Structure–function analyses driven by a crystal structure of the cytosolic domain of the Drp1 receptor MiD51 reveals a nucleotidyltransferase fold and nucleotide binding activity that is independent of its Drp1 binding activity.
Abstract
Mitochondrial fission is important for organelle transport, inheritance, and turnover, and alterations in fission are seen in neurological disease. In mammals, mitochondrial fission is executed by dynamin-related protein 1 (Drp1), a cytosolic guanosine triphosphatase that polymerizes and constricts the organelle. Recruitment of Drp1 to mitochondria involves receptors including Mff, MiD49, and MiD51. MiD49/51 form foci at mitochondrial constriction sites and coassemble with Drp1 to drive fission. Here, we solved the crystal structure of the cytosolic domain of human MiD51, which adopts a nucleotidyltransferase fold. Although MiD51 lacks catalytic residues for transferase activity, it specifically binds guanosine diphosphate and adenosine diphosphate. MiD51 mutants unable to bind nucleotides were still able to recruit Drp1. Disruption of an additional region in MiD51 that is not part of the nucleotidyltransferase fold blocked Drp1 recruitment and assembly of MiD51 into foci. MiD51 foci are also dependent on the presence of Drp1, and after scission they are distributed to daughter organelles, supporting the involvement of MiD51 in the fission apparatus.
Introduction
Mitochondria are highly dynamic organelles and undergo continuous fission and fusion events, a balance of which is vital for their function and cellular distribution (Westermann, 2010). Mutations in genes associated with mitochondrial dynamics have been linked with peripheral and optic neuropathies (Alexander et al., 2000; Delettre et al., 2000; Züchner et al., 2004; Davies et al., 2007) and fatal developmental abnormalities (Ishihara et al., 2009), whereas defects in mitochondrial dynamics have been implicated in Parkinson’s, Alzheimer’s, and Huntington’s disease (Wang et al., 2008; Arduíno et al., 2011; Reddy et al., 2011). The primary regulator of fission, controlled through both posttranslational modifications and interactions with mitochondrial adaptor proteins, is the dynamin family member Drp1 (Dnm1 in yeast; Bui and Shaw, 2013; Elgass et al., 2013). Cytosolic Drp1 is recruited to mitochondrial constrictions, often where the endoplasmic reticulum crosses (Friedman et al., 2011). There, it polymerizes into spirals and through GTP-dependent conformational changes it constricts the organelle leading to membrane scission (Lackner et al., 2009). The crystal structures of dynamin and Drp1 revealed interfaces involved in protein oligomerization along with a mechanism for polymer constriction (Faelber et al., 2011; Ford et al., 2011; Fröhlich et al., 2013). Unlike dynamin-1, Drp1 lacks a pleckstrin homology domain and is recruited to mitochondria through outer membrane receptors Fis1 (Mozdy et al., 2000; Yoon et al., 2003; Stojanovski et al., 2004), Mff (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010), along with chordate-specific MiD49 and MiD51 (Palmer et al., 2011; Zhao et al., 2011). Individually, Mff, MiD49, and MiD51 have been shown to be sufficient to recruit Drp1 to drive fission (Koirala et al., 2013; Losón et al., 2013; Palmer et al., 2013), whereas the role of Fis1 as a Drp1 receptor has been questioned (Otera et al., 2010). Recently, addition of stoichiometric amounts of MiD49 was found to increase Drp1-mediated constrictions of liposomes from 31 to 15 nm, a point at which membrane scission is possible (Koirala et al., 2013). This indicates that MiD49/51 may be actively involved in facilitating Drp1 scission similar to the unrelated yeast adaptors Mdv1/Caf4 that are absent in higher eukaryotes (Bui and Shaw, 2013). MiD49/51 lack significant sequence similarity to any other proteins and they do not possess characteristic sequence domains/motifs. Here we determine the crystal structure of the cytosolic domain of MiD51 and identify a critical region in MiD51 important for Drp1 recruitment and assembly of the fission apparatus.
Results and discussion
MiD51 belongs to the nucleotidyltransferase fold superfamily of proteins
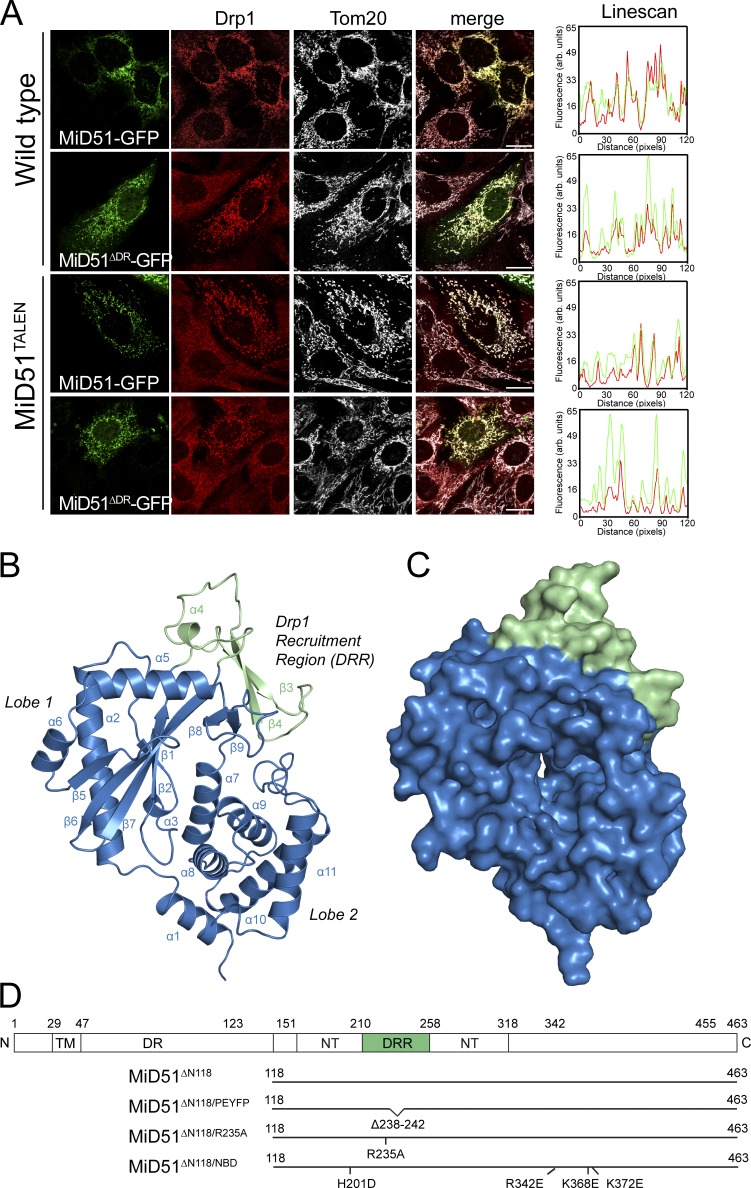
To search for a construct amenable to crystallization, we performed limited proteolysis on mouse MiD49 lacking its transmembrane domain. We identified a large (∼40 kD) trypsin-resistant fragment of MiD49 that lacks a predicted disordered region of the protein (Fig. S1 A). When human MiD51 lacking a homologous disordered region (residues 50–123) but still containing its N-terminal transmembrane anchor (termed MiD51ΔDR) was transiently expressed in mouse embryonic fibroblasts (MEFs), it was still able to recruit cytosolic Drp1 to mitochondria like full-length MiD51 (Fig. 1 A). We also confirmed this by expressing MiD51ΔDR in MEFs lacking endogenous MiD51 after TALEN-mediated gene disruption (Fig. 1 A and Fig. S1 B). These MEFs lacking MiD51 have more elongated mitochondria than control MEFs, consistent with a previous knockdown study (Palmer et al., 2011).
Figure 1.
Crystal structure of a MiD51 cytosolic domain sufficient for Drp1 recruitment. (A) MiD51-GFP and MiD51ΔDR-GFP were expressed in wild-type MEFs and MEFs lacking endogenous MiD51 (MiD51TALEN). Cells were subsequently immunostained for Drp1 and the mitochondrial marker protein Tom20 and visualized by fluorescence microscopy. (right) Linescans demonstrating colocalization of Drp1 with MiD51-GFP and MiD51ΔDR-GFP. Bars, 20 µm. (B) Structure of MiD51ΔN118 displaying the nucleotidyltransferase domain in blue and DRR domain in green. (C) Surface representation of MiD51ΔN118; view and coloring as in B. (D) Topology of MiD51 and illustration of mutant constructs used in this study. TM, transmembrane domain; DR, disordered region; NT, nucleotidyltransferase domain.
We crystallized recombinant, monomeric human MiD51ΔN118 (lacking its N-terminal transmembrane anchor and disordered region [Fig. S1 C]) and refined the native structure to Rwork and Rfree values of 0.223 and 0.263, respectively (Table 1). We observed clear and continuous electron density for residues 133–461 in all four copies of MiD51ΔN118 present in the asymmetric unit, with the remaining residues at the N terminus presumed to be disordered. Overall MiD51ΔN118 adopts a mixed α/β topology configured into two lobes separated by a deep cleft (Fig. 1, B and C; and Fig. S1 D). PISA analysis (Krissinel and Henrick, 2007) of the crystal packing identified an interface with complex formation significance scores ranging between 0.095 and 0.139, which may or may not imply an auxiliary role in complex formation, with complex formation significance scores of <0.1 considered not biologically relevant. A notable feature of this interface were pairs of equally charged acidic and basic residues, which would result in charge repulsion between two MiD51 monomers, suggesting that this interface is unlikely to have biological relevance. Structural comparisons reveal that MiD51 belongs to the nucleotidyltransferase fold superfamily of proteins. A DALI analysis (Holm and Rosenström, 2010) revealed that the closest homologues of MiD51 are cyclic GMP–AMP synthase (cGAS; PDB accession nos. 4K8V and 4JLX; Civril et al., 2013; Gao et al., 2013b), ILF2/NF45 (PDB accession no. 4AT8; Wolkowicz and Cook, 2012), and 2′–5′ oligoadenylate synthase 1 (PDB accession no. 1PX5; Hartmann et al., 2003). cGAS (Gao et al., 2013b) can be superimposed with MiD51 with a root mean square deviation (RMSD) of 2.4 Å over 300 Cα atoms (Fig. S1 E). cGAS binds cytosolic double-stranded DNA and synthesizes the second messenger cyclic GMP–AMP to trigger type I interferon induction as part of the innate immunity response (Sun et al., 2013). Both cGAS and MiD51 share an additional domain containing two β strands and a short helical turn that protrudes from the nucleotidyltransferase fold in lobe 1 (Fig. 1, B and D). Notable exceptions in MiD51 are (a) the absence of a zinc thumb that is present in cGAS and is involved in contacting double-stranded DNA and (b) the presence of a transmembrane anchor that is lacking in other known members of the superfamily.
Table 1.
Data collection and refinement statistics for crystals of the non-nucleotide–bound MiD51, the Xenon derivative used for SAD phasing, and various MiD51–nucleotide complexes
| Native MiD51 | Xe derivative | MiD51–GDP | MiD51–ADP | MiD51ΔPEYFP–GDP | MiD51ΔPEYFP–ADP | |
| Data collection | ||||||
| Wavelength (Å) | 0.9537 | 1.4586 | 0.9537 | 0.9537 | 1.4586 | 1.4586 |
| Space group | P1 | P1 | P1 | P1 | P43212 | P43212 |
| Cell dimensions | ||||||
| a, b, c (Å) | 72.72, 78.73, 79.36 | 73.14, 78.27, 80.42 | 72.34, 79.09, 80.05 | 72.57, 79.26, 79.38 | 57.72, 57.72, 255.40 | 57.53, 57.53, 253.75 |
| α, β, γ (°) | 66.26, 84.93, 64.07 | 65.17, 83.79, 62.39 | 65.81, 84.36, 64.08 | 65.44, 84.18, 63.35 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å)a | 40.67–2.12 (2.23–2.12) | 45.80–2.60 (2.74–2.60) | 40.00–2.30 (2.42–2.30) | 50.00–2.30 (2.42–2.30) | 50.00–2.55 (2.69–2.55) | 50.00–2.55 (2.69–2.55) |
| Rsym or Rmergea | 0.066 (0.500) | 0.059 (0.324) | 0.078 (0.447) | 0.082 (0.489) | 0.086 (0.499) | 0.083 (0.466) |
| I/σIa | 11.0 (2.8) | 16.6 (3.6) | 9.4 (2.2) | 10.8 (2.7) | 31.2 (7.9) | 28.7 (7.7) |
| Completeness (%)a | 97.3 (97.3) | 84.4 (44.2) | 98.1 (97.6) | 97.8 (97.3) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancya | 2.7 (2.8) | 3.5 (3.4) | 2.5 (2.5) | 3.5 (3.5) | 25.9 (27.5) | 22.3 (23.4) |
| Refinement | ||||||
| Resolution (Å)a | 39.01–2.12 (2.20–2.12) | – | 39.05–2.30 (2.33–2.30) | 39.19–2.30 (2.38–2.30) | 42.82–2.55 (2.71–2.55) | 42.62–2.55 (2.64–2.55) |
| No. of reflections | 79,549 | – | 63,108 | 62,135 | 15,007 | 14,827 |
| Rwork/Rfree | 0.2228/0.2626 | – | 0.1913/0.2375 | 0.2161/0.2477 | 0.2085/0.2452 | 0.1965/0.2414 |
| No. of atoms | ||||||
| Protein | 10,892 | – | 10,872 | 10,652 | 2,674 | 2,650 |
| Water | 527 | – | 356 | 341 | 114 | 94 |
| B factors | ||||||
| Protein | 48.81 | – | 48.03 | 45.70 | 56.05 | 52.13 |
| Water | 46.81 | – | 44.59 | 37.42 | 52.84 | 53.43 |
| RMSDs | ||||||
| Bond lengths (Å) | 0.004 | – | 0.005 | 0.006 | 0.003 | 0.005 |
| Bond angles (°) | 0.802 | – | 0.90 | 1.028 | 0.761 | 1.005 |
A single crystal was used for collection of each dataset.
Values in parentheses are for highest-resolution shell.
MiD51 specifically binds ADP and GDP
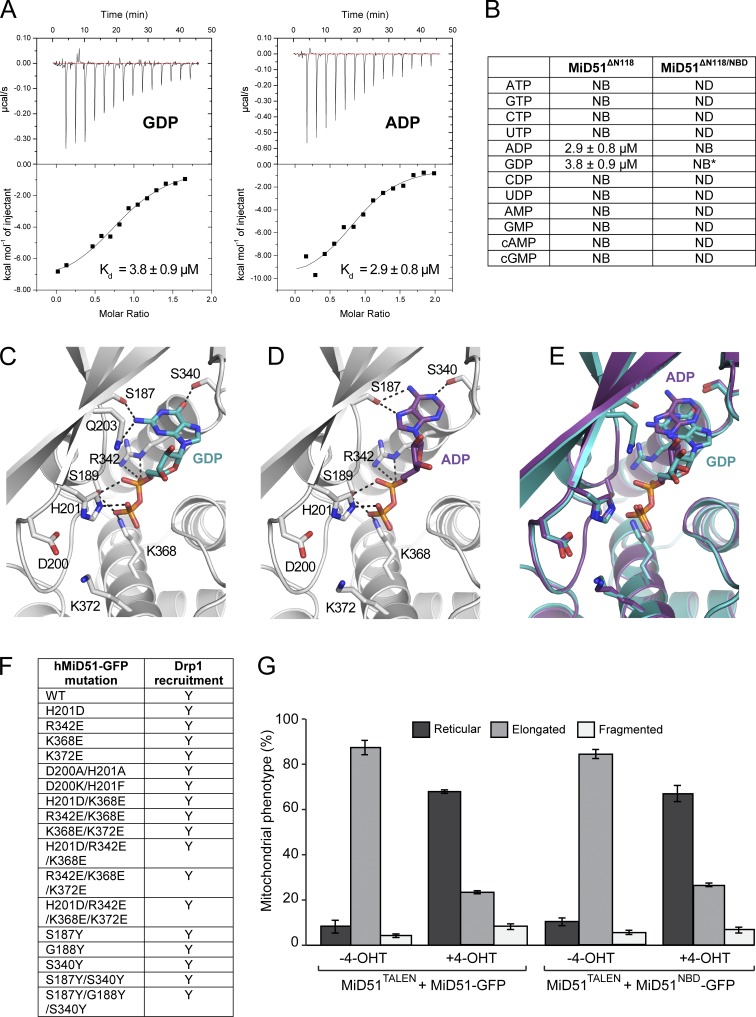
The region in MiD51 structurally homologous to the nucleotide binding site in cGAS and other structural homologues revealed additional electron density not associated with the polypeptide chain, which we were unable to assign (Fig. S2 A). Isothermal titration calorimetry (ITC) studies of MiD51 with various nucleotides showed specific binding to only ADP and GDP (Fig. 2, A and B) with affinities of 2.9 and 3.8 µM, respectively. We determined the crystal structures of MiD51 bound to either GDP or ADP (Fig. 2, C and D; and Table 1). Both structures revealed clear densities for a single nucleotide diphosphate (Fig. S2, B and C). Superimposition of the Cα backbone of MiD51 with MiD51–GDP or MiD51–ADP revealed no significant conformational changes, with RMSD variations ranging from 0.18 to 0.63 Å (Fig. S2 D). In the MiD51–ADP complex, Ser-187 and Ser-340 form hydrogen bonds with the purine, and Ser-189, His-201, Arg-342, and Lys-368 contact the phosphate groups (Fig. 2, C and D). In the MiD51–GDP complex, an additional hydrogen bond is formed by GDP with Gln-203 because of the purine group in GDP pointing inwards toward the back of the nucleotide binding pocket. It should be noted that the purine group of ADP is rotated away at a 180° angle compared with GDP, leading to an additional conformational shift in the ribose ring (Fig. 2 E).
Figure 2.
Analysis of MiD51 binding to GDP and ADP. (A) Raw heats of binding obtained using ITC by titration of MiD51ΔN118 with nucleotide. (B) Table of binding constants obtained by fitting ITC data to a single-state binding model. NB denotes no binding. Asterisk denotes n = 2. (C) Nucleotide binding pocket of MiD51 harboring GDP. Key MiD51ΔN118 residues are shown as sticks and GDP is shown in cyan. Hydrogen bonds are indicated by black dotted lines. (D) Nucleotide binding pocket of MiD51 harboring ADP. Key MiD51ΔN118 residues are shown as sticks and ADP is shown in magenta. Hydrogen bonds are indicated by black dotted lines. (E) Superposition of GDP-MiD51 (cyan) and ADP-MiD51 (magenta). (F) Summary of MiD51 mutants evaluated in immunofluorescence assays for Drp1 recruitment. (G) MiD51TALEN MEFs were induced to express MiD51-GFP or MiD51NBD-GFP with 4-OHT and mitochondrial morphology counted. Only MEFs with low-level GFP fluorescence were scored. n = 3; mean ± SEM; 100 cells counted/condition.
Catalytically active members of the nucleotidyltransferase family harbor a highly conserved set of acidic residues in their active sites ([DE]h[DE]h and h[DE]h), which coordinate divalent ions and activate hydroxyl acceptor groups in a substrate (Kuchta et al., 2009). In MiD51, only a single aspartic acid residue (Asp-200) is present in this region, indicating that MiD51 is unlikely to support nucleotide hydrolysis and transfer. Furthermore, no evidence for a catalytic Mg2+ ion site was discernible from the electron density, and cocrystallizing MiD51 with nucleotide and MnCl2 did not reveal bound Mn2+ ions in an anomalous difference Fourier map (Fig. S2, E and F). Thus MiD51 resembles a set of proteins, including NF45 and NF90 (Wolkowicz and Cook, 2012), which harbor a nucleotidyltransferase fold but have lost catalytic residues during evolution and therefore lack enzymatic activity.
We undertook extensive mutagenesis of residues involved in nucleotide binding (Fig. 2 F) and transiently expressed constructs in cells. In all cases, we found that Drp1 was still recruited to mitochondria (unpublished data). Simultaneous mutation of residues involved in contacting the phosphate backbone of ADP or GDP (K368E, K372E, R342E, and H201D) was found to disrupt nucleotide binding according to ITC measurements (Fig. 2 B), yet expression of this construct (termed nucleotide binding disruption, MiD51NBD) in MiD51TALEN MEFs still induced Drp1 recruitment to mitochondria (see Fig. 3 B). In addition, the mitochondrial network in MiD51TALEN MEFs shifted from an extended to a reticular phenotype after 4-hydroxytamoxifen (4-OHT) induced expression of either MiD51 or MiD51NBD (Fig. 2 G). Whether the nucleotide binding groove is physiologically relevant or whether mitochondria have simply adapted the fold for a novel purpose remains to be fully clarified.
Figure 3.
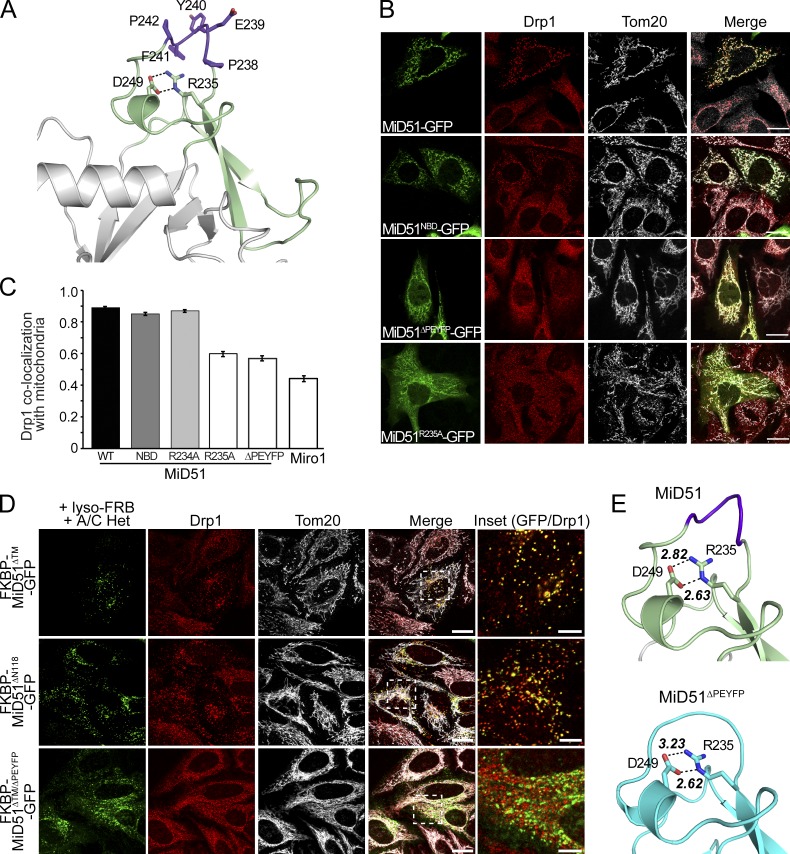
Identification of the MiD51 DRR. (A) Structure of the DRR (green) displaying residues deleted in MiD51ΔPEYFP (purple) and side chains of salt-bridging residues. (B) MiD51TALEN MEFs expressing GFP-tagged MiD51, MiD51ΔNBD, MiD51ΔPEYFP, and MiD51R235A (green) were fixed and immunostained for Drp1 (red) along with the mitochondrial marker Tom20 (gray). Bars, 20 µm. (C) Pearson correlation analysis of Drp1 colocalization on mitochondria with GFP-tagged MiD51WT, MiD51ΔNBD, MiD51ΔPEYFP, MiD51R234A, MiD51R235A, and Miro1 (control). n = 3; mean ± SEM; >20 linescans/experiment. (D) MiD51ΔPEYFP redirected to lysosomes cannot recruit Drp1. In the presence of the A/C Heterodimerizer (A/C Het), cytosolic FKBP-MiD51ΔTM-GFP, FKBP-MiD51ΔN118-GFP, and FKBP-MiD51ΔTM/ΔPEYFP-GFP are targeted to lysosomes by binding to lysosomal-targeted FRB (lyso-FRB). Bars: (merge) 20 µm; (inset) 5 µm. Cells were immunostained for Drp1 (red) and mitochondria (Tom20, gray). Images are shown in the merged panel and the inset is magnified in the rightmost panel, with GFP and Drp1 (red) shown. (E, top) Detailed view of the DRR (green) including key residues deleted in MiD51ΔPEYFP (purple), with black dotted lines indicating ionic interactions in a salt bridge and bond distances. (bottom) Crystal structure of the constricted DRR in MiD51ΔN118/PEYFP displaying salt-bridging residues and bond distances.
Identification of the Drp1 recruitment domain in MiD51
MiD51 contains an additional domain inserted into the nucleotidyltransferase fold (Fig. 1, B and D; and Fig. 3 A). To investigate if this region is involved in mediating mitochondrial fission, MiD51 mutants were expressed in MiD51TALEN MEFs. A MiD51-GFP fusion construct lacking a defined loop region corresponding to residues 238–242 (MiD51ΔPEYFP) was still targeted to mitochondria, yet, unlike MiD51-GFP, the active recruitment of Drp1 to the mitochondrial surface was blocked (Fig. 3 B). Similarly, a MiD51 mutant (MiD51R235A) that disrupts a salt bridge (R235-D249) located below this loop, was also unable to recruit Drp1 to mitochondria. Because of leaky GFP expression in this cell line, we repeated the expression in COS7 cells where such leakage was not observed (unpublished data). Again, Drp1 recruitment defects for MiD51ΔPEYFP and MiD51R235A were observed with these mutants resembling expression of the negative control protein Miro1 (Fig. 3 C). In contrast, an adjacent nonsalt bridge mutant (MiD51R234A) did not block Drp1 binding to mitochondria (Fig. 3 C). Similar findings were also observed when we induced the expression of untagged mouse MiD51 constructs in MiD51TALEN MEFs (Fig. S3, A and B).
We recently found that redirecting MiD51 to lysosomes by using a heterodimerizer assay led to the recruitment of Drp1 to that organelle (Palmer et al., 2013). This and other studies (Koirala et al., 2013; Losón et al., 2013) have shown that expression of MiD49 or MiD51 is sufficient for Drp1 recruitment and subsequent fission. We repeated this assay using MiD51ΔPEYFP and found that although this construct could be directed to lysosomes, Drp1 was not recruited (Fig. 3 D). In contrast, MiD51 constructs lacking only the transmembrane anchor (MiD51ΔTM) or additionally lacking the disordered region (MiD51ΔN118) were able to redirect Drp1 to lysosomes, hence underscoring the critical role of the β4–α4 loop in Drp1 recruitment.
To determine whether a loss of Drp1 recruitment by MiD51ΔPEYFP could be attributed to either misfolding or a conformational change in the Drp1 recruitment region (DRR), we determined the structures of MiD51ΔPEYFP bound to nucleotides (Fig. 3 E and Table 1). Superimposition of mutant and wild-type structures showed a distinct lack of conformational changes, with RMSD variation ranging from 1.09 to 1.32 Å (Fig. S3 C). Small differences are observed in helix α6 of MiD51ΔN118/PEYFP, which is located in a crystal contact, leading to the unwinding of two turns of helix. Loss of this secondary structure allows extension of α5 by five residues in MiD51ΔN118/PEYFP (residues 259–275) compared with MiD51ΔN118 (residues 259–270). Despite the shortened β4–α4 loop, the DRR in the deletion mutant exhibited a largely native conformation and the structurally important R235-D249 salt bridge was maintained (Fig. 3 E). A comparison of the electrostatic surface between wild type and MiD51ΔPEYFP revealed only small changes in the charge distribution between both structures (Fig. S3, D and E), suggesting that the topology of the β4–α4 loop is important for Drp1 recruitment. Notably, this loop is part of a region that is absent in all other structures determined for members of the nucleotidyltransferase fold superfamily with the exception of the recently determined structure of cGAS (Civril et al., 2013; Gao et al., 2013a). However, the β4–α4 loop is shorter in cGAS and the residues equivalent to PEYFP are located on the side of the loop rather than on the apex as is the case for MiD51.
Drp1 induces MiD51 assembly at foci that form scission sites
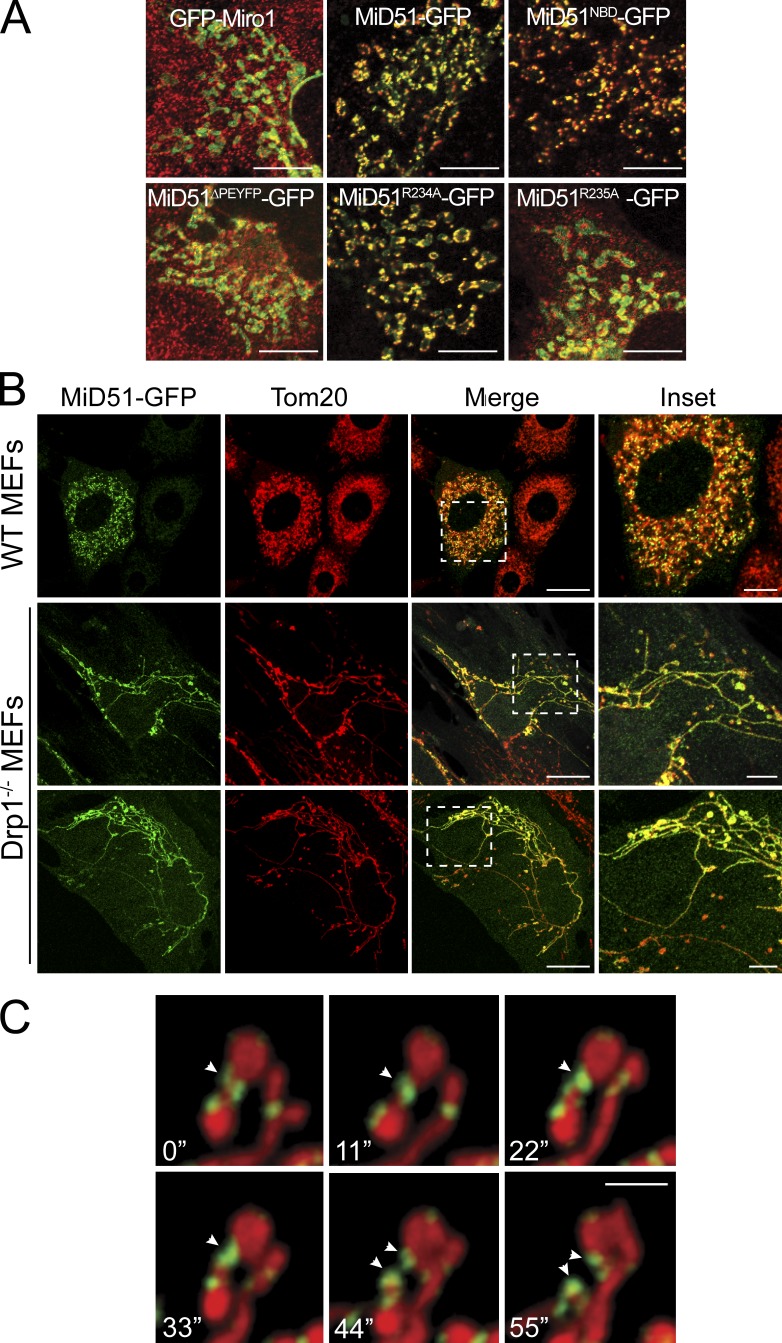
MiD51 and MiD49 can be found at foci at mitochondrial constriction sites, consistent with their role in fission (Palmer et al., 2011). Interestingly, MiD51 mutants still able to recruit Drp1 (e.g., MiD51NBD and MiD51R234A) also retained their ability to form foci at mitochondria, yet mutants MiD51ΔPEYFP and MiD51R235A that are deficient in Drp1 recruitment ability did not form foci and were instead diffusely distributed along the outer membrane like that of the control GFP-Miro1 (Fig. 4 A). We expressed MiD51-GFP in Drp1−/− MEFs (Wakabayashi et al., 2009) and found that it also did not assemble into foci and was instead distributed along the extended mitochondrial filaments (Fig. 4 B). We conclude that MiD49/51 assembly into foci is dependent on the presence of Drp1. Moreover, live-cell imaging of MiD51-GFP foci revealed that fission events indeed occurred at these sites and that a population of MiD51 in foci was inherited by each daughter organelle (Fig. 4 C and Videos 1 and 2). The distribution of MiD51 foci between organelles after a successful scission event also resembles that of yeast Drp1, Dnm1 (Bleazard et al., 1999). In yeast, Fis1 is a membrane receptor that helps assemble Mdv1p onto the mitochondrial surface where Mdv1p facilitates Dnm1 assembly into scission complexes (Tieu and Nunnari, 2000; Karren et al., 2005). Thus, MiD51 appears to act in a similar way to Mdv1p, yet it does not require an additional receptor because it is already anchored in the mitochondrial outer membrane. Both MiD51 and Mdv1p have different structural folds and hence the final scission complexes are likely to adopt different topologies. Consistent with this, it was recently proposed that variations in the sequence of the Insert B region of dynamin-related proteins accommodates diversity in adaptor binding (Ford et al., 2011; Bui and Shaw, 2013).
Figure 4.
Formation of MiD51 foci is dependent on Drp1. (A) MiD51TALEN MEFs expressing GFP-tagged Miro1, MiD51, MiD51NBD, MiD51ΔPEYFP, MiD51R234A, or MiD51R235A (green) were fixed and immunostained for Drp1 (red). Merged images show the absence or presence of foci at mitochondria. Bars, 10 µm. (B) Wild-type (WT) and Drp1−/− MEFs were transfected with MiD51-GFP (green) and fixed and immunostained for Tom20 (red). Bars: (merge) 20 µm; (inset) 5 µm. Dashed boxes represent insets shown in the far right panel. (C) Time-lapse imaging series of COS-7 cells transfected with MiD51-GFP (green) and stained for mitochondria with MitoTracker deep red. Arrowheads indicate the site of a fission event. Bar, 2 µm.
In conclusion, we found that MiD51 adopts a nucleotidyltransferase fold and can bind ADP and GDP. Considering the low micromolar affinities for both GDP and ADP, the ability of MiD51 to bind both nucleotides is intriguing, particularly in light of the absence of catalytic activity or ATP/GTP binding. Potential roles include nucleotide sensing or as an effector of Drp1’s GTPase activity. However, we found that fission events still occurred at foci formed by the MiD51 nucleotide binding mutant (Video 3). A more prosaic function such as nucleotides being a cofactor to stabilize MiD51 is therefore possible. The nucleotidyltransferase fold acts as a protein recruitment platform to assemble oligomers of the key mitochondrial fission regulator Drp1 into foci on the mitochondrial membrane. This unexpected functionality is mediated, at least in part, by a region inserted into the nucleotidyltransferase lobe that harbors the β4–α4 loop crucial for MiD51–Drp1 interactions. Intriguingly, it is not only the loss of the PEYFP motif in the loop, but also destabilization of the loop, that impairs Drp1 recruitment because mutation of R235, which forms a salt bridge directly below the β4–α4 loop, also abrogates Drp1 recruitment. This suggests that the loop topology may be a critical factor in Drp1 recruitment because loss of the loop does not adversely affect the protein fold and only minor changes in the charge distribution are seen after loop removal. Furthermore, the PEYFP sequence is not fully conserved in MiD49, which instead harbors a LEFCP motif in the β4–α4 loop, supporting the notion that it is loop topology rather than the precise amino acid sequence that is important. In a previous study, it was found that deletion of residues 160–169 in MiD51 also resulted in loss of Drp1 recruitment (Zhao et al., 2011). However, this region is a central part of the α2 helix and the deletion would most likely disrupt a key secondary structure element causing misfolding. Despite the identification of this critical loop element in MiD51 for Drp1 recruitment, the precise topology of the putative MiD51–Drp1 oligomer complex or its stoichiometry remains elusive. A previous study has suggested that MiD51 and Drp1 copolymerize, yet at least in vitro MiD51 does not appear to self-assemble into oligomers (Koirala et al., 2013). Whether or not the β4–α4 loop is the sole region of contact between MiD51 and Drp1 remains to be determined. Considering the proposed copolymerization of MiD51 and Drp1 it seems likely that other regions in MiD51 make direct contact with a Drp1 oligomer. Indeed, such contact surfaces may be necessary to achieve the reduction in diameter of Drp1 polymers from ∼31 to 15 nm to enable mitochondrial membrane scission. Additional contacts may also be mediated by other players of the fission machinery, including Mff, actin, and the endoplasmic reticulum (Friedman et al., 2011; Korobova et al., 2013). The interplay of these with MiD49/51 and Drp1 remains to be clarified.
Materials and methods
Accession numbers
Coordinates and structural factors have been deposited in the Protein Data Bank using accession numbers 4NXT, 4NXU, 4NXV, 4NXW, and 4NXX.
Expression and purification of MiD51
Mature MiD51 (UniProt accession no. Q9NQG6; aa 118–463) was cloned into the pGEX-4T1 expression vector (GE Healthcare). Plasmid DNA was transformed into chemically competent Escherichia coli (BL21(DE3)pLysS) as described previously (Sambrook and Russell, 2001) with positive transformants being selected for by carbenicillin addition to culture media. Recombinant protein was induced by addition of 0.5 mM IPTG at a culture density of OD600 ∼0.6, followed by 24-h expression at 22°C.
After harvesting and resuspension in GST binding buffer (0.1 M NaCl, 0.02 M Tris-Cl, pH 8.5, and 0.02 M EDTA), bacterial cells were lysed by freeze-thawing and sonication. Lysate was cleared by centrifugation (20,000 g for 30 min) and supernatant was applied to GST-resin (BD) loaded into a 20-ml gravity flow column. Resin was washed with 30 column volumes of wash buffer (PBS and 10% vol/vol glycerol) and eluted in 0.01 M of reduced glutathione and 0.05 M Tris, pH 8.5. Removal of the GST affinity tag occurred by in-solution cleavage with thrombin, followed by dialysis against PBS and collection of the flow-through upon reapplication to GST resin. Recombinant MiD51 was subjected to gel filtration (Superdex200 10/300 GL; GE Healthcare) where it eluted as a single peak and concentrated using Amicon Ultra 30,000 MWCO centrifugal filters (EMD Millipore) to a final concentration of 6 mg/ml. The identity of MiD51 was confirmed by mass spectrometry and protein concentration was determined using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). MiD51ΔN118/PEYFP and MiD51ΔN118/NBD were also purified using the protocol described.
Crystallization, derivatization, and structure determination of MiD51
Crystals of MiD51 and MiD51 complexes were obtained by hanging drop vapor diffusion in 16% (wt/vol) polyethylene glycol 8000, 0.10 M Hepes, pH 7.5, 10.0% (vol/vol) 2-propanol, and 0.2 M ammonium sulfate at 293 K by combining 1 µl of protein with 1 µl of crystallant over 500 µl of reservoir solution. Co-crystallization of nucleotides was performed by the addition of 10 mM nucleotide and 20 mM MnCl2, followed by flash-cooling in mother liquor supplemented with 15% glycerol, 0.02 M nucleotide, and 20 mM MnCl2. Xenon derivatization of native crystals was performed using a xenon chamber (Hampton) at the Australian Synchrotron. Native and derivative data were collected at the Australian Synchrotron, Beamline MX2, at 100 K and wavelengths of 0.9537 and 1.4586 Å, respectively. Initial experimental phases of native MiD51ΔN118 were obtained using Xe-SAD in PHENIX Autosol. Secondary structure elements were identified from the initial experimental map and modeled using polyalanine chains. Initial polyalanine models were used to improve xenon sites until the entire MiD51 chain could be traced. The polyalanine model was subsequently improved using automated building in Buccaneer (Cowtan, 2006, 2008), followed by manual rebuilding using Coot. The final model was refined to 2.0 Å with a final Rwork and Rfree of 0.218 and 0.266, respectively.
MiD51–GDP and MiD51–ADP complexes were solved by molecular replacement using nonnucleotide bound MiD51 as a search model. Crystallization of MiD51ΔN118/PEYFP in complex with GDP and ADP was achieved in 1.0 M sodium malonate, pH 6.0, 0.01 M nucleotide, and 0.02 M MnCl2. Crystals were cryoprotected in 15% glycerol and data were collected at 1.4586 Å for anomalous signal determination. The structure was solved by molecular replacement using a native MiD51 monomer as a search model. Refinement of all structures was performed using PHENIX (Adams et al., 2010). MolProbity validation revealed no residues in disallowed Ramachandran plot regions. Final crystallographic data and refinement statistics are summarized in Table 1.
ITC measurements
MiD51ΔN118 and MiD51ΔN118/NBD were diluted to 30 µM in buffer (0.02 M Tris-Cl, pH 8.0, 0.05 M NaCl, and 300 µM MgCl2) and subjected to calorimetry in an iTC200 system (MicroCal). Titrations of protein with nucleotides at 300 µM in buffer were performed at 25°C through 15 injections of 2.0 µl titrant. Fitting of experimental data to a single site binding model was analyzed using MicroCal Origin 7 SR4 (OriginLab). Three independent experiments were used to calculate binding affinities of ADP and GDP to MiD51ΔN118.
Antibodies, plasmids, and chemicals
Commercial antibodies used in this study were mouse anti-Drp1, mouse anti-OPA1 (BD), rabbit anti-MiD51(Proteintech), and rabbit anti-Tom20 (Santa Cruz Biotechnology, Inc.). Rabbit polyclonal antibodies against Mfn1 were provided by R. Youle and M. Lazarou (National Institutes of Health, Bethesda, MD). In-house polyclonal rabbit antibodies for Mff were raised against N-terminally GST-tagged human Mff (isoform 8; aa 2–222). Polyclonal rabbit antibodies for MiD49 were raised against N-terminally GST-tagged mouse MiD49 (aa 51–454). Polyclonal rabbit antibodies for Mfn2 and Fis1 were raised against human Mfn2 (aa 645–757) and human Fis1 (aa 2–122), respectively, as previously described (Stojanovski et al., 2004; Palmer et al., 2013).
MiD51-GFP and all MiD51 mutants were cloned into the GFP-N1 vector backbone (Takara Bio Inc.) with a CMV promoter at HindIII and BamHI to create in-frame GFP fusions. Miro1-GFP was cloned into the GFP-N1 vector backbone (Takara Bio Inc.) with a CMV promoter at NotI to create an in-frame GFP fusion. FKBP-containing plasmids (Lazarou et al., 2012) were supplied by R. Youle and M. Lazarou. 2× FKBP domain and MiD51ΔTM-GFP, MiD51ΔN118-GFP, or MiD51ΔTM/ΔPEYFP-GFP were PCR amplified and cloned into the pE-GFP backbone (Takara Bio Inc.) at BglII–HindIII, BamHI, and HindII, respectively, to create in-frame GFP fusion constructs under a CMV promoter. Lyso-FRB (LAMP1-FRB) was a gift from R. Youle and was constructed by inserting full-length rat LAMP-1 into the N terminus (EcoRI and XbaI sites) of the FRB domain in the pC4-RHE vector, containing a CMV promoter, replacing the C-terminal HA epitope tag or N-terminal methionine. A/C heterodimerizer (Takara Bio Inc.) was used at a final concentration of 250 nM for 3 h at 37°C. For lentiviral expression, the ORFs of human MiD51-GFP, human MiD51NBD-GFP, mouse MiD51-GFP, and mouse MiD51ΔPEYFP-GFP were cloned into the pF 5xUAS MCS SV40 vector using BglII and XbaI.
Cell culture and construction of MiD51TALEN MEFs
MEFs, COS-7, and HeLa cells were cultured as previously described (Palmer et al., 2011). Wild-type and Drp1−/− MEFs were previously obtained by Wakabayashi et al. (2009). Generation, selection, and induction of stable MiD51-inducible MEF cell lines were performed as previously described (Dunning et al., 2007). In brief, HEK293T cells were used to generate lentiviral particles and the resulting supernatants were obtained by transiently transfecting two plates of HEK293T cells using Lipofectamine 2000 (Invitrogen). Each plate was transfected with pCMVδR8.2 (packaging plasmid) and pGAG4 E20 (viral envelope). One plate was also transfected with pFU Gal4 (transcriptional activator), whereas the other was cotransfected with either pF-5xUAS-hMiD51-GFP, pF-5xUAS-hMiD51NBD-GFP, pF-5xUAS-mMiD51, or pF-5xUAS-mMiD51ΔPEYFP-GFP. After 24-h incubation, viral supernatants were harvested and filtered through a 0.45-µm membrane filter and incubated with MiD51TALEN MEFs along with 5 µg/ml polybrene (Sigma-Aldrich) for 48 h. After this the viral media was replaced with 10% (wt/vol) FCS DMEM containing 5 µg/ml puromycin and 200 µg/ml hygromycin.
To generate an MEF clonal cell line lacking MiD51 using TALEN technology, the initiation start codon in the first coding exon of mouse Smcr7l (GenBank accession no. NM_178719.5) encoding MiD51 was targeted using ZiFiT (Sander et al., 2007, 2010), yielding the TAL effector repeat-encoding DNA fragments mMiD51-L (NG NG NN NG HD NG NG HD NI NG NG NG HD HD NI NN NN NG) and mMiD51-R (NG NG HD HD HD NG NG NG NN HD NN HD NG HD NI NN NN NI). TAL effector repeat-encoding DNA fragments were assembled according to Reyon et al. (2012) and subcloned into the backbones pJDS78 and pJDS70 (Sander et al., 2011), generating pTALEN-mMiD51-L and pTALEN-mMiD51-R, respectively. Cells were transfected, grown, and sorted according to Stroud et al. (2013). Clonal populations were expanded and screened by SDS-PAGE and immunoblotting with antibodies specific to MiD51. To confirm gene disruption, the genomic region surrounding the target site was PCR amplified using the primers 5′-CTCGGATCCTTTCTTTTCCTGTCACGGGCT-3′ and 5′-GCGAAAGCTTGAAGCAAGCTGCCAGATGAG-3′. The polyallelic mixture was ligated into pGEM-4Z (Promega), after which eight individual clones were analyzed by Sanger sequencing, revealing biallelic disruption of the mouse Smcr7l initiation codon.
Transient transfections of cultured cells were performed using Lipofectamine LTX with Plus reagent (Invitrogen) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Mutagenesis of MiD51
MiD51 lacking the N-terminal transmembrane anchor and the disordered region (MiD51ΔN118) and MiD51 lacking the Drp1 recruitment domain (MiD51ΔPEYFP) were cloned downstream of the FKBP domain, followed by GFP. Other point mutations and truncation constructs were created using either extension overlap PCR or the Quick Change Site Directed Mutagenesis kit (Agilent Technologies). All MiD51 mutants directed to mitochondria still contained the N-terminal transmembrane anchor. All mutations and deletion constructs were verified by nucleotide sequencing.
Mitochondrial isolation and Western blotting
Mitochondrial isolation from cultured cells has been described previously (Palmer et al., 2011). In brief, cells were resuspended (20 mM Hepes, 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, and 0.5 mM PMSF, pH 7.6) and homogenized using a glass homogenizer and a drill-fitted pestle. The homogenate was then centrifuged at 800 g to pellet nuclear and cellular debris. The resulting supernatant was centrifuged at 10,000 g for 10 min at 4°C and the pellet was resuspended and centrifuged at 10,000 g for 10 min at 4°C to yield an enriched mitochondrial fraction. Proteins were separated by SDS-PAGE followed by Western blot analysis using relevant primary antibodies and horseradish peroxidase–conjugated secondary antibodies. Signals were detected using ECL substrate (GE Healthcare) and images were obtained using a G:Box Chemi XT system (Syngene).
Immunofluorescence assays
Immunofluorescence assays were performed as described previously (Palmer et al., 2011). Cells were fixed with 4% (wt/vol) paraformaldehyde in PBS, pH 7.4, and incubated with the primary antibody for 60 min at room temperature. Primary antibodies were labeled with Alexa Fluor 568– or Alexa Fluor 647–conjugated anti–mouse or anti–rabbit (Molecular Probes) secondary antibodies.
Microscopy
Cells were fixed, permeabilized, stained with the appropriate antibodies, mounted in mounting medium containing DABCO (Harlow and Lane, 1988), and then analyzed using a confocal microscope (LSM 510; Carl Zeiss) equipped with a ConfoCor 3 system containing an avalanche photodiode detector using a 100× oil immersion objective with a 1.4 numerical aperture (Fig. 1 A; Fig. 3, B and D; and Fig. 4, A and B). All confocal images were acquired using a CoolSnap HQ camera (Photometrics) with ZEN 2010 acquisition software (Carl Zeiss). Images were processed using ImageJ software (National Institutes of Health). Colocalization statistics were obtained from 30–35-µm line scans of images and are presented as Pearson correlation units (r). Images in all experimental groups were obtained with the same settings, except for detector gain adjustments that were performed to normalize saturation levels.
For 4-OHT–induced cells (Fig. S3 B), fixed cells were viewed using a fluorescence microscope (1X81; Olympus) using a 100× oil immersion objective with a 1.4 numerical aperture. Images were acquired using an F-view 2 camera (Olympus) and processed using Soft System SIS (Olympus).
For live-cell imaging (Fig. 4 C and Videos 1 and 2), COS-7 cells expressing MiD51-GFP were stained with 25 nM MitoTracker deep red (Molecular Probes), whereas for MiD51TALEN MEFs (Video 3), MiD51NBD-GFP was induced for 4 h with 100 nM 4-OHT and stained with 50 nM MitoTracker red. Cells in DMEM containing 5% (vol/vol) FBS incubated at 37°C and 5% CO2 were analyzed with a confocal microscope (LSM 510) fitted with an avalanche photodiode detector (40× oil objective with a 1.3 numerical aperture). Images were acquired at room temperature every 5.5 s to monitor rapid fission events. Cells were stitched together sequentially with ZEN 2009 software to create time-lapse videos. Contrast and brightness adjustment, gamma adjustment, region of interest selection, Gaussian smoothing, and movie making were conducted with ZEN lite 2011 software (Blue edition; Carl Zeiss).
Online supplemental material
Fig. S1 shows the generation of MiD51 soluble protein (A and B), a Western blot analysis of wild-type and MiD51TALEN mitochondria (B), a structural sequence alignment (C), and an alignment with cGAS (D). Fig. S2 shows structural detail of the MiD51 nucleotide binding pocket and RMSD values between different MiD51 structures. Fig. S3 shows an analysis of Drp1 recruitment in MiD51-inducible MEFs and the structural details of MiD51ΔPEYFP. Videos 1 and 2 show time-lapse imaging of fission events at MiD51-GFP foci in COS-7 cells and Video 3 shows fission events occurring at MiD51NBD-GFP in MiD51TALEN MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201311014/DC1.
Supplementary Material
Acknowledgments
We thank Santosh Panjikar, Janet Newman, Australian Synchrotron MX beamline staff, Sofia Caria, Grant Mills, Matt Perugini, and the CSIRO C3 Centre for assistance; Megan Maher, Begona Heras, and Mark Hulett for helpful discussions; and Richard Youle and Michael Lazarou for reagents.
This work was supported by grants from the Australian Research Council and National Health and Medical Research Council (to M.T. Ryan and M. Kvansakul) and National Institutes of Health (to H. Sesaki; grant number GM089853).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 4-OHT
- 4-hydroxytamoxifen
- cGAS
- cyclic GMP–AMP synthase
- DRR
- Drp1 recruitment region
- ITC
- isothermal titration calorimetry
- MEF
- mouse embryonic fibroblast
- RMSD
- root mean square deviation
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- Arduíno D.M., Esteves A.R., Cardoso S.M. 2011. Mitochondrial fusion/fission, transport and autophagy in Parkinson’s disease: when mitochondria get nasty. Parkinsons Dis. 2011:767230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1:298–304 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H.T., Shaw J.M. 2013. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr. Biol. 23:R891–R899 10.1016/j.cub.2013.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G., Hornung V., Hopfner K.P. 2013. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 498:332–337 10.1038/nature12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62:1002–1011 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- Cowtan K. 2008. Fitting molecular fragments into electron density. Acta Crystallogr. D Biol. Crystallogr. 64:83–89 10.1107/S0907444907033938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies V.J., Hollins A.J., Piechota M.J., Yip W., Davies J.R., White K.E., Nicols P.P., Boulton M.E., Votruba M. 2007. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet. 16:1307–1318 10.1093/hmg/ddm079 [DOI] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207–210 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- Dunning C.J., McKenzie M., Sugiana C., Lazarou M., Silke J., Connelly A., Fletcher J.M., Kirby D.M., Thorburn D.R., Ryan M.T. 2007. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26:3227–3237 10.1038/sj.emboj.7601748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgass K., Pakay J., Ryan M.T., Palmer C.S. 2013. Recent advances into the understanding of mitochondrial fission. Biochim. Biophys. Acta. 1833:150–161 10.1016/j.bbamcr.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Faelber K., Posor Y., Gao S., Held M., Roske Y., Schulze D., Haucke V., Noé F., Daumke O. 2011. Crystal structure of nucleotide-free dynamin. Nature. 477:556–560 10.1038/nature10369 [DOI] [PubMed] [Google Scholar]
- Ford M.G., Jenni S., Nunnari J. 2011. The crystal structure of dynamin. Nature. 477:561–566 10.1038/nature10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich C., Grabiger S., Schwefel D., Faelber K., Rosenbaum E., Mears J., Rocks O., Daumke O. 2013. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 32:1280–1292 10.1038/emboj.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., van der Bliek A.M. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N., Sun L., Chen Z.J. 2013a. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 341:903–906 10.1126/science.1240933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T., Serganov A.A., Liu Y., Jones R.A., Hartmann G., et al. 2013b. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 153:1094–1107 10.1016/j.cell.2013.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 726 pp [Google Scholar]
- Hartmann R., Justesen J., Sarkar S.N., Sen G.C., Yee V.C. 2003. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell. 12:1173–1185 10.1016/S1097-2765(03)00433-7 [DOI] [PubMed] [Google Scholar]
- Holm L., Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38(Web Server, Web Server issue):W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., et al. 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11:958–966 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- Karren M.A., Coonrod E.M., Anderson T.K., Shaw J.M. 2005. The role of Fis1p–Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 171:291–301 10.1083/jcb.200506158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S., Guo Q., Kalia R., Bui H.T., Eckert D.M., Frost A., Shaw J.M. 2013. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. USA. 110:E1342–E1351 10.1073/pnas.1300855110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V., Higgs H.N. 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339:464–467 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Kuchta K., Knizewski L., Wyrwicz L.S., Rychlewski L., Ginalski K. 2009. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37:7701–7714 10.1093/nar/gkp854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L., Horner J.S., Nunnari J. 2009. Mechanistic analysis of a dynamin effector. Science. 325:874–877 10.1126/science.1176921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Jin S.M., Kane L.A., Youle R.J. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 22:320–333 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O.C., Song Z., Chen H., Chan D.C. 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 24:659–667 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367–380 10.1083/jcb.151.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., Mihara K. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E., Ryan M.T. 2011. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12:565–573 10.1038/embor.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Elgass K.D., Parton R.G., Osellame L.D., Stojanovski D., Ryan M.T. 2013. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 288:27584–27593 10.1074/jbc.M113.479873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P.H., Reddy T.P., Manczak M., Calkins M.J., Shirendeb U., Mao P. 2011. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res. Brain Res. Rev. 67:103–118 10.1016/j.brainresrev.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. 2012. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30:460–465 10.1038/nbt.2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. 2001. Preparation and transformation of competent E. coli using calcium chloride. Molecular cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 1.116–1.118 [Google Scholar]
- Sander J.D., Zaback P., Joung J.K., Voytas D.F., Dobbs D. 2007. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 35(Web Server):W599–W605 10.1093/nar/gkm349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K., Dobbs D. 2010. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 38(Web Server):W462–W468 10.1093/nar/gkq319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J.D., Dahlborg E.J., Goodwin M.J., Cade L., Zhang F., Cifuentes D., Curtin S.J., Blackburn J.S., Thibodeau-Beganny S., Qi Y., et al. 2011. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods. 8:67–69 10.1038/nmeth.1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Koutsopoulos O.S., Okamoto K., Ryan M.T. 2004. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 117:1201–1210 10.1242/jcs.01058 [DOI] [PubMed] [Google Scholar]
- Stroud D.A., Formosa L.E., Wijeyeratne X.W., Nguyen T.N., Ryan M.T. 2013. Gene knockout using transcription activator-like effector nucleases (TALENs) reveals that human NDUFA9 protein is essential for stabilizing the junction between membrane and matrix arms of complex I. J. Biol. Chem. 288:1685–1690 10.1074/jbc.C112.436766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., Chen Z.J. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151:353–366 10.1083/jcb.151.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W., Iijima M., Sesaki H. 2009. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186:805–816 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Su B., Fujioka H., Zhu X. 2008. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am. J. Pathol. 173:470–482 10.2353/ajpath.2008.071208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11:872–884 10.1038/nrm3013 [DOI] [PubMed] [Google Scholar]
- Wolkowicz U.M., Cook A.G. 2012. NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Res. 40:9356–9368 10.1093/nar/gks696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Krueger E.W., Oswald B.J., McNiven M.A. 2003. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23:5409–5420 10.1128/MCB.23.15.5409-5420.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., Nistér M. 2011. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30:2762–2778 10.1038/emboj.2011.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J., et al. 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36:449–451 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.