Protein modification is extremely useful for both understanding of protein structure and function and the mechanism of the biological pathways that the protein is involved. For example, protein labeling can facilitate protein localization, binding partner identification, and purification both under native or denaturing conditions.[1] Furthermore, modification of protein can expand the proteins’ functional capacity, especially, for therapeutic proteins, which is very essential for enhancing their pharmacodynamic and pharmacokinetic properties.[2] The key point for a practical protein modification is to carry out site-specific chemistry so as to avoid upsetting the protein activity due to random modification since there is a diversity of chemically reactive functionalities in the protein in general. Particularly, site-specific attachment of two or more different functionalities to a protein is even more challenging than introducing a single modification but is highly sought after.[3] For example, introducing an analytical probe at one site for protein tracking and a functional moiety at the other site for expanding its functional capacity offers an enormous way to generate therapeutic proteins with enhanced biological activity and stability while affording efficient biological activity evaluation both in vitro and in vivo as well.
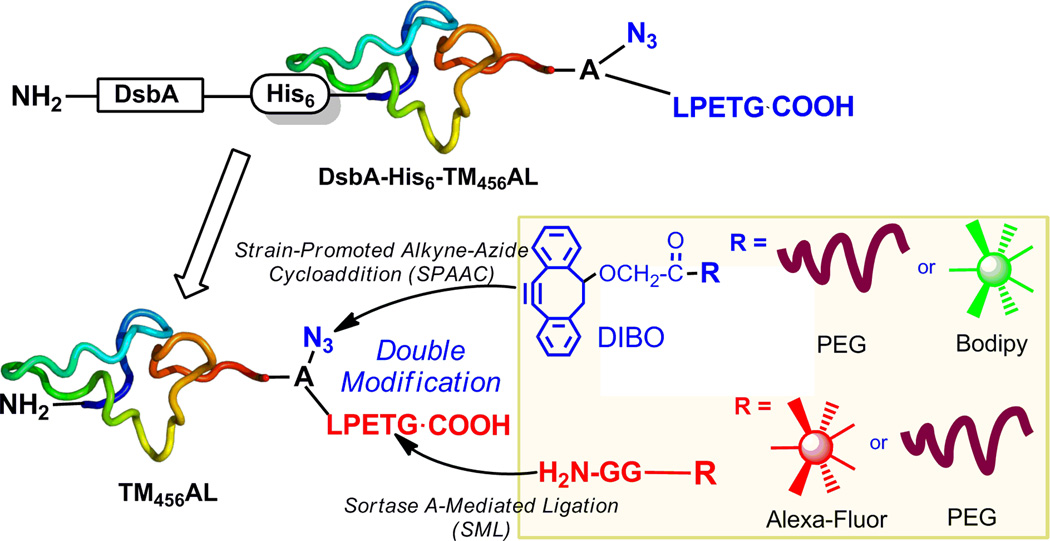
Bio-orthogonal chemistry has been developed for selective and efficient modification of proteins.[4] The most widely used approaches are azide-reactive Staudinger ligation, copper(I)-catalyzed click chemistry and strain-promoted alkyne-azide cycloaddition (SPAAC).[5] These approaches benefit from the azide functionality since it is small, bears no overall charge and can be introduced with relative ease to a wide variety of structures, often without disruption to biological behavior of proteins. On the other hand, enzymatic ligation has been explored for selective protein modification in native condition as well.[6] For example, sortase A-mediated ligation (SML) has been utilized to site-specifically modify proteins.[7] Specifically, SrtA recognizes a unique pentapeptide LPXTG, where X is variable, of the C-terminal domain of target proteins and transfers the carboxylic group of Thr to a substrate carrying an N-terminal glycine so to afford the transpeptidation products. Herein, we envision that the combination of SPAAC and SML will provide a practical strategy for site-specific double modification of proteins with specific purpose, such as site-specific poly(ethylene glycol) (PEG) modification and fluorescent labeling (Figure 1).
Figure 1.
Domain structures of recombinant TM456-derivertives and its site-specific double modification strategy via SPAAC and SML.
Thrombomodulin (TM), a membrane glycoprotein predominantly expressed by vascular endothelial cells, is involved in many biological processes such as thrombosis and inflammation.[8] Structurally, human TM consists of a single polypeptide chain of 557 amino acids with five distinct domains: a lectin-like domain, a domain with six epidermal growth factor (EGF)-like structures (EGF1-6), an O-glycosylation-rich domain, a transmembrane domain, and a cytoplasmic domain, each responsible for different biological functions.[9] It has been reported that EGF 4–6 (TM456) of the second domain is the key structure of TM involved in anticoagulant activity via a thrombin-mediated protein C activation mechanism.[10] Briefly, TM binds with thrombin via EGF56 to form a TM-thrombin complex, which then interacts with protein C via EGF4 to accelerate the activation of protein C. Recombinant TM456 has showed promising antithrombotic activity, however, the short half-life (6–9 min) limits its therapeutic application as an anticoagulant agent.[11] Modification with polymer such as PEG should be a choice to enhance the pharmacokinetics of recombinant TM456.[12] In addition, incorporation of a tag to the recombinant TM456 for subsequent detection or affinity purification will facilitate efficient biological evaluation for both in vitro and in vivo experiments. Herein, we report a straightforward and robust site-specific double modification of recombinant TM456, namely, PEGylation via CFCC and tagging with a variety of functionalities such as fluorescent dyes or affinity handles via SML concurrently (Figure 1).
As mentioned above, all three EGF domains of TM456 are critical for the interaction of TM with thrombin and protein C. Thus, we proposed a site-specific modification at the C-terminal of TM456 without diminishing its activity. In our previous study, we expressed a TM456 derivative with C-terminal LPETG tag for its end-point immobilization via SML, where the activity of immobilized TM456 was successfully retained.[13] In the present study, we designed a recombinant TM456 derivative with azidohomoalanine for SPAAC modification and LPETG tag for the recognition of SrtA both at the C-terminal of TM456 (TM456-Azide-LPETG, named as TM456AL) (Figure 1) (amino acid sequence see Supporting Information S1). In addition, since TM456 contains 9 pairs of disulfide bonds, for the proper folding of recombinant protein, a DsbA together with a His tag were fused to the N-terminal of TM456 protein (DsbA-His6-TM456AL), which was expressed in E. coli. B834. The targeted TM456AL was obtained after enzymatic digestion of DsbA-His6-TM456AL, followed by removal of DsbA-His6 with Nickel affinity column and further purification with HiTrap Q chromatography (Supporting Information S2). The resultant TM456AL was analyzed by SDS-PAGE (Figure 2A) and MALDI-TOF MS (Supporting Information S3), in which the molecular weight was exclusively consistent with the azide-incorporated target protein.
Figure 2.
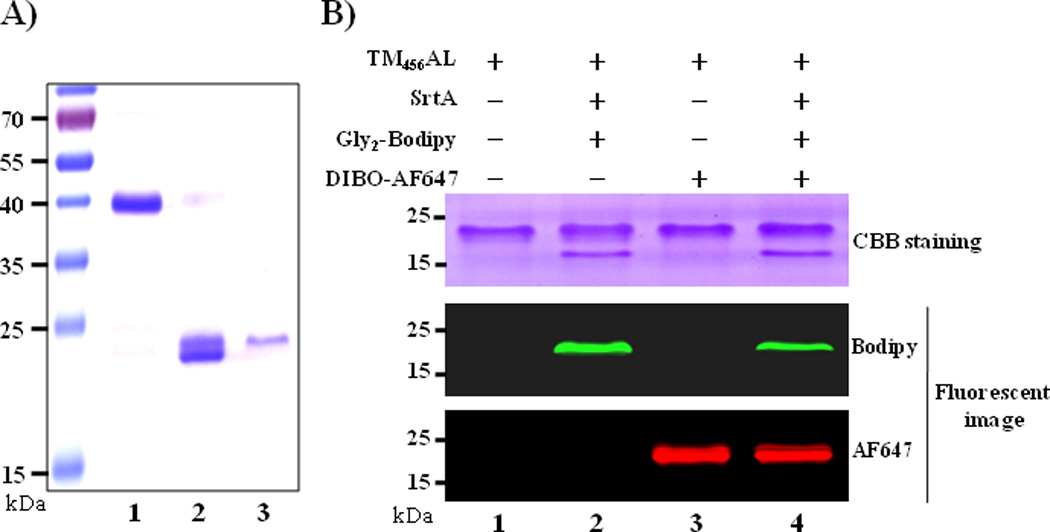
(A) SDS-PAGE analysis of purified recombinant TM proteins: Lane 1. DsbA-His6-TM456AL fusion protein, lane 2. Thrombin treated DsbA-His6-TM456AL, lane 3. Purified TM456AL from HiTrap Q column eluate; (B) SDS-PAGE analysis and in-gel fluorescence analysis of the double labeling reactions of TM456AL (10 µM) with DIBO-AF647 (10 µM) or/and Gly2-Bodipy (200 µM) in the presence of SrtA (2 µM) in 20 mM HEPEs (pH 7.4), 150 mM NaCl and 5 mM CaCl2 at 37 °C for 1 h.
With the purified TM456AL in hand, we first examined site-specific fluorescent labeling via SPAAC and SML, respectively. Two fluorescent probes with different wavelengths were used in order to selectively confirm the successful labeling. A commercial DIBO-Alexa Fluor 647 (DIBO-AF647) (Invitrogen) was used for SPAAC labeling, while a synthesized Gly2-Bodipy (Supporting Information S4) was used for SML labeling of TM456AL. As shown in the SDS-PAGE gel resulting from Coomassie blue staining and fluorescent imaging, the two fluorescent probes were successfully conjugated onto the TM456AL via SPAAC and SML, respectively (Figure 2B, lane 2 and 3). Next, double labeling of TM456AL was performed and the fluorescent intensity for both ligations were observed and appeared comparable (Figure 2B, lane 4). These results indicated that the one-pot SPAAC and SML reaction can be applied for site-specific double labeling of the TM456AL.
Studies show that SPAAC is very efficient in protein modification,[14] while SML is a reversible reaction[15] and thus requires further optimization to increase the overall efficiency of the proposed double-modification. A previous kinetic study of SML suggested some solutions to enhance its efficiency, such as using high concentration of LPXTG tagged substrate and excess of oligo-glycine nucleophile, as well as removal of product during reaction.[16] Therefore, we evaluated the reaction conditions that satisfy both ligation methods in the one-pot reaction of SPAAC and SML. First, a stable TM456AL concentration of 10 µM was optimized for all its modifications below since higher concentrations induce precipitations in the reaction solutions, which is most likely due to its rich cysteine residues that form multiple intermolecular disulfide bonds easily at high concentration. Then, the molar ratio between TM456AL and reactant dyes was investigated for both SPAAC and SML labeling (Supporting Information Figure S6A). As summarized in Table 1, 5 equivalents of DIBO-AF647 were required for a sufficient SPAAC-mediated labeling, while at least 20 equivalents of Gly2-Bodipy were required for SML to obtain a satisfactory labeling efficiency. In addition, the pH value was investigated for both reactions. As a result, pH value does not affect the SPAAC labeling, while slightly basic conditions (pH 7–9) benefit the SML reaction (Supporting Information S6B). By using the reaction conditions optimized above, reaction kinetics of both SML and SPAAC were investigated. Based on the fluorescence intensities of one-pot reaction products (Figure 3B), SPAAC-mediated labeling reached an approximate maximum in 2 hours, whereas, it took 3 hours for SML. It must be noted that the reaction time should not last indefinitely since SrtA irreversibly hydrolyzes the LPXTG motifs of both diglycine substrate and product as previously reported.[15] Taken together, an optimized reaction condition, 50 µM alkyne reactant, 400 µM diglycine nucleophiles and 5 µM SrtA in Tris buffer pH 8.0 with 5 mM Ca2+ for 3 hours, was used in the following one-pot double modification of TM456AL.
Table 1.
Optimized one-pot double modification reaction conditions*
| Factors | One-pot reaction condition | |
|---|---|---|
| SML | SPAAC | |
| Gly2-R1 : TM456AL (molar) | ≥ 20:1 | NA |
| Alkyne-R2 : TM456AL (molar) | NA | ≥5:1 |
| SrtA : TM456AL (molar) | 0.5–1 | NA |
| pH | 7.0 to 9.0 | |
| Reaction time | ≥ 3 hours | |
| Temperature | RT to 37 °C | |
Detail experiments see Supporting Information S6
Figure 3.

Optimization of reaction time of one-pot double labeling of TM456AL via SML and SPAAC: TM456AL (10 µM) was incubated with Gly2-Bodipy (400 µM), SrtA (5 µM) and DIBO-AF647 (50 µM) in 20 mM Tris (pH 8.0), 150 mM NaCl and 5 mM CaCl2 at 37 °C for different time (1–240 min): (A) SDS-PAGE followed by fluorescent scanning (Bodipy: Ex/Em=488/526 nm; AF647: Ex/Em = 633/670 nm) and (B) in-gel fluorescence intensities calculated and compared by Image J. The highest intensity of SML or SPAAC was set as 100%. Data points represent the mean ± SD from three independent experiments.
As mentioned above, PEGylation is a common method to stabilize protein and fluorescent dye labeling facilitates protein tracking in biological system. In the present study, we tested the feasibility of one-pot PEGylation and fluorescent labeling of TM456AL via SPAAC and SML. The first combination was SPAAC-mediated PEGylation with DBCO-PEG5000-OMe (Click-Chemistry Tools) and SrtA-mediated fluorescent labeling with Gly2-Bodipy. As shown in SDS-PAGE and western blot by using anti-human TM mAb (Figure 4A and 4B), double modification of TM456AL with PEG and Bodipy afforded a new band with a 7 kDa molecular weight increase, which corresponds to the molecular weight of DBCO-PEG5000-OMe. More than 90% TM456AL was PEGylated within 3 hours compared to the unreacted TM band in the control experiment, proving an excellent yield for SPAAC in one-pot reaction. Furthermore, a monoclonal Ab E11 (Academia Sinica), which specifically recognizes the backbone of PEG molecule, was used in western blot to confirm the PEG conjugates (Figure 4D) and fluorescent imaging was carried out to confirm Gly2-Bodipy anchored onto TM456 (Figure 4C), respectively. Moreover, the double modified product (Figure 4, lane 4) exhibited comparable signal strength to the individual reactions (Figure 4, lane 2 and 3), demonstrating that the SML and SPAAC could take place in one-pot reaction without interfering with each other.
Figure 4.

One-pot PEGylation via SPAAC and fluorescent labeling via SML of TM456AL: (A) SDS-PAGE Coomassie blue staining, (B) western blot analysis using anti-human TM mAb, (C) fluorescent image (Ex/Em = 488/526 nm), and (D) western blot analysis using anti-PEG mAb E11 of. The reactions of TM456AL (10 µM), SrtA (5 µM), Gly2-Bodipy (400 µM) and DBCO-PEG5000-OMe were taken place in 20 mM Tris (pH 8.0), 150 mM NaCl and 5 mM CaCl2 at 37°C for 3 h.
Alternatively, we examined the reverse set of the double modification, in which PEGylation with Gly2-PEG5000-OMe (Supporting Information S5) via SML and fluorescent labeling with DIBO-AF647 via SPAAC were performed. The PEGylated product around 35 kDa was confirmed by western blot using anti-PEG mAb (Figure 5A and 5B), the molecular weight being similar to that observed for PEGylation via SPAAC. However, this PEGylation product presented in low yield (less than 10%, when compared with control sample) (Figure 5A, lane2). In addition, a 40 kDa product was observed in a considerable amount, which was identified as the complex of TM456AL and SrtA (Figure 5A, lane 3 and 4). In SML, Cys184 first reacts with LPXTG tagged substrate to form LPXT-SrtA, a covalent acyl-enzyme intermediate, which is subsequently approached by the oligo-glycine nucleophile to form the conjugate of two substrates.[17] Our result indicated that the 40 kDa product was the acyl-SrtA intermediate and barely reacted with the second substrate Gly2-PEG5000-OMe, which was probably due to the steric issues resulting from the size of the target substrate. To confirm this speculation, SrtA-mediated modification of TM456AL with molecules of different sizes containing N-terminal diglycines were tested. Herein, FITC-labeled SrtA (Supporting Information S8) was used for easy confirmation of the intermediates in SDS-PAGE. As a result, large quantities of intermediates formed in the absence of oligo-glycine substrate (Figure 5C, lane 2), as well as in the presence of Gly2-PEG5000-OMe (Figure 5C, lane 6). In contrast, in the group of small nucleophiles, such as pentaglycine, Gly2-Biotin and Gly2-Dansyl, there was significantly less intermediates observed in the fluorescent image (Figure 5C, lane 3–5) but more conjugates formed (Supporting Information S7). These results indicated that small nucleophilic substrate resolves the acyl-enzyme intermediate more efficiently than large size substrates such as PEGs due to the steric issues.[18] Therefore, to cost effectively produce double-modified conjugates, we utilized SPAAC for PEGylation and SML for linking of diverse small molecules in larger scale reactions.
Figure 5.
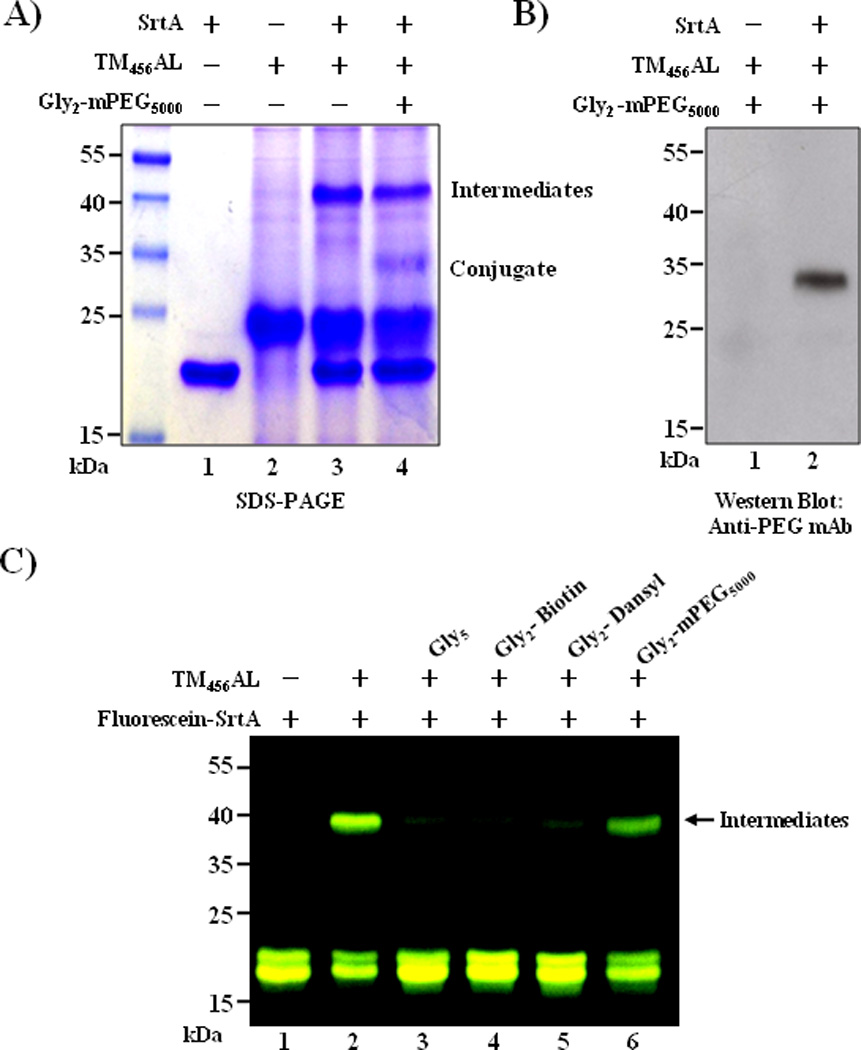
PEGylation of TM456AL via SML with Gly2-PEG5000-OMe: (A) SDS-PAGE analysis of reactions between SrtA (5 µM), TM456AL (10 µM) and Gly2-PEG5000-OMe (400 µM); (B) western blot analysis using anti-PEG mAb E11; (C) in-gel fluorescent image of reactions between FITC-labeled SrtA ((Ex/Em = 488/526 nm, 5 µM), TM456AL (10 µM) and diglycine nucleophiles (400 µM). All reactions were taken place in 20 mM Tris (pH 8.0), 150 mM NaCl and 5 mM CaCl2 at 37 °C for 3 h.
After confirming the proper ligation strategy, we further tested the influence of the double-modification on the activity of TM456AL modified with DBCO-PEG5000-OMe via SPAAC and three diglycine functionalities (Bodipy, Biotin and Dansyl) via SML. In this study, protein C (PC) activation activities of these conjugates (Supporting Information S9) were measured and compared with the unmodified TM456AL. The absorptions of activated protein C (APC) cleaved chromogenic substrates were measured at 405 nm and then compared. The rate of activating PC by thrombin is quite slow in the absence of TM456AL (Figure 6, column 1), while in the presence of TM456AL and those double-modified conjugates, the generation of APC was significantly enhanced (Figure 6, column 2–6). There was no obvious change in PC activation activities of dual-modified conjugates by compared to unmodified TM, indicating that the one-pot double modification does not hamper the TM456 activity. It has been reported that the activity decline of immobilized TM may be caused by random modification,[19] whereas in our previous work,[13] the activity of TM was fully retained after the site-specific functionalization and immobilization of TM via SML. Our study here further demonstrated that it is feasible to immobilize TM and introduce other functionalities at the same time without affecting its activity, which will be used in anticoagulant materials with multi-functions in the future.
Figure 6.
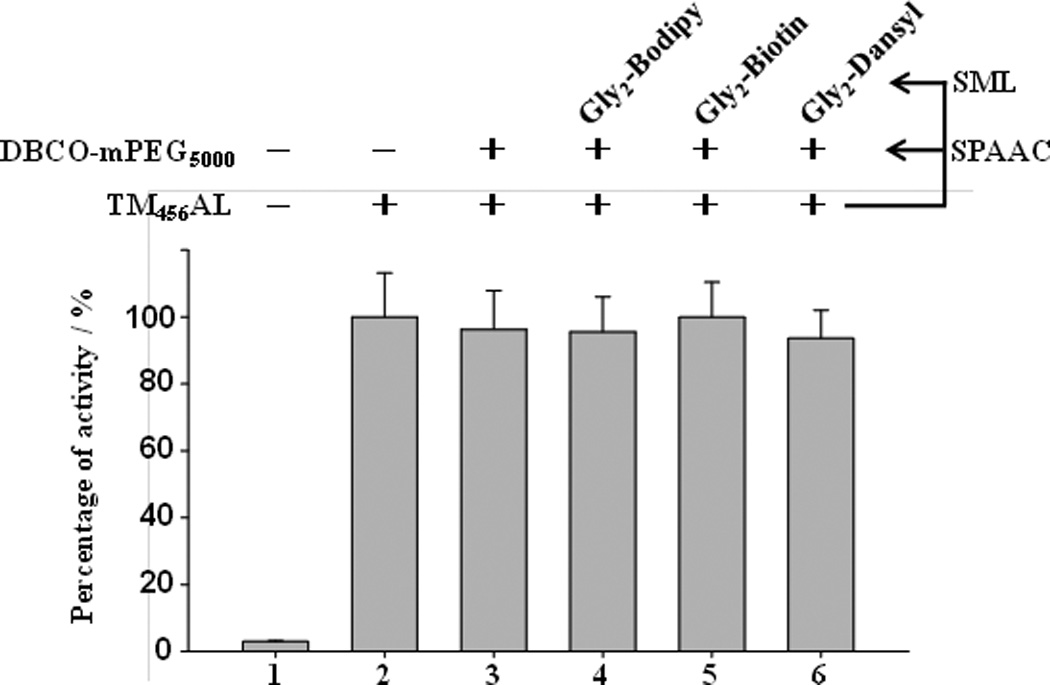
Activated protein C (APC) generation by TM456AL and its derivatives via SPAAC and SML modification. 100% was defined for APC generation by TM456AL and all others were compared with TM456AL. Data points represent the mean ± SD, for three independent measurements. Detail assay conditions are described in the Supporting Information S8.
In conclusion, we have developed a one-pot/site-specific strategy for double modification of recombinant thrombomodulin via strain-promoted alkyne-azide cycloaddition (SPAAC) and sortase A-mediated ligation (SML) without upsetting its anticoagulatory activity. Both reactions have advantages of mild reaction conditions and occurring in a single step without prior chemical modification of the target protein. We found that SPAAC is more efficient for modification of TM with both large and small substrates. However, SML requires high concentration of protein-LPXTG and diglycine containing substrate to reach a satisfying reaction yield. Particularly, our results suggest that SML is more suited for protein modification with small molecules, such as fluorescent dye and biotin. In the case of some large substrates or the substrates with poor solubility, SML generates excess intermediate byproduct and gave low yield of product. In this study, we took the advantages of each ligation reaction to doubly modify the C-terminal of TM456 via one-pot reaction. We anticipate that the double-modified antithrombotic rTM456 with PEGylated and fluorescence-labeled will show an enhanced pharmacokinetic property with easy fluorescent probing feature in both in vitro and in vivo assay. We believe that this strategy is viable for modification of a variety of proteins with different molecules for elevated activity and stability, novel functions, easy bioassay capacity, and other specific properties of interest as well.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH (5R01HL102604-04, X.-L. Sun), National Science Foundation MRI Grant (CHE-1126384, X.-L. Sun) and Cleveland State University Faculty Research Development Grant.
References
- 1.Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solá RJ, Griebenow K. Biodrugs. 2010;24:9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heal WP, Wright MH, Thinon E, Tate EW. Nat. Protoc. 2011;7:105–117. doi: 10.1038/nprot.2011.425. [DOI] [PubMed] [Google Scholar]
- 4.Willems LI, van der Linden WA, Li N, Li KY, Liu N, Hoogendoorn S, van der Marel GA, Florea BI, Overkleeft HS. Acc Chem Res. 2011;44:718–729. doi: 10.1021/ar200125k. [DOI] [PubMed] [Google Scholar]
- 5.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS. Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 6.Wu Z, Guo Z. J. Carbohydr. Chem. 2012;31:48–86. doi: 10.1080/07328303.2011.635251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukiji S, Nagamune T. Chem BioChem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 8.Morse J. Current Drug Targets. 2012;13:421–431. doi: 10.2174/138945012799424606. [DOI] [PubMed] [Google Scholar]
- 9.Weiler H, Isermann BH. J. Thromb. Haemost. 2003;1:1515–1524. doi: 10.1046/j.1538-7836.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 10.Zushi M, Gomi K, Yamamoto S, Maruyama I, Hayashi T, Suzuki K. J. Biol. Chem. 1989;264:1035110353. [PubMed] [Google Scholar]
- 11.Suzuki M, Mohri M, Yamamoto S. Thromb Haemost. 1998;79:417–422. [PubMed] [Google Scholar]
- 12.Cazalis CS, Haller CA, Sease-Cargo L, Chaikof EL. Bioconjug. Chem. 2004;15:1005–1009. doi: 10.1021/bc049903y. [DOI] [PubMed] [Google Scholar]
- 13.Jiang R, Weingart J, Zhang H, Ma Y, Sun XL. Bioconjug. Chem. 2012;23:643–649. doi: 10.1021/bc200661w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang S, Sachin K, Lee HJ, Kim DW, Lee HS. Bioconjugate Chem. 2012;23:2256–2261. doi: 10.1021/bc300364z. [DOI] [PubMed] [Google Scholar]
- 15.Yamamura Y, Hirakawa H, Yamaguchi S, Nagamune T. Chem. Commun. 2011;47:4742–4744. doi: 10.1039/c0cc05334a. [DOI] [PubMed] [Google Scholar]
- 16.Pritz S, Wolf Y, Kraetke O, Klose J, Bienert M, Beyermann M. J. Org. Chem. 2007;72:3909–3912. doi: 10.1021/jo062331l. [DOI] [PubMed] [Google Scholar]
- 17.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Proc. Natl. Acad. Sci. U S A. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antos JM, Miller GM, Grotenbreg GM, Ploegh HL. J. Am. Chem. Soc. 2008;130:16338–16343. doi: 10.1021/ja806779e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh HY, Lin JC. J. Biomater. Sci. Polym. Ed. 2009;20:807–819. doi: 10.1163/156856209X426952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.