A: The N-terminal portion of cytotoxic-associated genes, CagA, the effector protein injected into the host cell through type IV secretion system. The three domains (residues 24-221, blue; 303-644, yellow; 645-824, salmon; coordinates from PDB 4DVY) are shown in different colors; B: CagD dimer. The two monomers are linked together by a disulfide bridge between the two C-terminal β-strands. Coordinates from PDB 3CWX.
The cag-PAI includes some structural components and other accessory proteins necessary for its assembly, totaling approximately fifteen polypeptides[39,40]. We currently lack a three-dimensional structure of the T4SS core complex of H. pylori, but the structures either from X-ray diffraction[47] or electron microscopy[35] of the core portion of the T4SS from the plasmid pKM101 that crosses the inner and outer membranes can be used to figure out the architecture of H. pylori T4SS. In addition, several studies on the localization, protein topology predictions and determinations, localization and functional studies of the cag-PAI VirB homologs have suggested an overall model of the T4SS[48-51]. In particular, there is a general consensus that the external pilus is made by a large number of copies of CagC (VirB2), plus some CagL (VirB5), while the trans-membrane core complex is formed by CagY, CagT and CagX (VirB7, VirB9 and VirB10, respectively)[40,52]. The composition of the cytoplasmic side of the T4SS is less clear, as several components with various functions are included: three ATPases (Cagα/VirB11, Cagβ/VirD4 and CagE/VirB3/4), CgaV/VirB8 and CagW/VirB6. The exact role and localization of the other components (CagM, Cagδ, CagN, CagU, CagH) is less clear, but still they appear to be essential components of the T4SS[40]. Two hypotheses have been proposed regarding the activation of pilus formation and its binding to the host cell receptor: that the binding to integrin β1 is mediated by CagL[53] or by CagA, CagI and CagY[54].
The number of genes included in the cag-PAI is greater than that required for the assembly and operation of T4SS. The role of the other genes is sometimes defined (for example, Cagγ is the peptidoglycan hydrolase that allows for the insertion of the system in the periplasm), but for others it is not. For example, CagF is essential for CagA secretion[55], similar to the complex Cagβ-CagZ[56]. In other cases, the role of the protein is ambiguous. An example of the latter is represented by CagD, a protein that is present mainly in the periplasmic space. CagD is essential for CagA translocation, but it only reduces the induction of IL-8 without totally abolishing it[57]. Finally, some cag-PAI components, including CagS, are not essential for CagA translocation[58].
The crystal structures of four proteins of the H. pylori cag-PAI are known: Cagα[59,60], CagZ[61], CagS[62], and CagD[57] (Figure 2). A molecular model of some other proteins can be constructed by homology modeling using the structure of orthologs from other species. To summarize, we are beginning to develop a clear picture of the structural aspects of H. pylori T4SS, but we still lack most of the functional and structural data on this secretion system, which is more complex and sophisticated than the classical T4SS in other gram negative bacteria. In particular, our knowledge regarding the role of the accessory components of the cag-PAI is quite limited, and much remains to be discovered.
FLAGELLA COMPONENTS
Flagella are of paramount importance for H. pylori, as they are necessary for survival of the bacterium in the stomach, particularly during the initial phases of infection[63,64]. In fact, for the bacterium to survive and colonize the host, it has to avoid the very acidic milieu of the stomach lumen and its periodic mechanical clearance. H. pylori is able to swim through the mucus layer and adhere to gastric epithelial cells thanks to flagella and to the presence of several outer membrane adhesins. In contrast to many other Gram-negative bacteria, Helicobacter (and Campylobacter) species possess an unusual velocity in viscous media, possibly due to their helical shapes and to the presence of exclusively polar flagella[65]. Flagella are complex organelles composed of approximately 30 different proteins, but many other proteins are necessary for flagella expression and assembly. At least 45 proteins (listed in Table 3) can be identified as members of flagella or as necessary for flagella assembly in H. pylori.
Table 3.
Flagella components
| Protein symbol | Symbol | Name or proposed function | UniProtKB code | Protein Data Bank code or homology model |
| FlaB | HP0115 | Flagellin subunit | Q07911 | |
| FliR | HP0173 | Export component | O24978 | |
| FlgJ | HP0245 | Rod capping protein; muramidase | P64657 | |
| FlgI | HP0246 | P-ring protein; part of bushing; internal disulfide bridge | O25028 | |
| FliJ | HP0256 | General chaperone | O25037 | |
| FlgL | HP0295 | Hook-associated protein 3; second hook-filament junction protein | Model | |
| FlgH | HP0325 | L-ring protein; part of bushing; lipoprotein | O25092 | |
| FliF | HP0351 | MS-ring protein; mounting flange for rotor/ switch and rod; housing for export apparatus | O25118 | Model |
| FliG | HP0352 | Rotor/switch protein; torque generation; strong interaction with MS ring | O25119 | 4FQ0, 3USY, 343 |
| FliH | HP0353 | Negative regulator of FliI | O25120 | Model |
| FliO | HP0583 | Export component | O25305 | |
| FliN | HP0584 | C ring; rotor/switch protein | O25306 | Model |
| FlaA | HP0601 | Flagellin subunit. Polymerizes with FlaB | ||
| FliP | HP0685 | Export component | O25394 | |
| FlgR | HP0703 | Transcriptional activator of flagellar proteins | O25408 | Model |
| FliD | HP0752 | HAP2; filament-capping protein; flagellin folding chaperone | P96786 | |
| FliS | HP0753 | FliC-specific chaperone | O25448 | 3IQC |
| FliT | HP0754 | FliD-specific chaperone | O25449 | |
| FlhB | HP0770 | Export component; substrate specificity switch; target for soluble export complex | P56416 | Model |
| MotA | HP0815 | Stator protein; exerts torque against rotor/switch | P65410 | |
| MotB | HP0816 | Stator protein; converts proton energy into torque | P56427 | 3S02, 3S03, 3S06, 3S0H, 3S0W, 3S0Y, 3CYP, 3CYQ |
| FlgE | HP0870 | Hook protein | P50610 | |
| FliK | HP0906 | Hook-length-control protein | Q4FEW5 | |
| FlgD | HP0907 | Hook-capping protein | O25565 | Model |
| FliM | HP1031 | C ring; rotor/switch protein; target for CheY-P binding | 4GC8 | |
| ylxH | HP1034 | Flagellum site-determining protein YlxH | O25678 | Model |
| FlhF | HP1035 | Flagellar biosynthesis protein | O25679 | Model |
| FlhA | HP1041 | Export component; target for soluble export complex | O06758 | 3MYD |
| FliS-chaperon | HP1076 | 3K1H, 3K1I | ||
| FlgG_1 | HP1092 | O25724 | Model | |
| FlgK | HP1119 | HAP1; first hook-filament junction protein | O25744 | Model |
| FliW1 | HP1154 | Flagellar assembly factor FliW1 | O25769 | Model |
| pFlA | HP1274 | Paralysed flagella protein | O25864 | |
| FliW2 | HP1377 | Flagellar assembly factor FliW2 | O25929 | Model |
| FliQ | HP1419 | Export component | POAO53 | |
| FliI | HP1420 | ATPase; drives type III flagellar export | O07025 | Model |
| FliC | HP1450 | Filament protein; flagellin | ||
| FlgN | HP1457 | FlgK-, FlgL-specific chaperone | O25995 | |
| FlgA | HP1477 | Chaperone for P-ring protein | O26012 | Model |
| FliE | HP1557 | MS-ring rod junction protein; export gate | P67708 | |
| FlgC | HP1558 | Rod protein; transmission shaft | O26080 | |
| FlgB | HP1559 | Rod protein; transmission shaft | O26081 | |
| FlgG | HP1585 | Distal rod protein; transmission shaft | O26104 | |
| FlgF | HP1585? | Rod protein; transmission shaft |
The first column refers to the protein symbol, the second to the symbol used for strain ACT 700392/26695, the third to the protein name, the fourth to the UniProtKB code, and the fifth to the Protein Data Bank code, if a three-dimensional structure exists, or to the model if it is possible to build an homology-based molecular model.
The structural organization and control of flagella in Gram-negative bacteria have been thoroughly studied[65-69]. A flagellum can be divided in two main portions, the hook-basal body and the extracellular filament. The former, in turn, can be divided into three substructures: (1) the base, localized in the inner membrane and spanning to the cytoplasm; (2) the rod and ring structures, located in the periplasm; and (3) the hook, present on the surface. An exhaustive description of the organelle is outside of the scope of this review, and the reader is directed to more specialized papers. Here, we want to indicate that the three-dimensional structures of only 5 proteins of H. pylori flagella (including flagellar chaperons) have been determined, but another 16 molecular models can be constructed by homology modeling through, for example, an automatic server[70], thanks to the structures of homologs from other species.
As a final comment in this section, we want to indicate that not all of the proteins that are flagella components are listed as “secreted” proteins in Table 1, similar to proteins of the cag-PAI. This result can be explained in different ways: (1) some of the polypeptide chains are part of a larger complex and are secreted along with a component that bears a secretion signal; (2) some proteins were never detected as secreted until now; and (3) some proteins are secreted through a secretion system that has yet to be identified, different from Sec. Regardless of the explanation, Table 1 does not include all of the H. pylori proteins that are actually exported into the external space but most likely includes some proteins that are not actually secreted (see below).
BINDING AND TRANSPORT PROTEINS
Few H. pylori proteins that can be classified as binding or transport proteins are secreted. A significant example is HP1286, a protein that presents a secretion signal at the N-terminus[23] and has been found in the culture medium in three different studies[20,71,72]. This protein is overexpressed under acidic stress conditions, along with other virulence factors[73]. The crystal structure has been determined[74], demonstrating that HP1286 belongs to the lipocalin family, a group of binding proteins that are characterized by the presence of a molecular core formed by an eight-stranded β-barrel that forms a cavity where the ligand is eventually hosted (Figure 3). In the crystal, the HP1286 cavity is occupied by erucamide, the amide of erucic acid. The latter is quite common in nature, being a component of several edible oils. Erucic acid is suitable for human consumption, but only at relatively low doses because a high amount can be toxic for humans. Because the protein for crystallization was expressed in a heterologous system, it cannot be stated that the chemical compound found in the protein cavity is the natural ligand, but the shape and the electrostatic properties of the cavity suggest that the natural ligand is a fatty acid or an amide with a linear chain of approximately 22 carbon atoms. In addition, the protein in the crystal is a dimer, and the buried surface suggests that it is a physiological dimer. This fact is quite unusual, as all of the structurally characterized lipocalins are monomeric, and HP1286 represents the first dimeric member of the family. Because HP1286 is secreted and is involved in the adaptation to the acidic environment, we can speculate that its function could be sequestering specific fatty acids from the environment, but the exact physiological function remains elusive.
Figure 3.
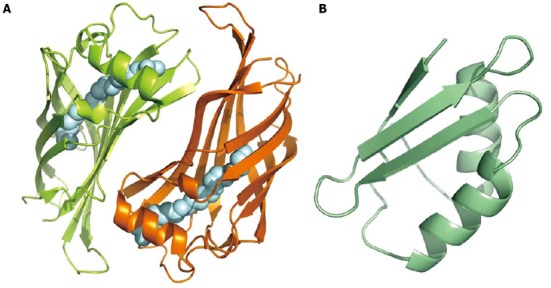
Binding and transport proteins. A: Cartoon model of HP1286 lipocalin dimer. The two monomers, related by a two-fold axis, bind in the inner cavity a molecule of erucamide (silver spheres; PDB 3HPE); B: Nuclear magnetic resonance structure of apo-CopP, a copper binding regulatory protein of 66 amino acid residues (PDB 1YG0).
hp1561 and hp1562 are two homologous genes that code for proteins belonging to the CeuE family. These proteins are designated as CeuE1 and CeuE2, and they possess 335 and 333 amino acids, respectively, with a sequence identity of 86%. Both are annotated in the UniProt database (http://www.uniprot.org/) as “Iron (III) ABC transporter periplasmic iron-binding protein”. These proteins have a signal sequence for export to the periplasm. Crystallographic data, binding assays and in vivo studies indicate that CeuE1 is able to bind and transport specific metals complexed by exogenous metallophores. The protein is not secreted in the external medium, but it is located in the periplasmic space, and its function is to transport the metal ion to an adenosine-triphosphate binding cassette (ABC) transporter, corresponding to HP0888/HP0889, also designated FecD. Note that HP0888 is listed among the secreted proteins (Table 1) and, in fact, is considered highly immunogenic[75], whereas HP0889 does not bear a secretion signal. A similar situation is represented by the pair HP0298/HP1252. The former is annotated as a periplasmic dipeptide-binding protein (PgbA), whereas the second is classified as an ABC transporter, OppA, corresponding to the substrate-binding domain of an oligopeptide transporter (OppABCD)[76].
The hp0970 and hp1073 genes are involved in metal homeostasis. HP1073 belongs to a cluster of three proteins, CnxC (HP0971), CnzB (HP0970), and CnzA (HP0969), which form a Czc-type metal export pump and are required for cadmium, zinc and nickel resistance. In addition, these proteins are involved in urease modulation and gastric colonization[77]. HP1073, CopP, is a copper-binding regulatory protein of 66 amino acid residues (Figure 3). This protein binds Cu (I) through a classic CXXC motif[78].
The last secreted binding-protein, HP0508, is a polypeptide chain of 452 amino acids annotated as the plasminogen-binding protein PgbA. The recombinant protein has been expressed in E. coli and binds plasminogen. It has been hypothesized that the enzyme may allow H. pylori to coat its surface with plasminogen that, once bound to plasmin, could enhance the virulence of the bacterium[79].
VACA AND OTHER TOXINS
The VacA (HP0289) protein represents one of the major secreted virulence factors of H. pylori. The name VacA is derived from its capability to be internalized and to induce the formation of intracellular vacuoles as the result of the osmotic swelling of late endocytic compartments. A large variety of additional cytotoxic functions has been attributed to VacA in the last 10 years of extensive characterization, such as altering the endosomal function, inhibiting T-cell proliferation, internalizing and damaging mitochondria, and inducing apoptosis, among others (Reviewed in[80,81]). While the mature form of the toxin corresponds to a 88-kDa species, the pro-protein (140 kDa) accounts for further N-terminal and C-terminal fragments that undergo multiple processing steps that are required for the export and maturation of VacA. Indeed, similar to other Gram-negative bacterial proteins that are secreted by the type V pathway, VacA includes both an N-terminal signal peptide to cross the inner membrane through a Sec machinery and a C-terminal β-barrel motif that is required for insertion into the outer membrane. The latter allows for the auto-export of the so called autotransporter passenger domain, characterized by a right-handed β-helix topology, highly conserved despite the fact that the corresponding sequences show poor homology among the family members. The active secreted VacA toxin might be further processed into two main subunits, an amino-terminal 33-37 kDa fragment (p33) and the subsequent 55 kDa autotransporter passenger domain (p55); the two associate into hetero-oligomers composed of 6-7 subunits forming two rings with a “snowflake” shape, as demonstrated by electron microscopy[82]. Such multimerization confers to VacA the capability of being inserted into lipid bilayers and to form anion-selective pores at low pH, a feature that strongly correlates with its vacuolization effects. The p33 subunit defines the core of the membrane channel, while p55 localizes to the peripheral arms. The latter, which corresponds to the so called autotransporter passenger domain, has been characterized by X-Ray diffraction studies (PDB code 2QVE); it is organized as a right-handed parallel β-helix with a small (75 amino acids) globular C-terminal appendage[83] (Figure 4).
Figure 4.
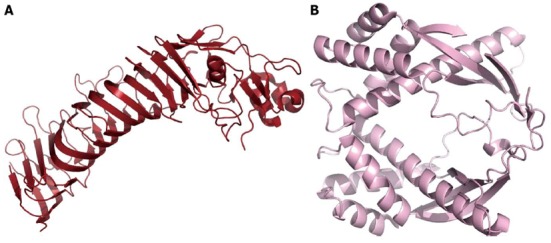
Toxins. A: Cartoon model of the p55 domain of vacuolating toxin (VacA) (coordinates from PDB 2QV3). The structure is a predominantly right-handed parallel β-helix, and the domain mediates the binding of VacA to the host cell; B: Cartoon model of a truncated form of tumor-necrosis-factor α (TNFα) inducing protein, a virulence factor that enters gastric cells and stimulates both the production of TNFα and the nuclear factor kappa B pathway (coordinates PDB 2WCR).
Three proteins (ImaA/HP0289, FaaA/HP0609-10, and VlpC/Hp0887), exported by the type V autotransporter machinery and localized to the bacterial cell surface, have been identified as VacA-like proteins. Similar to the vacuolating toxin A, these proteins are classified among the largest multidomain H. pylori proteins (larger than 250 kDa), and they share multiple copies of a VacA-conserved motif (pfam03077) and the typical N- and C-terminal export motifs for the type V export pathway. A mouse model of infection demonstrated that all of the Vac-like proteins are upregulated in vivo, and the corresponding mutants show a clear deficiency in colonization and persistence compared to the wild type bacteria[84]. In particular, FaaA enriches in the flagellar sheath and contributes to the stability, functionality and proper localization of the flagella. ImaA (immunomodulatory autotransporter protein A) expression is upregulated under acidic stress conditions and stimulates the induction of IL-8 chemokine as well as TNF-α in AGS cell culture[85].
HP0569 has been identified within the pull of secreted and highly immunogenic factors of H. pylori. HP0569 dimers (38 kDa) localize to the periplasm, attached to the inner membrane and to the secreted medium. This protein is capable of penetrating gastric cells and possibly enters the nucleus. HP0569 has been named tumor-necrosis-factor α inducing protein (Tipα) because it strongly stimulates the expression of the cytokine TNF-α as well as nuclear factor kappa B (NF-KB) and multiple chemokines in gastric cell lines. Such a cascade of pro-inflammatory events, which correlates with cancer progression, convinced the authors to classify Tipα as a carcinogenic factor[86].
The crystal structure of Tipα (Figure 4) was determined in 2009[87,88] at two different pH values, indicating that full length Tipα is a dimer and adopts a novel fold consisting of an α-β sandwich combined with a four-helix bundle domain. Tipα’s dimeric association seems to involve the presence of a disulfide bridge only under acidic pH levels, but not at higher pH values. The structure can be divided into three main motifs that suggest a propensity to protein-protein or protein-DNA interactions: an N-terminal flexible extension, a dodecin-like domain and a SAM-like domain.
ENZYMES
Several secreted proteins with enzymatic activities were identified and are listed in Table 1. In this chapter, the description of these proteins has been organized according to their functional similarities.
Proteins involved in antioxidant systems
Oxidative stress is experienced by essentially all living systems, and, for H. pylori, it represents a very serious problem. The bacterium, in fact, is permanently and massively exposed to oxidative damage, as its presence in the host stomach induces an increase in phagocytic cells, macrophages and polymorphonuclear leukocytes at the site of infection. As a consequence, the oxidative burst produces reactive oxygen species, such as superoxide anions, hydrogen peroxide and hydroxyl radicals. To maintain a long-term persistence in the host, the bacterium fights oxidative stress using a battery of diverse antioxidant systems, including superoxide dismutase (SOD), catalase, thioredoxins and peroxiredoxins, NADPH quinone reductase, and many others[89,90].
Some of these proteins are predicted to be secreted or have been found in the external medium (Table 1). Among them, the thioredoxins-thioredoxin reductase pair represents one of the most classical examples of reductant system[90], particularly relevant in H. pylori, as the bacterium lacks the glutathione-/glutathione-dependent enzymes and relies mostly on thioredoxins for its defense against ROS. In H. pylori the electron transfer process proceeds according to NADPH → TrxB → Trx → Peroxiredoxin → ROOH.
Two thioredoxins, TrxA and TrxC, and one thioredoxin reductase (TrxR or TrxB) are coded in the H. pylori genome and are all secreted[20,23,71,91]. Several studies have characterized the properties of the Trx/TrxB system, and chemically, the two appear similar to the homologs in E. coli. In contrast, the functional difference between the two thioredoxins, TrxA and TrxC, which share a sequence identity of only 33%, is not well defined and awaits better characterization. The crystal structure of H. pylori TrxB has been determined in both oxidized and reduced forms (Figure 5)[92], while the structures of TrxA and TrxC are not present in the PDB, but a molecular model can be easily built by homology modeling using the structure of the E. coli ortholog. The comparison of the two H. pylori molecules with the corresponding E. coli pair shows that residue substitutions that change the shape of the surface in both molecules may account for the specificity of the Trx-TrxB interaction[92].
Figure 5.
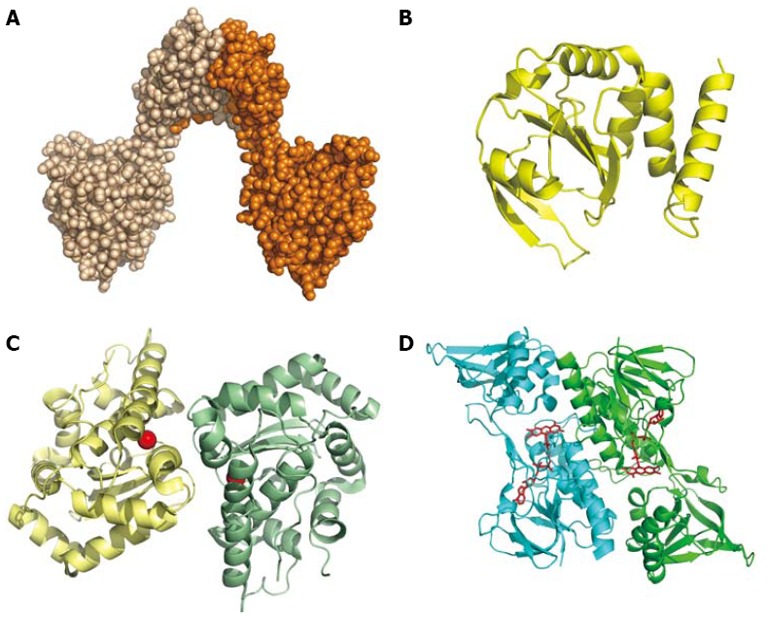
Redox proteins. A: Space-filling model of the dimer of DsbG (HP0231; PDB 3TDG); B: Cartoon of DsbC (HP0377; Coordinates PDB 4FYC), an enzyme with a thioredoxin-like fold possibly involved in cytochrome c assembly; C: Cartoon of the dimeric Fe-superoxide dismutase (Coordinates PDB 3CEI). The iron ion is represented by a red sphere; D: Dimeric thioredoxin reductase (Coordinates PDB 3ISH). The FAD bound is shown as a ball-and-stick model.
The bacterium codes for three peroxiredoxins, alkyl hydroperoxide reductase[93], Tpx (or TagD, Hp0390) and Bcp (HP0136)[89,94], none of which appear to be secreted except for Tpx, which has been associated with the outer membrane fraction in few cases. In contrast, methionine sulfoxide reductase (Msr), another substrate of Trx, is secreted.
Another two proteins that are involved in disulfide bond formation or exchange are also secreted: DsbG (HP0231) and DsbC (HP0377). The latter presents a thioredoxin-like fold (Figure 5) and has been proposed to perform multiple reductase roles in the bacterium[95]. In contrast, DcbG, which catalyzes disulfide bond formation[96], presents a totally different architecture: the molecule is a V-shaped homo-dimer, where each monomer is formed by two globular domains connected by a long α-helix[97] (Figure 5A). DsbG functions in vitro as a reductase against HP0518, a putative L,D-transpeptidase with a catalytic cysteine residue[97]. Due to its strong immunogenicity[98], DsbG has also been proposed as a potential candidate for a vaccine against H. pylori[99].
Among other antioxidant proteins, both catalase (HP0785) and a catalase-like protein (HP0485) present a secretion signal. Catalase is a widespread enzyme that protects against reactive oxygen species, as it catalyzes the conversion of hydrogen peroxide to molecular oxygen and water. The crystal structure of HP0785 has been determined at a 1.6 Å resolution[100,101], and its overall fold is similar to that of other enzymes of the family: the enzyme is a tetramer with 222 symmetry, where each monomer includes an N-terminal arm, an antiparallel eight-stranded β-barrel, a long loop and a C-terminal α-helical domain. A heme is bound to each subunit, while H. pylori catalase does not bind NADPH, similar to the other enzymes of the clade III group. It must be noted that the crystal structure does not offer any clue as to the possible localization of the enzyme on the external surface of the cell membrane. Catalase was, in fact, detected in the supernatant along with SOD[102], but the conclusion was that the amount of secreted proteins was too low to justify for the enzyme a role of scavenging oxidants from injured mucosa. In contrast, the presence of Msr (HP0224), a methionine repair enzyme, among the secreted proteins appears significant, as a synergistic role of Msr and GroEL (another secreted factor; see the considerations in the Section “Other proteins”) in repairing oxidant-damaged catalase has been demonstrated[103]. Finally a catalase-like protein that is much shorter (314 amino acids) and with a low degree of similarity to catalase (62 identical and 102 similar amino acids) is also predicted to be secreted, but its function has not yet been characterized.
H. pylori, in contrast to other Gram-negative bacteria such as E. coli, produces a single iron-dependent superoxide dismutase (SOD) that is required for bacterial colonization[104]. The crystal structure (Figure 5C) of this Fe-SOD shows that its fold is quite similar to that of other Fe-SOD, the most significant difference being an extended C-terminal tail, which has been hypothesized to be responsible for the interaction with the external cell membrane and possibly for phosphorylation[105].
Some of the proteins discussed above present an export signal to the periplasmic space, while others do not have a signal peptide but rather have been experimentally detected in the external medium of an in vitro bacterial culture, and for a few others both conditions apply. The most likely hypothesis is that these proteins play their protective roles against oxidation in the periplasm and are eventually released into the external medium, perhaps due to leaks in the external membrane. It is, in fact, unlikely that the bacterium tries to protect itself from oxidizing agents acting directly in the external space, where a very large concentration of enzymes should be necessary. On the contrary, the periplasm, being confined and spatially limited, appears as an ideal first trench. A similar situation holds for pH, which is buffered mainly in the periplasm to better preserve the cytoplasmic state.
Some of the members of the electron transport chain are listed among the secreted proteins. In Gram-negative bacteria, these proteins are localized to the inner membrane (F0F1 ATP synthase and NADH ubiquinone reductase) or to the periplasm (formate dehydrogenase cytochrome c553)[106]. Other enzymes, such as aspartate aminotransferase, that are involved in ATP biosynthesis, which in eukaryotic cells localize to mitochondria, in bacteria should localize to the cytoplasm or periplasm, close to the inner membrane. Specific studies on the previous systems have not been performed in H. pylori and are annotated based on sequence homology.
H. pylori flavodoxin (FldA, HP1161) functions as an acceptor of the electrons that are generated by pyruvate oxidation[107]: pyruvate + CoA → acetyl-CoA + CO2 + 2e-.
In addition, patients affected by MALT lymphoma tested positive for antibodies against this protein. The HP1161 crystal structure is similar to that of flavodoxins from other bacteria, with the exception of a minor difference at the active site[108]. Because flavodoxin is essential for bacterial survival, its inhibitors could be of pharmacological relevance.
Other proteins, such as ketol-acid reductoisomerase (HP0330) and cytochrome C551 peroxidase (HP1461), have not yet been characterized. HP0330 has been detected in a larger complex that includes GroEL/ES and UreA, but the real meaning of this complex has not been further investigated[109].
Putative solenoid proteins
Solenoid proteins, characterized by a modular architecture, include members of the Sel1-like repeat (SLR) and tetratricopeptide repeat families. These proteins present low sequence similarity and possibly different functions and are mostly characterized by their three-dimensional structure. Although eukaryotic SLR proteins can perform diverse functions, from adaptor proteins for the assembly of macromolecular complexes to ER-associated protein degradation, bacterial SLR are considered mediators of the interactions between bacterial and eukaryotic host cells[110]. Nine secreted proteins from H. pylori are classified as belonging to the SLR family: five of them, HP0160, HP0211, HP0235, HP0336 and HP1098, are classified as β-lactamases or cysteine-rich proteins and labeled HcpAD, HcpA, HcpE, HcpB, and HcpC, respectively (Figure 6)[111,112]. HcpC, HcpA and HcpB bind 6-amino-penicillic acid in vitro, but antibiotic resistance does not appear to represent their in vivo function, as resistance against the β-lactam antibiotic is HcpA-independent[113]. Their involvement in cell wall biosynthesis is also not viable, as an HcpA deletion mutant grows normally[114]. The most likely function of these proteins seems to be the binding to and recognition of different specific peptides, possibly explaining their involvement in the host immune response. For example, HcpA induces the release of a complex cytokine pattern[114]. It is not clear if all of the Hcps proteins are involved in the pro-inflammatory pathway or if they perform different functions despite a similar architecture. More recently, HcpC has been shown to directly interact with human proteins such as Nek9, Hsp90, and Hsv71[115]. Finally, it has been proposed that their modular architecture could be used for host adaptation[110].
Figure 6.
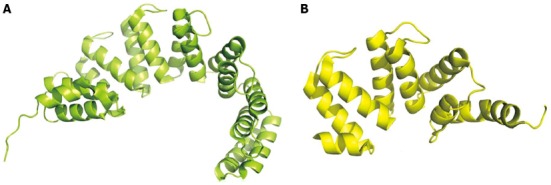
Two examples of solenoid-class proteins. A: HcpB (HP0336, coordinates PDB1KLX); B: HcpC (HP1098, coordinates PDB 1OUV).
Other SLR secreted proteins include HP0519, HP0628, and HP1117. Despite their exact role being unknown, the Sel-like genes have been used to characterize the geographic partitioning of H. pylori populations in a phylogenetic analysis[116] and to trace human migrations[117].
The last secreted member of the solenoid protein family is HP1124, classified based on sequence similarity into the tetratricopeptide-like repeat family protein, similar to Sel1-like repeat proteins.
Proteases
Few secreted proteins can be classified as proteases. Among them are HP0657 and HP1012, the former of which is predicted to be an inactive domain of a processing protease, the second a Zn-dependent protease. Both functions were inferred from sequence similarity, but no literature exists supporting these functions. In contrast, the HP1037 aminopeptidase function has been experimentally tested[118]. Despite being annotated as two genes, HP1018 and HP1019 sequences belong to a single gene called htrA (high-temperature requirement A) encoding a periplasmic 50-kDa protease. Indeed, HtrA has been discovered in the secreted H. pylori proteome to play both the role of protein quality control chaperone and trypsin-like serine protease. Up-regulated under bacterial stress conditions, HtrA further contributes to epithelial barrier disruption by destroying the cell adhesion junctions through the cleavage of the cell adhesion protein E-cadherin. A similar virulence mechanism guarantees persistent colonization and pathogenesis to other enteropathogens, such as E. coli, Shigella flexneri, and Campylobacter jejuni. Due to its relevant and prevalent role in many Gram-negative bacteria, HtrA has been proposed to be a novel candidate for therapeutic intervention strategies, as a lead compound developed to specifically inhibit HtrA was proved to impair E-cadherin proteolysis and the intercellular penetration of H. pylori[119]. HP0570 is also predicted to be an aminopeptidase and is enriched in the extracellular fraction[22] but is annotated as a cytosolic aminopeptidase. Finally, HP1350 is predicted to be a serine protease of the CtpA (carboxy-terminal protease) family.
Other enzymes
Other enzymes not belonging to the previous classes have been identified as secreted. Among them are the two subunits of the urease enzyme, UreA and UreB. It is interesting to note that the latter has been identified as secreted in several studies, while the UreA subunit was identified as secreted in only in one case[17] (Table 1). Because UreA and UreB are members of the same complex, it appears unlikely that only UreB is secreted. Urease is considered to be active mainly in the cytoplasm, but it cannot be excluded that some pH buffering activity also occurs in the extracellular space, considering that its enzymatic activity is unaffected by the pH 3[120].
Another essential component of the pH buffering system in H. pylori is α-carbonic anhydrase (HP1186), which plays a role in the periplasm. This enzyme catalyzes the conversion of CO2, generated in cytoplasm by the degradation of urea by urease, into HCO3-. The buffering of pH in the periplasm depends, in fact, not only on the efflux of NH3 from the cytoplasm but also from the production of hydrogen carbonate[121]. HP1186 expression is regulated by the two-component system ArsRS in response to low environmental pH[122].
Another enzyme involved in ammonia production is HP1118, a protein that has been identified as secreted in four different experiments[20,22,23,123]. This enzyme exhibits γ-glutamyltranspeptidase activity[124,125], but its main function in Helicobacter is most likely related to ammonia production in function of pH stabilization in the periplasm as a by-product of glutamine breakdown[126]. As a secondary effect, HP1118 induces the apoptosis of the host cells. The inhibition of HP1118 resulted in a complete loss of apoptotic activity, and in an isogenic mutant deficient strain, this activity was significantly lower compared to the parent strain[127]. This role of HP1118 is quite relevant, as H. pylori infection induces apoptosis in gastric epithelial cells[128]. HP1118 represents the first case of a γ-glutamyl transpeptidase with pro-apoptotic activity, and the former is necessary to induce the latter[127].
HP0392 is annotated as histidine kinase CheA and is involved in chemotaxis. Bacterial chemotaxis is fundamental for optimal colonization, as it controls flagellar rotation directing the bacterium towards nutrients or to safer places, detecting chemical cues in the environment[129]. The H. pylori chemoreceptor system significantly differs from the better characterized system of E. coli; in addition to CheA, it includes CheY (HP1067), CheW (HP0391), a CheZ-homolog (HP0170), and three CheVs (HP0019, HP0393, and HP0616)[130]. CheYs accept phosphate from CheA, and the three CheVs mediate the dephosphorylation of CheA[131]. Only CheA is predicted to be secreted, while all of the other proteins that are involved in chemotaxis are not secreted.
Two nucleases, HP0275 and HP0323, are predicted secreted proteins. HP0323 is a cation-independent nuclease, NucT, associated with the membrane that preferentially cleaves single-stranded DNA[132]. NucT most likely performs DNA processing and uptake similar to the well-known process in gram-positive bacteria. HP0275 is predicted to be an ATP-dependent nuclease, AddB, but no further characterization has been performed.
Peptidoglycan deacetylase (PgdA, HP0310) is the enzyme responsible for a peptidoglycan modification that counteracts the host immune response[118]. The protein has been crystallized and its structure determined[133]. Despite the overall fold being similar to that of other deacetylases, H. pylori PgdA does not exhibit a solvent-accessible polysaccharide-binding grove, indicating that the enzyme binds a smaller substrate at the active site.
Two genes encoding aliphatic amidases have been identified, hp0294 and hp1238. The corresponding proteins are called AmiE and AmiF, respectively[134,135]. Aliphatic amidases are usually cytoplasmic enzymes that catalyze the hydrolysis of short-chain amides to produce ammonia, and their presence in H. pylori is most likely justified by the fact that urease activity alone is not sufficient to buffer the bacterium pH when that of the extracellular medium becomes very low[136]. Only AmiE has been identified as secreted, and we speculate that it acts in the periplasm, while AmiF is active in the cytoplasm.
Finally, some of the enzymes reported in Table 1, including citrate synthase (HP0026), glutamate dehydrogenase (HP0380) and enolase (HP0154), are clearly cytoplasmic enzymes and were identified as secreted in only one study[22]. These enzymes were detected in the extracellular fraction, but using a more stringent criterion, they should have been excluded; therefore, their presence in the table can be considered an artifact. A related enzyme is triosephosphate isomerase (TIM), a well-characterized enzyme of the glycolytic pathway[137]. This enzyme is predicted to contain a secretion signal, but it appears unlikely that the enzyme is secreted. The role of HP1178, a purine nucleoside phosphorylase[138], and of HP1375, an UDP-N-acetylglucosamine acyltransferase, in the secretion of other enzymes, including HP0672, a member of the PLP-dependent aminotransferase superfamily clan, is not evident.
The existence of H. pylori-related lipase and phospholipase activities against the gastric mucus layer were reported for the first time by Slomiany et al[157]. Later, a cytoplasmic enzyme (HP0739) with lipolytic activity was discovered and characterized. Because this enzyme exhibits the typical behavior of a carboxylesterase, with a clear preference for short chains, it has been grouped into the bacterial lipase family V and called EstV. However, none of the studies aimed at identifying secreted enzymes have been able to detect either the EstV protein or any other lipase in the H. pylori secretome, thus suggesting a different role for EstV, which could degrade lipids taken up by the bacterium from the medium after their release due to the catalysis of host hydrolases. Similarly, H. pylori secretome studies have never detected the outer membrane phospholipase OMPLA (HP0499). OMPLA has been proposed to be involved in the variation in the H. pylori membrane lipid composition, relevant for gastric environment adaptation. Indeed, the phase variations in the corresponding gene have been correlated with the necessity for a reversible and spontaneous response in terms of lysophospholipid enrichment, as these variations represent an advantage for bacterial persistence and epithelial cell adherence under acidic conditions[158]. In line with its absence in the secreted protein studies reviewed in this paper, the phospholipase activity of the OMPLA/HP0499 protein was detected in vitro only if the bacteria were lysed by sonication, indicating that this protein is not exposed to the cell envelope. The authors further suggest that the catalytic activity of this protein could be responsible for the degradation of membrane phospholipids, perturbing the bacterial membrane and consequently facilitating the release of virulence factors such as urease.
OTHER PROTEINS
Proteins that were not classified in the previous groups are listed in the “Others” section in Table 1. Among them, HP0166 is the ArsR (also called OmpR) member of the two-component signal transduction system ArsRS that regulates the acid-induced expression of α-carbonic anhydrase, an enzyme present in the periplasm (see the “Other enzymes” section)[122,139]. Interestingly, ArsR was detected among the factors related to gastric cancer, and in the same group, the rod shape-determining protein HP0743 is also present[140]. Another secreted protein is HP1126, TolB, a member of the Tol/Pal system. The latter has not yet been characterized in Helicobacter, but it is well known in other Gram-negative species. Because the Tol/Pal system plays a role in bacterial envelope integrity and is associated with the peptidoglycan[141], it is likely to assume that HP1126 is present in the periplasm.
A special case is represented by the chaperone GroEL (or Hsp60, HP0010). In a recent paper[142], the authors show that GroEl from H. pylori is also able to bind iron, a property that is not shared with GroEL from other bacteria, including E. coli. Despite the absence of an export peptide signal, the protein was identified as secreted, and the authors claim that GroEL is secreted as a heme scavenger to supply iron to the bacterium. In parallel, a synergistic role of GroEL with two enzymes, Msr and catalase, has been observed[103]; the in vitro addition of GroEL to a mixture of catalase and Msr in the presence of an antibacterial oxidant strongly enhances the recovery of catalase activity, indicating that the presence of the chaperonin helps the enzyme be repaired by Msr to recover its native folding. Interestingly, all three of the proteins involved in the process (catalase, Msr and GroEL) have been identified as secreted (see the paragraph on proteins involved in redox processes). We speculate that, when catalase is secreted to protect the bacterium against oxidative stress, the presence of both Msr, another enzyme acting against the oxidative stress mostly localized in the bacterial membranes[143], and GroEL helps in recovering catalase activity.
Finally, for those proteins that have been identified in vitro as secreted by H. pylori[71] but that play a clear function in the cytoplasm and do not bear any signal peptide, it is hard to hypothesize whether such secretion occurs in vivo. These proteins include two ribosomal proteins (HP1201 and Hp1202), a histone-like DNA-binding protein and an ssDNA-binding precursor. The ssDNA-binding precursor three-dimensional structure in solution suggests the presence of a possible RNA-binding site[144].
HYPOTHETICAL UNCHARACTERIZED PROTEINS
The group of hypothetical uncharacterized proteins is one of the most crowded: 28 genes encode proteins that are predicted to be secreted but whose function remains undefined (Table 1). Of these proteins, 23 are without any predicted function, as they have no orthologs in other species whose function is known, or they are unique to Helicobacter. Three of these proteins (HP0122, HP0135, and HP0720) are very short peptides, ranging from 44 to 52 amino acids. The second, HP0135, is predicted by bioinformatic analysis to be a lipoprotein. For the remaining five putative proteins, some function can be hypothesized, albeit with limited confidence. HP0659 is predicted to be SurA, a chaperone of outer membrane proteins. HP0169 is predicted to be a secreted collagenase that is essential for colonization (Kavermann, 2003). HP0721 is a very special case, as we have much information, including its three-dimensional structure. Being a relatively small protein of 152 amino acids, it has been proposed to be a new sialic acid-binding protein[145] and/or a factor that is involved in nickel homeostasis[146]. This protein belongs to the group of H. pylori proteins that are S-nitrosylated[147]. The fold of the crystal structure of the core domain of the protein is that of an orthogonal α-helical bundle, with a hydrophobic cavity in the center[148]. Despite all of these data available, it is not possible to assign a defined role to HP0721. HP1285 is a protein of 230 amino acids that, according to its amino acid sequence, resembles the LppC from Streptococcus equisimilis and other lipoprotein members of the e (P4) family (LppA from Streptococcus pyogenes, OplA from Flavobacterium meningosepticum, HeI from Haemophilus influenzae[149]). LppC acts as an acid phosphatase, while the functions of the other members of the family remain unknown. Finally, HP1580 is uncharacterized, but in some strains it is annotated on bioinformatics bases as a member of the phosphatidic acid phosphatase (PAP2) protein family[150].
CONCLUSION
In this paper, we include 163 H. pylori proteins that have been experimentally identified as or are predicted to be secreted. Some of these proteins exert their role in the periplasm, some are embedded in the internal or external membrane, and others are secreted into the extracellular space where they perform their function. Often, it is not easy to determine whether a protein is present in the periplasm or in the extracellular milieu, as proteins present in the periplasm are often experimentally detected in vitro in the external medium, perhaps due to the weakness of the external membrane. However, in vitro experiments aimed at detecting secreted proteins are performed under conditions that may not reflect the different states of the bacterium in vivo. Consequently, some proteins that could have been secreted are not expressed or are eventually not secreted under the experimental conditions used.
The secretome includes proteins belonging to different classes; the most populated classes are those of the outer membrane proteins and the enzymes; others include components of flagella, of cag-PAI and of toxins. Note that 28 proteins, approximately 17% of the total proteins secreted are hypothetical proteins whose functions are unknown. Finally, from a structural point of view, only 15 among the secreted proteins, approximately 9% of the total, have been structurally characterized. This number is consistent with our present knowledge of the bacterium, as 148 unique H. pylori structures (This number includes unreleased entries at that date. The total number of files was 340) were present in the PDB in April 2013, corresponding to approximately 9% of the total proteome.
The previous considerations suggest that, despite the mass of data accumulated until now on H. pylori secretome, much work is required to reach a full comprehension of the system. This comprehension is fundamental because this knowledge is not only relevant for the comprehension of the physiology of the bacterium, but, above all, because secreted or exposed bacterial proteins directly contact the host and may directly influence the outcome of the pathology. We cannot exclude that future discoveries may add new secreted factors and significantly change our view of the interaction of this pathogen with its host.
Footnotes
Supported by the University of Padua grant “Progetto di Ateneo 2011” and by PRIN 2010-2011 “Unraveling structural and functional determinants behind Helicobacter pylori pathogenesis and persistence”
P- Reviewers: Buzas GM, Slomiany BL S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
References
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin CS, Mendall MM, Northfield TC. Helicobacter pylori infection. Lancet. 1997;349:265–269. doi: 10.1016/S0140-6736(96)07023-7. [DOI] [PubMed] [Google Scholar]
- 3.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 5.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 6.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Rieder G, Fischer W, Haas R. Interaction of Helicobacter pylori with host cells: function of secreted and translocated molecules. Curr Opin Microbiol. 2005;8:67–73. doi: 10.1016/j.mib.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Kao JY, Rathinavelu S, Eaton KA, Bai L, Zavros Y, Takami M, Pierzchala A, Merchant JL. Helicobacter pylori-secreted factors inhibit dendritic cell IL-12 secretion: a mechanism of ineffective host defense. Am J Physiol Gastrointest Liver Physiol. 2006;291:G73–G81. doi: 10.1152/ajpgi.00139.2005. [DOI] [PubMed] [Google Scholar]
- 10.Gerhard M, Schmees C, Voland P, Endres N, Sander M, Reindl W, Rad R, Oelsner M, Decker T, Mempel M, et al. A secreted low-molecular-weight protein from Helicobacter pylori induces cell-cycle arrest of T cells. Gastroenterology. 2005;128:1327–1339. doi: 10.1053/j.gastro.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Zanotti G, Papinutto E, Dundon W, Battistutta R, Seveso M, Giudice G, Rappuoli R, Montecucco C. Structure of the neutrophil-activating protein from Helicobacter pylori. J Mol Biol. 2002;323:125–130. doi: 10.1016/s0022-2836(02)00879-3. [DOI] [PubMed] [Google Scholar]
- 12.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 13.Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, Kelleher D, Rappuoli R, Montecucco C, Rossi F. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med. 2000;191:1467–1476. doi: 10.1084/jem.191.9.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amedei A, Cappon A, Codolo G, Cabrelle A, Polenghi A, Benagiano M, Tasca E, Azzurri A, D’Elios MM, Del Prete G, et al. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Invest. 2006;116:1092–1101. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Elios MM, Codolo G, Amedei A, Mazzi P, Berton G, Zanotti G, Del Prete G, de Bernard M. Helicobacter pylori, asthma and allergy. FEMS Immunol Med Microbiol. 2009;56:1–8. doi: 10.1111/j.1574-695X.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 16.Teneberg S, Miller-Podraza H, Lampert HC, Evans DJ, Evans DG, Danielsson D, Karlsson KA. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J Biol Chem. 1997;272:19067–19071. doi: 10.1074/jbc.272.30.19067. [DOI] [PubMed] [Google Scholar]
- 17.Carlsohn E, Nyström J, Karlsson H, Svennerholm AM, Nilsson CL. Characterization of the outer membrane protein profile from disease-related Helicobacter pylori isolates by subcellular fractionation and nano-LC FT-ICR MS analysis. J Proteome Res. 2006;5:3197–3204. doi: 10.1021/pr060181p. [DOI] [PubMed] [Google Scholar]
- 18.Phadnis SH, Parlow MH, Levy M, Ilver D, Caulkins CM, Connors JB, Dunn BE. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 20.Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, Meyer TF, Jungblut PR. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect Immun. 2002;70:3396–3403. doi: 10.1128/IAI.70.7.3396-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim N, Weeks DL, Shin JM, Scott DR, Young MK, Sachs G. Proteins released by Helicobacter pylori in vitro. J Bacteriol. 2002;184:6155–6162. doi: 10.1128/JB.184.22.6155-6162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith TG, Lim JM, Weinberg MV, Wells L, Hoover TR. Direct analysis of the extracellular proteome from two strains of Helicobacter pylori. Proteomics. 2007;7:2240–2245. doi: 10.1002/pmic.200600875. [DOI] [PubMed] [Google Scholar]
- 23.Müller SA, Findeiß S, Pernitzsch SR, Wissenbach DK, Stadler PF, Hofacker IL, von Bergen M, Kalkhof S. Identification of new protein coding sequences and signal peptidase cleavage sites of Helicobacter pylori strain 26695 by proteogenomics. J Proteomics. 2013;86:27–42. doi: 10.1016/j.jprot.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 24.Odenbreit S. Adherence properties of Helicobacter pylori: impact on pathogenesis and adaptation to the host. Int J Med Microbiol. 2005;295:317–324. doi: 10.1016/j.ijmm.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Odenbreit S, Swoboda K, Barwig I, Ruhl S, Borén T, Koletzko S, Haas R. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect Immun. 2009;77:3782–3790. doi: 10.1128/IAI.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA. 2011;108:5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bina J, Bains M, Hancock RE. Functional expression in Escherichia coli and membrane topology of porin HopE, a member of a large family of conserved proteins in Helicobacter pylori. J Bacteriol. 2000;182:2370–2375. doi: 10.1128/jb.182.9.2370-2375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–4168. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahdavi J, Borén T, Vandenbroucke-Grauls C, Appelmelk BJ. Limited role of lipopolysaccharide Lewis antigens in adherence of Helicobacter pylori to the human gastric epithelium. Infect Immun. 2003;71:2876–2880. doi: 10.1128/IAI.71.5.2876-2880.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, McGee DJ, Rhoads RE, Slepenkov S, Testerman TL. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun. 2011;79:3106–3116. doi: 10.1128/IAI.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaoka Y, Ojo O, Fujimoto S, Odenbreit S, Haas R, Gutierrez O, El-Zimaity HM, Reddy R, Arnqvist A, Graham DY. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55:775–781. doi: 10.1136/gut.2005.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mobley HLT, Mendz GL, Hazell SL. Overview. In: Mobley HLT, Mendz GL, Hazell SL, editors. Source Helicobacter pylori: Physiology and Genetics. Chapter 1. Washington D.C. ASM Press; 2001. [PubMed] [Google Scholar]
- 34.Backert S, Selbach M. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol. 2008;10:1573–1581. doi: 10.1111/j.1462-5822.2008.01156.x. [DOI] [PubMed] [Google Scholar]
- 35.Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Z, Censini S, Bayeli PF, Telford JL, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Covacci A, Falkow S, Berg DE, Rappuoli R. Did the inheritance of a pathogenicity island modify the virulence of Helicobacter pylori? Trends Microbiol. 1997;5:205–208. doi: 10.1016/S0966-842X(97)01035-4. [DOI] [PubMed] [Google Scholar]
- 39.Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190–1202. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer W. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 2011;278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 41.Cascales E. The type VI secretion toolkit. EMBO Rep. 2008;9:735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Mémet S, Huerre MR, Coyle AJ, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 43.Segal ED, Lange C, Covacci A, Tompkins LS, Falkow S. Induction of host signal transduction pathways by Helicobacter pylori. Proc Natl Acad Sci USA. 1997;94:7595–7599. doi: 10.1073/pnas.94.14.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163–176. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 45.Hayashi T, Senda M, Morohashi H, Higashi H, Horio M, Kashiba Y, Nagase L, Sasaya D, Shimizu T, Venugopalan N, et al. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe. 2012;12:20–33. doi: 10.1016/j.chom.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Kaplan-Türköz B, Jiménez-Soto LF, Dian C, Ertl C, Remaut H, Louche A, Tosi T, Haas R, Terradot L. Structural insights into Helicobacter pylori oncoprotein CagA interaction with β1 integrin. Proc Natl Acad Sci USA. 2012;109:14640–14645. doi: 10.1073/pnas.1206098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol. 2008;190:2161–2171. doi: 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Backert S, Fronzes R, Waksman G. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 2008;16:409–413. doi: 10.1016/j.tim.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Buhrdorf R, Förster C, Haas R, Fischer W. Topological analysis of a putative virB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int J Med Microbiol. 2003;293:213–217. doi: 10.1078/1438-4221-00260. [DOI] [PubMed] [Google Scholar]
- 51.Andrzejewska J, Lee SK, Olbermann P, Lotzing N, Katzowitsch E, Linz B, Achtman M, Kado CI, Suerbaum S, Josenhans C. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J Bacteriol. 2006;188:5865–5877. doi: 10.1128/JB.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terradot L, Waksman G. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 2011;278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- 53.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 54.Jiménez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, et al. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5:e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Couturier MR, Tasca E, Montecucco C, Stein M. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun. 2006;74:273–281. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurik A, Hausser E, Kutter S, Pattis I, Prassl S, Weiss E, Fischer W. The coupling protein Cagbeta and its interaction partner CagZ are required for type IV secretion of the Helicobacter pylori CagA protein. Infect Immun. 2010;78:5244–5251. doi: 10.1128/IAI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cendron L, Couturier M, Angelini A, Barison N, Stein M, Zanotti G. The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion. J Mol Biol. 2009;386:204–217. doi: 10.1016/j.jmb.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 58.Fischer W, Püls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 59.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 60.Terradot L, Durnell N, Li M, Li M, Ory J, Labigne A, Legrain P, Colland F, Waksman G. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol Cell Proteomics. 2004;3:809–819. doi: 10.1074/mcp.M400048-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Cendron L, Seydel A, Angelini A, Battistutta R, Zanotti G. Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a type IV secretion system. J Mol Biol. 2004;340:881–889. doi: 10.1016/j.jmb.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 62.Cendron L, Tasca E, Seraglio T, Seydel A, Angelini A, Battistutta R, Montecucco C, Zanotti G. The crystal structure of CagS from the Helicobacter pylori pathogenicity island. Proteins. 2007;69:440–443. doi: 10.1002/prot.21576. [DOI] [PubMed] [Google Scholar]
- 63.Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. Int J Med Microbiol. 2002;291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 64.Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. Architecture of the flagellar rotor. EMBO J. 2011;30:2962–2971. doi: 10.1038/emboj.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 70.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 71.Vanet A, Labigne A. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect Immun. 1998;66:1023–1027. doi: 10.1128/iai.66.3.1023-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, Meng FL, He LH, Zhang JZ. Secreted protein HP1286 of Helicobacter pylori strain 26695 induces apoptosis of AGS cells. Biomed Environ Sci. 2012;25:614–619. doi: 10.3967/0895-3988.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 73.Toledo H, Valenzuela M, Rivas A, Jerez CA. Acid stress response in Helicobacter pylori. FEMS Microbiol Lett. 2002;213:67–72. doi: 10.1111/j.1574-6968.2002.tb11287.x. [DOI] [PubMed] [Google Scholar]
- 74.Sisinni L, Cendron L, Favaro G, Zanotti G. Helicobacter pylori acidic stress response factor HP1286 is a YceI homolog with new binding specificity. FEBS J. 2010;277:1896–1905. doi: 10.1111/j.1742-4658.2010.07612.x. [DOI] [PubMed] [Google Scholar]
- 75.Spreng S, Gentschev I, Goebel W, Mollenkopf H, Eck M, Müller-Hermelink HK, Schmausser B. Identification of immunogenic antigens of Helicobacter pylori via the Escherichia coli hemolysin secretion system (1) FEMS Microbiol Lett. 2000;186:251–256. doi: 10.1111/j.1574-6968.2000.tb09113.x. [DOI] [PubMed] [Google Scholar]
- 76.Weinberg MV, Maier RJ. Peptide transport in Helicobacter pylori: roles of dpp and opp systems and evidence for additional peptide transporters. J Bacteriol. 2007;189:3392–3402. doi: 10.1128/JB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stähler FN, Odenbreit S, Haas R, Wilrich J, Van Vliet AH, Kusters JG, Kist M, Bereswill S. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect Immun. 2006;74:3845–3852. doi: 10.1128/IAI.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park SJ, Jung YS, Kim JS, Seo MD, Lee BJ. Structural insight into the distinct properties of copper transport by the Helicobacter pylori CopP protein. Proteins. 2008;71:1007–1019. doi: 10.1002/prot.21957. [DOI] [PubMed] [Google Scholar]
- 79.Jönsson K, Guo BP, Monstein HJ, Mekalanos JJ, Kronvall G. Molecular cloning and characterization of two Helicobacter pylori genes coding for plasminogen-binding proteins. Proc Natl Acad Sci USA. 2004;101:1852–1857. doi: 10.1073/pnas.0307329101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montecucco C, de Bernard M. Molecular and cellular mechanisms of action of the vacuolating cytotoxin (VacA) and neutrophil-activating protein (HP-NAP) virulence factors of Helicobacter pylori. Microbes Infect. 2003;5:715–721. doi: 10.1016/s1286-4579(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 81.Boquet P, Ricci V. Intoxication strategy of Helicobacter pylori VacA toxin. Trends Microbiol. 2012;20:165–174. doi: 10.1016/j.tim.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 82.Chambers MG, Pyburn TM, González-Rivera C, Collier SE, Eli I, Yip CK, Takizawa Y, Lacy DB, Cover TL, Ohi MD. Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin. J Mol Biol. 2013;425:524–535. doi: 10.1016/j.jmb.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci USA. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Radin JN, Gaddy JA, González-Rivera C, Loh JT, Algood HM, Cover TL. Flagellar localization of a Helicobacter pylori autotransporter protein. MBio. 2013;4:e00613–e00612. doi: 10.1128/mBio.00613-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sause WE, Castillo AR, Ottemann KM. The Helicobacter pylori autotransporter ImaA (HP0289) modulates the immune response and contributes to host colonization. Infect Immun. 2012;80:2286–2296. doi: 10.1128/IAI.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suganuma M, Yamaguchi K, Ono Y, Matsumoto H, Hayashi T, Ogawa T, Imai K, Kuzuhara T, Nishizono A, Fujiki H. TNF-alpha-inducing protein, a carcinogenic factor secreted from H. pylori, enters gastric cancer cells. Int J Cancer. 2008;123:117–122. doi: 10.1002/ijc.23484. [DOI] [PubMed] [Google Scholar]
- 87.Tosi T, Cioci G, Jouravleva K, Dian C, Terradot L. Structures of the tumor necrosis factor alpha inducing protein Tipalpha: a novel virulence factor from Helicobacter pylori. FEBS Lett. 2009;583:1581–1585. doi: 10.1016/j.febslet.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 88.Jang JY, Yoon HJ, Yoon JY, Kim HS, Lee SJ, Kim KH, Kim do J, Jang S, Han BG, Lee BI, et al. Crystal structure of the TNF-alpha-Inducing protein (Tipalpha) from Helicobacter pylori: Insights into Its DNA-binding activity. J Mol Biol. 2009;392:191–197. doi: 10.1016/j.jmb.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 89.Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61:847–860. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 90.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 91.Windle HJ, Fox A, Ní Eidhin D, Kelleher D. The thioredoxin system of Helicobacter pylori. J Biol Chem. 2000;275:5081–5089. doi: 10.1074/jbc.275.7.5081. [DOI] [PubMed] [Google Scholar]
- 92.Gustafsson TN, Sandalova T, Lu J, Holmgren A, Schneider G. High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori. Acta Crystallogr D Biol Crystallogr. 2007;63:833–843. doi: 10.1107/S0907444907026303. [DOI] [PubMed] [Google Scholar]
- 93.Papinutto E, Windle HJ, Cendron L, Battistutta R, Kelleher D, Zanotti G. Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori. Biochim Biophys Acta. 2005;1753:240–246. doi: 10.1016/j.bbapap.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 94.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoon JY, Kim J, An DR, Lee SJ, Kim HS, Im HN, Yoon HJ, Kim JY, Kim SJ, Han BW, et al. Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori. Acta Crystallogr D Biol Crystallogr. 2013;69:735–746. doi: 10.1107/S0907444913001236. [DOI] [PubMed] [Google Scholar]
- 96.Roszczenko P, Radomska KA, Wywial E, Collet JF, Jagusztyn-Krynicka EK. A novel insight into the oxidoreductase activity of Helicobacter pylori HP0231 protein. PLoS One. 2012;7:e46563. doi: 10.1371/journal.pone.0046563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoon JY, Kim J, Lee SJ, Kim HS, Im HN, Yoon HJ, Kim KH, Kim SJ, Han BW, Suh SW. Structural and functional characterization of Helicobacter pylori DsbG. FEBS Lett. 2011;585:3862–3867. doi: 10.1016/j.febslet.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 98.Haas G, Karaali G, Ebermayer K, Metzger WG, Lamer S, Zimny-Arndt U, Diescher S, Goebel UB, Vogt K, Roznowski AB, et al. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics. 2002;2:313–324. doi: 10.1002/1615-9861(200203)2:3<313::aid-prot313>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 99.Sabarth N, Hurwitz R, Meyer TF, Bumann D. Multiparameter selection of Helicobacter pylori antigens identifies two novel antigens with high protective efficacy. Infect Immun. 2002;70:6499–6503. doi: 10.1128/IAI.70.11.6499-6503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Loewen PC, Carpena X, Rovira C, Ivancich A, Perez-Luque R, Haas R, Odenbreit S, Nicholls P, Fita I. Structure of Helicobacter pylori catalase, with and without formic acid bound, at 1.6 A resolution. Biochemistry. 2004;43:3089–3103. doi: 10.1021/bi035663i. [DOI] [PubMed] [Google Scholar]
- 101.Alfonso-Prieto M, Borovik A, Carpena X, Murshudov G, Melik-Adamyan W, Fita I, Rovira C, Loewen PC. The structures and electronic configuration of compound I intermediates of Helicobacter pylori and Penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations. J Am Chem Soc. 2007;129:4193–4205. doi: 10.1021/ja063660y. [DOI] [PubMed] [Google Scholar]
- 102.Mori M, Suzuki H, Suzuki M, Kai A, Miura S, Ishii H. Catalase and superoxide dismutase secreted from Helicobacter pylori. Helicobacter. 1997;2:100–105. doi: 10.1111/j.1523-5378.1997.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 103.Mahawar M, Tran V, Sharp JS, Maier RJ. Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. J Biol Chem. 2011;286:19159–19169. doi: 10.1074/jbc.M111.223677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seyler RW, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001;69:4034–4040. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Esposito L, Seydel A, Aiello R, Sorrentino G, Cendron L, Zanotti G, Zagari A. The crystal structure of the superoxide dismutase from Helicobacter pylori reveals a structured C-terminal extension. Biochim Biophys Acta. 2008;1784:1601–1606. doi: 10.1016/j.bbapap.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 106.Kern M, Simon J. Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim Biophys Acta. 2009;1787:646–656. doi: 10.1016/j.bbabio.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 107.Kaihovaara P, Höök-Nikanne J, Uusi-Oukari M, Kosunen TU, Salaspuro M. Flavodoxin-dependent pyruvate oxidation, acetate production and metronidazole reduction by Helicobacter pylori. J Antimicrob Chemother. 1998;41:171–177. doi: 10.1093/jac/41.2.171. [DOI] [PubMed] [Google Scholar]
- 108.Freigang J, Diederichs K, Schäfer KP, Welte W, Paul R. Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori. Protein Sci. 2002;11:253–261. doi: 10.1110/ps.28602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pyndiah S, Lasserre JP, Ménard A, Claverol S, Prouzet-Mauléon V, Mégraud F, Zerbib F, Bonneu M. Two-dimensional blue native/SDS gel electrophoresis of multiprotein complexes from Helicobacter pylori. Mol Cell Proteomics. 2007;6:193–206. doi: 10.1074/mcp.M600363-MCP200. [DOI] [PubMed] [Google Scholar]
- 110.Mittl PR, Schneider-Brachert W. Sel1-like repeat proteins in signal transduction. Cell Signal. 2007;19:20–31. doi: 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 111.Mittl PR, Lüthy L, Reinhardt C, Joller H. Detection of high titers of antibody against Helicobacter cysteine-rich proteins A, B, C, and E in Helicobacter pylori-infected individuals. Clin Diagn Lab Immunol. 2003;10:542–545. doi: 10.1128/CDLI.10.4.542-545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dumrese C, Slomianka L, Ziegler U, Choi SS, Kalia A, Fulurija A, Lu W, Berg DE, Benghezal M, Marshall B, et al. The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype. FEBS Lett. 2009;583:1637–1643. doi: 10.1016/j.febslet.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwon DH, Dore MP, Kim JJ, Kato M, Lee M, Wu JY, Graham DY. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47:2169–2178. doi: 10.1128/AAC.47.7.2169-2178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deml L, Aigner M, Decker J, Eckhardt A, Schütz C, Mittl PR, Barabas S, Denk S, Knoll G, Lehn N, et al. Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent. Infect Immun. 2005;73:4732–4742. doi: 10.1128/IAI.73.8.4732-4742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roschitzki B, Schauer S, Mittl PR. Recognition of host proteins by Helicobacter cysteine-rich protein C. Curr Microbiol. 2011;63:239–249. doi: 10.1007/s00284-011-9969-2. [DOI] [PubMed] [Google Scholar]
- 116.Ogura M, Perez JC, Mittl PR, Lee HK, Dailide G, Tan S, Ito Y, Secka O, Dailidiene D, Putty K, et al. Helicobacter pylori evolution: lineage- specific adaptations in homologs of eukaryotic Sel1-like genes. PLoS Comput Biol. 2007;3:e151. doi: 10.1371/journal.pcbi.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kersulyte D, Kalia A, Gilman RH, Mendez M, Herrera P, Cabrera L, Velapatiño B, Balqui J, Paredes Puente de la Vega F, Rodriguez Ulloa CA, et al. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One. 2010;5:e15076. doi: 10.1371/journal.pone.0015076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choi HP, Juarez S, Ciordia S, Fernandez M, Bargiela R, Albar JP, Mazumdar V, Anton BP, Kasif S, Ferrer M, et al. Biochemical Characterization of Hypothetical Proteins from Helicobacter pylori. PLoS One. 2013;8:e66605. doi: 10.1371/journal.pone.0066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoy B, Löwer M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schröder P, Sewald N, Backert S, Schneider G, et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010;11:798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dunn BE, Grütter MG. Helicobacter pylori springs another surprise. Nat Struct Biol. 2001;8:480–482. doi: 10.1038/88527. [DOI] [PubMed] [Google Scholar]
- 121.Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J Bacteriol. 2005;187:729–738. doi: 10.1128/JB.187.2.729-738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J Bacteriol. 2007;189:2426–2434. doi: 10.1128/JB.01492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schmees C, Prinz C, Treptau T, Rad R, Hengst L, Voland P, Bauer S, Brenner L, Schmid RM, Gerhard M. Inhibition of T-cell proliferation by Helicobacter pylori gamma-glutamyl transpeptidase. Gastroenterology. 2007;132:1820–1833. doi: 10.1053/j.gastro.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 124.McGovern KJ, Blanchard TG, Gutierrez JA, Czinn SJ, Krakowka S, Youngman P. gamma-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect Immun. 2001;69:4168–4173. doi: 10.1128/IAI.69.6.4168-4173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Boanca G, Sand A, Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori gamma-glutamyltranspeptidase. J Biol Chem. 2006;281:19029–19037. doi: 10.1074/jbc.M603381200. [DOI] [PubMed] [Google Scholar]
- 126.Ki MR, Yun NR, Hwang SY. Glutamine-induced production and secretion of Helicobacter pylori gamma-glutamyltranspeptidase at low pH and its putative role in glutathione transport. J Microbiol Biotechnol. 2013;23:467–472. doi: 10.4014/jmb.1210.10035. [DOI] [PubMed] [Google Scholar]
- 127.Shibayama K, Kamachi K, Nagata N, Yagi T, Nada T, Doi Y, Shibata N, Yokoyama K, Yamane K, Kato H, et al. A novel apoptosis-inducing protein from Helicobacter pylori. Mol Microbiol. 2003;47:443–451. doi: 10.1046/j.1365-2958.2003.03305.x. [DOI] [PubMed] [Google Scholar]
- 128.Moss SF, Calam J, Agarwal B, Wang S, Holt PR. Induction of gastric epithelial apoptosis by Helicobacter pylori. Gut. 1996;38:498–501. doi: 10.1136/gut.38.4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Terry K, Go AC, Ottemann KM. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol Microbiol. 2006;61:871–882. doi: 10.1111/j.1365-2958.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- 130.Lam KH, Ling TK, Au SW. Crystal structure of activated CheY1 from Helicobacter pylori. J Bacteriol. 2010;192:2324–2334. doi: 10.1128/JB.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jiménez-Pearson MA, Delany I, Scarlato V, Beier D. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology. 2005;151:3299–3311. doi: 10.1099/mic.0.28217-0. [DOI] [PubMed] [Google Scholar]
- 132.O'Rourke EJ, Pinto AV, Petroni EA, Tolmasky ME, Ielpi L. Evidence for the active role of a novel nuclease from Helicobacter pylori in the horizontal transfer of genetic information. J Bacteriol. 2004;186:2586–2593. doi: 10.1128/JB.186.9.2586-2593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shaik MM, Cendron L, Percudani R, Zanotti G. The structure of Helicobacter pylori HP0310 reveals an atypical peptidoglycan deacetylase. PLoS One. 2011;6:e19207. doi: 10.1371/journal.pone.0019207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Skouloubris S, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997;25:989–998. doi: 10.1111/j.1365-2958.1997.mmi536.x. [DOI] [PubMed] [Google Scholar]
- 135.Skouloubris S, Labigne A, De Reuse H. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol Microbiol. 2001;40:596–609. doi: 10.1046/j.1365-2958.2001.02400.x. [DOI] [PubMed] [Google Scholar]
- 136.Zanotti G, Cendron L. Functional and structural aspects of Helicobacter pylori acidic stress response factors. IUBMB Life. 2010;62:715–723. doi: 10.1002/iub.382. [DOI] [PubMed] [Google Scholar]
- 137.Chu CH, Lai YJ, Huang H, Sun YJ. Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori. Proteins. 2008;71:396–406. doi: 10.1002/prot.21709. [DOI] [PubMed] [Google Scholar]
- 138.Liechti G, Goldberg JB. Helicobacter pylori relies primarily on the purine salvage pathway for purine nucleotide biosynthesis. J Bacteriol. 2012;194:839–854. doi: 10.1128/JB.05757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bury-Moné S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 140.Lin YF, Wu MS, Chang CC, Lin SW, Lin JT, Sun YJ, Chen DS, Chow LP. Comparative immunoproteomics of identification and characterization of virulence factors from Helicobacter pylori related to gastric cancer. Mol Cell Proteomics. 2006;5:1484–1496. doi: 10.1074/mcp.M600111-MCP200. [DOI] [PubMed] [Google Scholar]
- 141.Abergel C, Bouveret E, Claverie JM, Brown K, Rigal A, Lazdunski C, Bénédetti H. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Structure. 1999;7:1291–1300. doi: 10.1016/s0969-2126(00)80062-3. [DOI] [PubMed] [Google Scholar]
- 142.González-López MA, Velázquez-Guadarrama N, Romero-Espejel ME, Olivares-Trejo Jde J. Helicobacter pylori secretes the chaperonin GroEL (HSP60), which binds iron. FEBS Lett. 2013;587:1823–1828. doi: 10.1016/j.febslet.2013.04.048. [DOI] [PubMed] [Google Scholar]
- 143.Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:1397–1406. doi: 10.1111/j.1365-2958.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- 144.Jang SB, Ma C, Lee JY, Kim JH, Park SJ, Kwon AR, Lee BJ. NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site. J Biochem. 2009;146:667–674. doi: 10.1093/jb/mvp105. [DOI] [PubMed] [Google Scholar]
- 145.Bennett HJ, Roberts IS. Identification of a new sialic acid-binding protein in Helicobacter pylori. FEMS Immunol Med Microbiol. 2005;44:163–169. doi: 10.1016/j.femsim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 146.Sun X, Ge R, Chiu JF, Sun H, He QY. Identification of Proteins Related to Nickel Homeostasis in Helicobater pylori by Immobilized Metal Affinity Chromatography and Two-Dimensional Gel Electrophoresis. Met Based Drugs. 2008;2008:289490. doi: 10.1155/2008/289490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Qu W, Zhou Y, Sun Y, Fang M, Yu H, Li W, Liu Z, Zeng J, Chen C, Gao C, et al. Identification of S-nitrosylation of proteins of Helicobacter pylori in response to nitric oxide stress. J Microbiol. 2011;49:251–256. doi: 10.1007/s12275-011-0262-7. [DOI] [PubMed] [Google Scholar]
- 148.Cioci G, Terradot L, Dian C, Mueller-Dieckmann C, Leonard G. Crystal structure of HP0721, a novel secreted protein from Helicobacter pylori. Proteins. 2011;79:1678–1681. doi: 10.1002/prot.22988. [DOI] [PubMed] [Google Scholar]
- 149.Malke H. Cytoplasmic membrane lipoprotein LppC of Streptococcus equisimilis functions as an acid phosphatase. Appl Environ Microbiol. 1998;64:2439–2442. doi: 10.1128/aem.64.7.2439-2442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gutiérrez-Martínez E, Fernández-Ulibarri I, Lázaro-Diéguez F, Johannes L, Pyne S, Sarri E, Egea G. Lipid phosphate phosphatase 3 participates in transport carrier formation and protein trafficking in the early secretory pathway. J Cell Sci. 2013;126:2641–2655. doi: 10.1242/jcs.117705. [DOI] [PubMed] [Google Scholar]
- 151.Hoy B, Geppert T, Boehm M, Reisen F, Plattner P, Gadermaier G, Sewald N, Ferreira F, Briza P, Schneider G, et al. Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J Biol Chem. 2012;287:10115–10120. doi: 10.1074/jbc.C111.333419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. 2012;2:92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liechti G, Goldberg JB. Outer membrane biogenesis in Escherichia coli, Neisseria meningitidis, and Helicobacter pylori: paradigm deviations in H. pylori. Front Cell Infect Microbiol. 2012;2:29. doi: 10.3389/fcimb.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsuge H, Tsurumura T, Utsunomiya H, Kise D, Kuzuhara T, Watanabe T, Fujiki H, Suganuma M. Structural basis for the Helicobacter pylori-carcinogenic TNF-alpha-inducing protein. Biochem Biophys Res Commun. 2009;388:193–198. doi: 10.1016/j.bbrc.2009.07.121. [DOI] [PubMed] [Google Scholar]
- 155.Rathinavelu S, Kao JY, Zavros Y, Merchant JL. Helicobacter pylori outer membrane protein 18 (Hp1125) induces dendritic cell maturation and function. Helicobacter. 2005;10:424–432. doi: 10.1111/j.1523-5378.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 156.Lüthy L, Grütter MG, Mittl PR. The crystal structure of Helicobacter cysteine-rich protein C at 2.0 A resolution: similar peptide-binding sites in TPR and SEL1-like repeat proteins. J Mol Biol. 2004;340:829–841. doi: 10.1016/j.jmb.2004.04.055. [DOI] [PubMed] [Google Scholar]
- 157.Slomiany BL, Piotrowski J, Slomiany A. Anti-Helicobacter pylori activities of ebrotidine. A review of biochemical and animal experimental studies and data. Arzneimittelforschung. 1997;47:475–482. [PubMed] [Google Scholar]
- 158.Tannaes T, Dekker N, Bukholm G, Bijlsma JJ, Appelmelk BJ. Phase variation in the Helicobacter pylori phospholipase A gene and its role in acid adaptation. Infect Immun. 2001;69:7334–7340. doi: 10.1128/IAI.69.12.7334-7340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


