Abstract
Introductiont
The increased disparity between organ supply and need has led to the use of extended criteria donors (ECD) and donation-after-cardiac-death (DCD) donors with other comorbidities.
Methods
We have examined the pre-implantation transcriptome of 112 kidney transplant recipient (KTRs) samples from 100 deceased donor (DD) kidneys by microarray profiling. Subject groups were segregated based on estimated glomerular filtration rate at 1-month post-transplantation (post-KTx): the GFR-high group (N=74) included patients with eGFR >45 mL/min/1.73m2 while the GFR-low group (N=35) included patients with eGFR ≤45 mL/min/1.73m2.
Results
Gene expression profiling identified higher expression of 160 probesets (140 genes) in the GFR-low group while expression of 37 probesets (33 genes) was higher in the GFR-high group (p<0.01, FDR<0.2). Four genes (CCL5, CXCR4, ITGB2, and EGF) were selected based on fold change and p-value and further validated using an independent set of samples. A random forest analysis identified three of these genes (CCL5, CXCR4, and ITGB2) as important predictors of graft function post-transplant.
Conclusions
Inclusion of pre-transplant molecular gene expression profiles in donor quality assessment systems may provide the necessary information for better donor organ selection and function prediction. These biomarkers would further allow a more objective and complete assessment of procured renal allografts at pre transplantation time.
Keywords: pre-implantation, donor quality, DGF, CAD with IF/TA, gene expression
INTRODUCTION
The increased disparity between organ supply and need has led to the use of extended criteria donors (ECD) and donation-after-cardiac-death (DCD) donors in the last decades. Unfortunately, the use of these higher risk donors may be associated with an increased risk of renal graft dysfunction early and late post-KTx (1). As consequence, evaluation of organ quality at time of transplantation, as a predictor of graft performance, represents a critical clinical challenge. Different pre-operative donor scoring systems have been developed to provide a tool for organ quality assessment of DD kidney allografts. The Donor Risk Score (DRS), proposed by Schold et al (2) uses 10 risk factors of graft survival including donor age and race, cold ischemia time (CIT) and HLA mismatches among others. However, upon validation the DRS had a modest predictive value with a c-statistic of 0.67 in predicting eGFR at 12-months (3). The Kidney Donor Risk Index (KDRI), calculates a risk score for graft failure using a combination of donor and recipient risk factors. After cross-validation the KDRI was found to have a c-statistic of 0.62 when predicting 12-month outcome (4). Other scoring systems such as the Deceased Donor Score (DDS), and the expanded criteria donor (ECD) system have had similar predictive performance (5). Histological evaluation of pre-implantation biopsies has also been proposed as a method to assess donor organ quality. Mazzucco et al (6) demonstrated that histological evaluation of pre-transplant biopsies provided reliable data on the state of the organ. However, data on the predictive value of such biopsies remains inconsistent. While several studies have reported an association between glomerulosclerosis (GS) at pre-implantation with short- and long-term outcomes, other studies have not found such an association (7). Moreover, studies have reported that histological evaluation can significantly vary between observers (8).
Improving the prediction and selection capability of donor organ scoring systems is critical for improving organ allocation and achieving better long-term graft survival. Molecular profiling goes beyond histopathology, is objective and quantitative. Studies have shown that molecular profiles can distinguish between LD and DD kidneys while histological evaluations do not (9). Gene expression profiles of pre-implantation biopsies may provide the information necessary for a more complex understanding of donor organ quality and development of better organ quality assessment methods that could optimize the use of available grafts.
We recently reported the identification of a subset of delayed graft function (DGF) kidney graft with increased expression of a number of genes involved in antigen processing and presentation and selection/activation of T-cell mediated cytotoxicity in pre-implantation biopsies (10). Although, these patients recovered function, first year post-transplant eGFR remained nearthe 45 mL/min/1.73m2 cut-off and did not significantly improve. We also identified a subset of non-DGF patients with similar graft performance during the same period of time. Here, we expand on our previously published results (10), by increasing the number of subjects as well as length of clinical follow-up and investigate the molecular basis behind this occurrence in order to identify potential molecular biomarkers for use in pre-transplant donor quality assessment.
RESULTS
Sample cohort characteristics
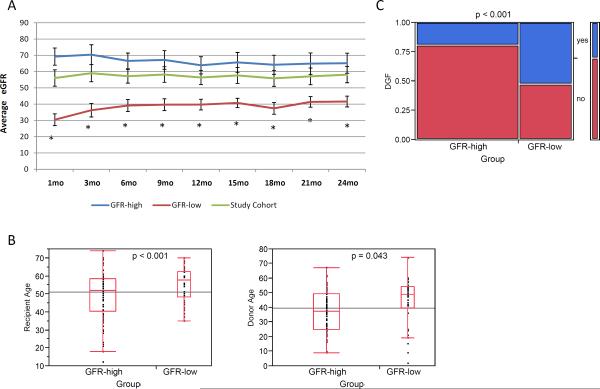
Average follow up time was 32 ± 16 months with a median of 29 months. Patient pre-implantation samples were divided into two groups based on the patient's eGFR at 1-month post-KTx following the criteria described by Kainz et al. (11): the GFR-high group (N=74) included patients with eGFR >45 mL/min/1.73m2 while the GFR-low group (N=38) included patients with eGFR ≤45 mL/min/1.73m2 (Figure 1). Estimated GFR values were statistically significant (p <0.001) between patient groups at each examined time point (Figure 1A).
Figure 1.
A. Estimated GFR in the patient sample groups. Average eGFR ± 95% confidence interval for the two patient groups. The asterisk indicates a statistically significant difference p<0.001 between the 2 groups. B. Box plots of the donor and recipient age distribution in the two patient groups. The horizontal line within the box indicates the median. C. Distribution of DGF patients within the GFR-high and GFR-low groups. A higher incidence of DGF occurred within the GFR-low group.
By 24-months post-KTx, 3 patients had passed away (all with functioning grafts), while 6 patients had suffered allograft loses. Three individuals were part of the GFR-low group (lost at 364, 371 and 22 days post-transplant) and three were part of the GFR-high group (lost at 350, 494, and 635 days post-transplant).
Donor, recipient and transplant characteristics can be found in Table 1. The two patient groups were significantly different with respect to recipient (p=0.011) and donor (p=0.043) age, with the GFR-low group generally consisting of older donors and recipients as well as older donor-recipient pairs (Figure 1B and SDC-Figure 1). HLA-A mismatch was marginally significant between the subject groups (p = 0.036) and HLA-total mismatch was significant (p=0.042) when grouping the total number of mismatches (0–2 vs. 3–4 vs. 5–6) but not without grouping. Spearman's rank correlation between 1- and 24-month eGFR values was 0.773 and between 1- and 12-month values was 0.728 both with p-values<0.001 (SDC-Figure 2). The incidence of DGF was also proportionally higher in the GFR-low group (p<0.001) (Figure 1C). DGF diagnoses can be found in SDC-Table 1. No statistical differences were observed in the rates of acute rejection between the two groups.
Table 1.
Sample cohort demographics. Statistical significance was calculated using a paired t-test unless otherwise indicated. AVG = average; STD = standard deviation.
| GFR-low | GFR-high | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| AVG | STD | AVG | STD | p-value | ||
| Recipient Data | ||||||
| Avg. Age | 55.0 | 10.0 | 49.1 | 13.6 | 0.011 | |
| Gender | M (F) | 23 (12) | 46 (28) | NS† | ||
| Race | C | 11 | 13 | NS† | ||
| AA | 25 | 58 | ||||
| Other | 2 | 3 | ||||
| HCV status (pos) | 2 | 0 | NS‡ | |||
| CMV Disease (pos) | 0 | 0 | NS‡ | |||
| CMV status (pos) | 0 | 0 | NS‡ | |||
| Donor Data | ||||||
| Avg. Donor Age | 44.0 | 17.1 | 37.4 | 14.1 | 0.043 | |
| Gender | M (F) | 18 (17) | 47 (23) | NS† | ||
| Race | C | 21 | 49 | NS† | ||
| AA | 14 | 24 | ||||
| Other | 3 | 1 | ||||
| HBV cAb (pos) | 1 | 6 | NS‡ | |||
| HBV sAb (pos) | 1 | 5 | NS‡ | |||
| HCV Ab (pos) | 2 | 11 | NS‡ | |||
| CMV status (pos) | 23 | 44 | NS‡ | |||
| Donor Type | SCD | 23 | 55 | NS† | ||
| ECD | 6 | 10 | ||||
| DCD | 9 | 9 | ||||
| Transplant | ||||||
| Avg. CIT (min) | 1222.1 | 389.5 | 1253.2 | 529.0 | NS | |
| Avg. WIT (min) | 31.1 | 7.0 | 30.6 | 7.9 | NS | |
| Avg. PPP Time (min) | 824.7 | 310.8 | 778.8 | 343.1 | NS | |
| PPP (% of group) | 55.3 | 60.8 | NS‡ | |||
| Avg. Last Donor SCr. | 1.1 | 0.6 | 1.1 | 0.5 | NS | |
| HLA-A (0 / 1 / 2 MM*) | 5 / 14 / 16 | 9 / 15 / 53 | 0.036† | |||
| HLA-B (0 / 1 / 2 MM) | 3 / 9 / 23 | 4 / 20 / 53 | NS† | |||
| HLA-DR (0 / 1 / 2 MM) | 8 / 14 / 13 | 6 / 38 / 33 | NS† | |||
| HLA MM total | 0–2 | 5 | 5 | 0.042† | ||
| 3–4 | 15 | 22 | ||||
| 5–6 | 15 | 50 | ||||
| PRA at transplant - T | 38.2 | 35.4 | 40.6 | 35.7 | NS | |
| PRA at transplant - B | 18.8 | 31.0 | 20.7 | 30.6 | NS | |
| Post-transplant Data | ||||||
| Cases with ACR | 2 | 4 | NS | |||
| Cases with DGF | 20 | 14 | <0.001 | |||
| Avg. eGFR (mL/mm/1.73m2) | 1mo | 30.4 | 11.4 | 69.2 | 23.1 | <0.001 |
| 3mo | 36.3 | 12.7 | 70.4 | 26.5 | <0.001 | |
| 6mo | 39.1 | 11.3 | 66.6 | 20.5 | <0.001 | |
| 9mo | 39.7 | 10.5 | 67.2 | 23.8 | <0.001 | |
| 12mo | 39.7 | 9.3 | 63.9 | 22.4 | <0.001 | |
| 15mo | 40.7 | 7.6 | 65.7 | 23.5 | <0.001 | |
| 18mo | 37.4 | 9.7 | 64.2 | 23.5 | <0.001 | |
| 21mo | 41.4 | 8.5 | 64.9 | 24.2 | <0.001 | |
| 24mo | 41.6 | 8.5 | 65.1 | 24.2 | <0.001 | |
ACR = acute cellular rejection
DGF = delayed graft frunction
NS = not significant
MM = mismatch(es)
Chi-square test
Fisher's Exact test
Gene expression profiling of pre-implantation biopsies
When comparing profiles between the two subject groups we identified 197 differentially expressed probesets (FDR<0.2). Expression of 160 probesets (140 genes) was higher in the GFR-low group whereas expression of 37 probesets (33 genes) was higher in the GFR-high group (SDC-Table 2). Top scoring genes (p<0.01, FDR<0.05) included CCL5, TRBC1, CXCL6, S100A9, AIF1, ITGB2, CD52, and CD48 (Table 2).
Table 2.
Top differentially expressed genes. Top scoring probesets identified as differentially expressed between GFR-low and GFR-high patients (p-value <0.01, FDR < 0.05).
| Affymetrix ID | Gene Symbol | Entrez ID | pvalue | FDR | Up-regulated in |
|---|---|---|---|---|---|
| 1405_i_at | CCL5 | 6352 | 1.15E-06 | 0.0207 | GFR-low |
| 204655_at | CCL5 | 6352 | 4.94E-06 | 0.0357 | GFR-low |
| 210915_x_at | TRBC1 | 28639 | 6.08E-06 | 0.0357 | GFR-low |
| 206336_at | CXCL6 | 6372 | 8.48E-06 | 0.0357 | GFR-low |
| 203535_at | S100A9 | 6280 | 1.17E-05 | 0.0357 | GFR-low |
| 213095_x_at | AIF1 | 199 | 1.27E-05 | 0.0357 | GFR-low |
| 202803_s_at | ITGB2 | 3689 | 1.39E-05 | 0.0357 | GFR-low |
| 34210_at | CD52 | 1043 | 1.73E-05 | 0.0388 | GFR-low |
| 204118_at | CD48 | 962 | 2.42E-05 | 0.0482 | GFR-low |
Functional analysis and interaction networks
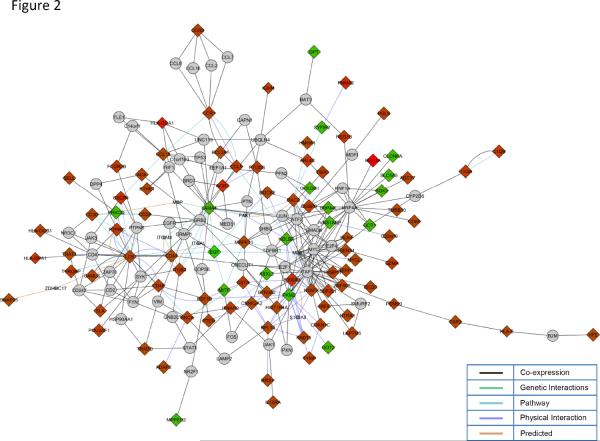
Top biological processes identified exclusively from genes up-regulated in the GFR-low group included regulation of immune system process (p=5.18E-15), antigen processing and presentation (p=5.26E-09), and T-cell activation (p=2.82E-07) among others (SDC-Table 3). No significant biological processes were identified from genes up-regulated in the GFR-high group (down-regulated in GFR-low samples). Differentially expressed genes were up-loaded into Cytoscape (12) and a gene interaction network generated using the MiMI and GeneMania plug-ins. Only first-neighbors shared by more than two differentially expressed genes were retained in the final network (Figure 2). Since the network consisted primarily of up-regulated genes, biological processes were then identified solely from these genes and mapped using the BiNGO cytoscape plug-in (SDC-Figure 3).
Figure 2. Gene interaction network.
Cytoscape was used to create a gene interaction network using the identified differentially expressed genes. Diamonds represent genes in the input list (differentially expressed genes), circles represent first neighbors shared by more than 2 input genes. The red and green coloring indicates whether the gene was found to be up-regulated or down-regulated respectively. Lines connecting the various molecules are color coded to represent the type of interaction identified.
Ingenuity Pathway Analysis (IPA, www.ingenuity.com) was also used to identify gene interactions networks. The top two gene networks identified by IPA included genes involved in inflammation (network 1, score of 39), antigen presentation (network 2, score of 39), cell-to-cell signaling (networks 1 and 2), and hematological system development (networks 1 and 2). SDC-Figure 4 shows a merging of the two top scoring networks. All three annotation methods used identified immune response, inflammation T-cell activation, and antigen presentation as the most prominent biological processes.
Validation of Microarray Results
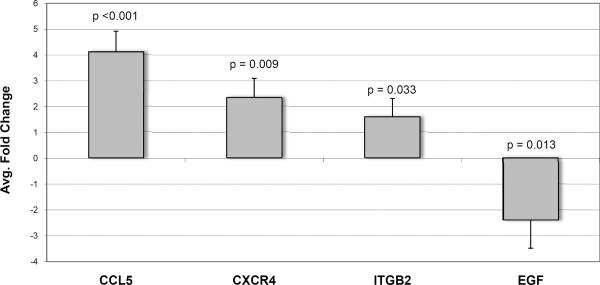
Four genes (CCL5, CXCR4, ITGB2 and EGF) were selected for validation in an independent set of pre-implantation biopsies by real-time QPCR. Differential expression between GFR-low (N=19) and GFR-high (N=13) of all four genes selected was confirmed (Figure 3). Relative expression (fold change) was calculated using the ΔΔCt method by comparing GFR-low group samples to GFR-high group (control) samples.
Figure 3. Validation of microarray results.
Real-time quantitative PCR results of genes selected for confirmation from the microarray results. CCL5, CXCR4 and ITGB2 were up-regulated while EGF was down-regulated in GFR-low samples. GAPDH was used as the normalizing internal control. Statistical significance was tested using a paired t-test comparing GFR-high (control) samples to GFR-low samples.
Random Forest (RF) and proportional hazards (PH) ratio
We used a random forest algorithm to identify a small set of gene expression profiles and/or clinical variables that could be used to accurately predict eGFR status. The random forest algorithm identified several probesets and clinical variables with high importance in prediction of eGFR (high vs. low) with an average prediction accuracy of 68% (SDC-Figure 5). Of the top 30 identified variables of high importance, 27 were probesets, including CD52, CD69, CXCR4, CCL5, ITGB2, CXCL6 and AIF1, and 3 were clinical variables (donor CMV, WIT and PPP time).
We next used a proportional hazards (PH) model to test short and long-term effects. When fitting an intercept only PH cure model, cure was significant for graft survival (p=0.00012). When examining time-to-effect on graft survival, only 2 probesets were significant at p<0.05. Due to the small number of significant probesets, Cox proportional hazards were fit ignoring cure resulting in 310 probesets (275 genes) significantly associated with graft survival at the p<0.01 level (SDC-Table 4). Twenty-six of the identified genes were also identified as differentially expressed between the two groups of patients including CD52, CD48, EGF and AIF1.
Three-month post-transplant biopsies
Gene expression from 3-month biopsies (K3) was also available from a subset of the enrolled patients (N=48). Comparison of the two eGFR-based groups (GFR-high N=29; GFR-low N=19) identified 783 probesets (657 genes) differentially expressed genes between the two groups (p<0.01, FDR <0.05). When comparing PI and K3 gene expression profiles, 56 genes were found to overlap, including CXCR4, CXCL6, CD44, CD52, CD69 and EGF. Gene ontology analyses of PI and K3 signatures shows overrepresentation of similar biological processes, including regulation and activation of immune responses, T-cell activation, and inflammatory response (data not shown). Moreover, when comparing these GE profiles to a published IF/TA signature (13), 42 genes were found in all three signatures while 98 genes identified at PI were also identified in the IF/TA signature (SDC-Figure 6).
DISCUSSION
We have identified a set of genes with elevated expression including a number of pro-inflammatory genes, as CCL5, CXCR4, CXCL6, CD52, CD48, TRBC1 and AIF1, in pre-implantation biopsies of kidney graft recipients with associated persistent lower graft function post-KTx (SDC-Table 2). This subgroup of patients also showed a higher incidence of DGF suggesting this pre-existing inflammatory state may be a contributing factor to the development of DGF. More importantly, regardless of whether DGF occurred or not, post-transplant graft performance remained persistently low in these patients.
Identification of these genes in pre-transplant graft biopsies may have important clinical implications. Transcripts from several inflammatory chemokines, including CCL5, have been shown to be significantly increased in allografts with subclinical and acute rejection (14) as well as in CAD with IF/TA (13). CCL5 is chemotactic for T-cells, eosinophils and basophils playing an active role in recruitment of these cells to sites of inflammation (15). Increased expression of CCL5 has also been reported in chronic proteinuric glomerulopathies (15) and has been correlated with poorer 1-year graft function in both LD and DD kidney transplant recipients (16). CXCR4 plays a role in neutrophil chemotaxis (17) and has been found to be up-regulated in CAD with IF/TA biopsies and acute rejection (13).
CD52 is an antigen expressed in thymocytes, lymphocytes, monocytes, and granulocytes while CD48 expression is restricted to lymphocytes, macrophages, and dendritic cells; both molecules are involved in T-cell activation (18). TRBC1 (also known as MICB, MHC class I-related chain B) is a stress induced polymorphic gene expressed on epithelial cells whose up-regulation is associated with post-transplant development of DGF (19). Allograft inflammatory factor 1 (AIF1), has been shown to be expressed in podocytes, promote T-cell infiltration and increase production of cytokines (20). Additionally, an increase of AIF1-activated macrophages has been observed during acute cellular rejection (20). Finally, CXCL6 has been shown to be a potent inflammatory mediator that binds chemokine receptors CXCR1 and CXCR2 (21). However, its specific role in renal transplantation remains unexplored.
The findings reported here are consistent with previous reports. The presence of inflammatory molecular profiles and activation of the complement cascade has been reported in apparently histological normal allografts and has been associated with lower graft function at 1-year post-KTx (11,22). Hauser et al (22) identified 48 genes that classified cadaveric donor kidneys according to their post-transplant course (delayed vs. immediate graft function). Similarly, Kainz et al (11) also identified a small number of genes up-regulated in pre-implantation biopsies of patients with low post-KTx graft function categorized into functional classes of immunity and defense, signal transduction, and oxidative stress response. Unfortunately, both of these studies were limited by their sample size (Hauser et al, N=29; Kainz et al, N=31). In the Kainz et al report, donor age appeared to be a confounding factor suggesting that the gene expression changes identified may have been the results of age-related changes. In our current study cohort, subjects in the GFR-low group tended to be older and had received allografts primarily from older donors, however subjects of similar ages were also found in the GFR-high group. Elevated expression of CD69 (also found in our study), HLA-DRB1, and NKG2D was identified in zero-hour biopsies and reported to be indicative of decreased graft function (<45 mL/min/1.73m2) at 6- and 12-months post-KTx (23). However, this study combined LD and DD, known to have different transcriptional profiles post-KTx (9), likely influencing the ability of the system to detect DD specific genes. Our present study only included recipients of DD renal allografts allowing the identification of such genes. Six genes identified in this study have previously been associated with lower graft function at 3 months (REG1A, REG1B, IGJ and IGKC) (11), or an associated higher risk of late allograft dysfunction (NNMT and CXCL6) (24). These gene expression biomarkers might be useful for LD but remain to be tested.
Identification of the causes for the observed pre-transplant immune/inflammatory activation and subsequent poor post-transplant performance seems critical to the future development of new therapeutic strategies. Suggested contributing factors include age, race and gender, CIT, the amount of PRA at transplant, and donor cause of death (COD) (25). Of these, only age (donor and recipient) were found to be statistically different between the two patient groups in our study (Table 1). It can be anticipated that CIT would play a role in the initiation of inflammatory signals; however, CIT was not found to be statistically different between the two sample groups. Regardless, early immune and inflammatory activation within the allograft and/or prolonged post-transplant activation of these signaling pathways may lead to long-term subclinical inflammation with detrimental effects on graft function. The data presented here suggest that the presence of pre-implantation pro-inflammatory and other signals may be responsible at least in part for the observed lower post-transplant eGFR in a subgroup of patients. Early adjustment of the immunosuppression therapy or future therapies may help to reduce or block inflammation pathways and potnetialy to improve the graft function.
The increasing demand for renal allografts warrants the development of a donor organ standard scoring system that can help objectively identify optimal from sub-optimal quality organs prior to implantation. Previous donor scoring systems, although successful at predicting short-term outcome, fall short when predicting log-term outcome beyond 12-months post-KTx. This is mainly due to the inability of such scoring systems to factor in later events, such as acute rejection episodes that affect overall graft performance even after resolution. A study by Kaplan and Schold (26), showed that prediction of long-term graft survival (5 years) is possible when considering demographic (i.e. recipient and donor age), clinical (i.e. episodes of acute rejection) and pharmacological (i.e. immunosuppression) factors.
Both the random forest and PH analyses identified TRBC1, AIF1, CD48 and CD52 as significant predictors of graft function and having an association with increased risk of graft loss warranting a closer evaluation of their pre-transplant and follow-up expression and the recipient's graft outcome. Moreover, of the four validated genes in this study, three (CCL5, CXCR4, and ITGB2) were also identified by the Random Forest algorithm as important in predicting graft function. Inclusion of selected pre-transplant molecular biomarkers to donor quality scoring systems may provide the necessary information for better donor organ selection.
Finally, this biomarker-donor characteristics score combination would not only allow accurate assessment of procured renal allografts early on but would also provide insight into the future long-term performance of the allografts and therapy opportunities.
MATERIALS AND METHODS
Enrolled population
The study included 112 kidney transplant recipient (KTRs) samples from 100 DD kidneys. Patients were enrolled and consented between January, 2006 and November, 2011. The study was conducted at Virginia Commonwealth University and at the University of Virginia after Institutional Review Board (IRB) approval was obtained at both institutions (VCU#HM11454, UVA#14849). No living donors (LD), HIV positive or re-transplant patients were included. Only patients between the ages of 18 and 70 were enrolled. Biopsies from kidneys preserved using cold and pump perfusion preservation (PPP) were included. All enrolled subjects were treated post-KTx with triple immunosuppressant therapy consisting of tacrolimus, mycophenolate mofetil and prednisone. Estimated GFR (eGFR) was calculated using the abbreviated Modification of Diet in Renal Disease (MDRD) formula (27). DGF was defined as the need for dialysis during the first 7 days post-KTx.
A total of 160 renal allograft biopsies were collected at pre-implantation (PI, N=112) and at 3-months post-KTx (K3, N=48). Allograft tissue was obtained through an ultrasound guided 18-gauge biopsy needle and immediately placed in RNAlater (Life Technologies, Grand Island, NY). Total RNA was isolated using Trizol (Life Technologies) following the manufacturer's recommended protocol and cleaned of impurities using the RNeasy mini kit (Qiagen, Valencia, CA) RNA Clean-up protocol. RNA integrity and quality was checked using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Quality control criteria has been described previously (13).
Microarray data analysis
Quality of the hybridized arrays was assessed by examining the average background, scaling factor, percent of probe sets called present by the detection call algorithm, and the 3':5' ratio for GAPDH and ACTIN. Control probesets and probesets considered absent in all samples were removed; the remaining probesets were normalized by quantile normalization and summarized with median polish summarization using the Robust Multiarray Average method (28). For each probeset, a moderated t-test was used to compare the two groups of renal transplant recipients classified as GFR-high or GFR-low. To adjust for the multiple hypothesis tests, the p-values were used in estimating the false discovery rate using the Benjamini and Hochberg method (29). Probesets having an FDR <0.20 were considered significant.
Random Forest (RF)
Random forest modeling was implemented using the randomForest package in R (30). Gene expression data only for the probes identified as differentially expressed was combined with the available clinical and phenotype variables. For each analysis using, 500 regression trees were fit. For each tree, rather than using the entire set of gene expression and clinical variables as possible predictors, the algorithm was set to choose a small random subset of m variables to use instead of all p predictors. Empirical studies have shown that m=p/3 is optimal for regression problems, so this value was used in our analysis (30).
Proportional hazards (PH)
We used a PHPH regression model to accommodate the presence of long-term graft survival. For each probe set, a PHPH model was fit using the nltm package in R (31). The p-values associated with long-term effect, risk of graft failure, and short-term effect-were examined for each probe set by modeling time to event. To adjust for the multiple hypothesis tests, an α level of 0.01 was used.
Additional Materials and Methods can be found in the supplemental digital content SDC-Materials and Methods.
Supplementary Material
ACKNOWLEDGEMENTS
The research results included in this report were supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant, R01DK080074.
ABBREVIATIONS
- CAD
Chronic Allograft Dysfunction
- CIT
cold ischemia time
- CMV
cytomegalo virus
- DCD
donation-after-cardiac-death
- DD
deceased donor
- DDS
Deceased Donor Score
- DGF
delayed graft function
- DRS
Donor risk score
- ECD
extended criteria donors
- eGFR
estimated Glomerular Filtration Rate
- FDR
False Discovery Rate
- GS
glomerulosclerosis
- HBV
Hepatitis B Virus
- HCV
Hepatitis C Virus
- HIV
Human Immunodeficiency Virus
- HLA
Human Leukocyte Antigen
- IF
Interstitial Fibrosis
- IPA
Ingenuity Pathway Analysis
- KDRI
Kidney Donor Risk Index
- KTRs
kidney transplant recipient
- LD
living donors
- MDRD
Modification of Diet in Renal Disease
- PH
Proportional Hazards
- post-KTx
post-trasnplantation
- PPP
pump perfusion preservation
- PRA
panel reactive antibody
- RF
Random Forest
- TA
Tubular atrophy
- WIT
warm ischemia time
Footnotes
Author contributions: Participated in the research design: MJS, VRM, DGM
Participated in the performance of the research: VRM, MJS, SDT, KJA, JLS, KGD
Participated in the data analysis: MJS, KJA, SDT, VRM
Participated in the manuscript preparation: MJS, VRM, DGM
Final approval of the version to be published: MJS, DGM, KJA, SDT, JLS, KGD, ALK, MPP, KLB, VRM
DISCLOSURE STATEMENT The authors of this manuscript have no conflicts of interest to disclose as described by the journal Transplantation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sung RS, Guidinger MK, Christensen LL, et al. Development and current status of ECD kidney transplantation. 2005. p. 37. [PubMed] [Google Scholar]
- 2.Schold J, Kaplan B, Baliga R, Meier Kriesche H. The broad spectrum of quality in deceased donor kidneys. American journal of transplantation. 2005;5(4):757. doi: 10.1111/j.1600-6143.2005.00770.x. [DOI] [PubMed] [Google Scholar]
- 3.Moore J, Ramakrishna S, Tan K, et al. Identification of the optimal donor quality scoring system and measure of early renal function in kidney transplantation. Transplantation. 2009;87(4):578. doi: 10.1097/TP.0b013e3181949e71. [DOI] [PubMed] [Google Scholar]
- 4.Rao P, Schaubel D, Guidinger M, et al. A comprehensive risk quantification score for deceased donor kidneys: the kidney donor risk index. Transplantation. 2009;88(2):231. doi: 10.1097/TP.0b013e3181ac620b. [DOI] [PubMed] [Google Scholar]
- 5.Nyberg SL, Baskin-Bey ES, Kremers W, Prieto M, Henry ML, Stegall MD. Improving the prediction of donor kidney quality: deceased donor score and resistive indices. Transplantation. 2005;80(7):925. doi: 10.1097/01.tp.0000173798.04043.af. [DOI] [PubMed] [Google Scholar]
- 6.Mazzucco G, Magnani C, Fortunato M, Todesco A, Monga G. The reliability of pre-transplant donor renal biopsies (PTDB) in predicting the kidney state. A comparative single-centre study on 154 untransplanted kidneys. Nephrology, Dialysis, Transplantation. 2010;25(10):3401. doi: 10.1093/ndt/gfq166. [DOI] [PubMed] [Google Scholar]
- 7.Lu AD, Desai D, Myers BD, Dafoe DC, Alfrey EJ. Severe glomerular sclerosis is not associated with poor outcome after kidney transplantation. The American journal of surgery. 2000;180(6):470. doi: 10.1016/s0002-9610(00)00502-x. [DOI] [PubMed] [Google Scholar]
- 8.Furness PN, Taub N, Assmann KJ, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am.J.Surg.Pathol. 2003;27(6):805. doi: 10.1097/00000478-200306000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Mueller TF, Reeve J, Jhangri GS, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am.J.Transplant. 2008;8(1):78. doi: 10.1111/j.1600-6143.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- 10.Mas VR, Scian MJ, Archer KJ, et al. Pretransplant transcriptome profiles identify among kidneys with delayed graft function those with poorer quality and outcome. Mol.Med. 2011;17(11–12):1311. doi: 10.2119/molmed.2011.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kainz A, Perco P, Mayer B, et al. Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation. 2007;83(8):1048. doi: 10.1097/01.tp.0000259960.56786.ec. [DOI] [PubMed] [Google Scholar]
- 12.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scian MJ, Maluf DG, Archer KJ, et al. Gene expression changes are associated with loss of kidney graft function and interstitial fibrosis and tubular atrophy: diagnosis versus prediction. Transplantation. 2011;91(6):657. doi: 10.1097/TP.0b013e3182094a5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo D, Weaver T, Kleiner D, et al. Chemokines and their receptors in human renal allotransplantation. Transplantation. 2011;91(1):70. doi: 10.1097/TP.0b013e3181fe12fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krensky AM, Ahn YT. Mechanisms of disease: regulation of RANTES (CCL5) in renal disease. Nat.Clin.Pract.Nephrol. 2007;3(3):164. doi: 10.1038/ncpneph0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dikow R, Becker LE, Schaier M, Waldherr R, Gross ML, Zeier M. In Renal Transplants With Delayed Graft Function Chemokines and Chemokine Receptor Expression Predict Long-Term Allograft Function. Transplantation. 2010 doi: 10.1097/TP.0b013e3181f009ef. [DOI] [PubMed] [Google Scholar]
- 17.Gouwy M, Struyf S, Catusse J, Proost P, Van Damme J. Synergy between proinflammatory ligands of G protein-coupled receptors in neutrophil activation and migration. J.Leukoc.Biol. 2004;76(1):185. doi: 10.1189/jlb.1003479. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Campo PM, Almeida J, Matarraz Sde Santiago M, Sanchez ML, Orfao A. Quantitative analysis of the expression of glycosylphosphatidylinositol-anchored proteins during the maturation of different hematopoietic cell compartments of normal bone marrow. Cytometry B.Clin.Cytom. 2007;72(1):34. doi: 10.1002/cyto.b.20143. [DOI] [PubMed] [Google Scholar]
- 19.Quiroga I, Salio M, Koo DDH, et al. Expression of MHC class I-related Chain B (MICB) molecules on renal transplant biopsies. Transplantation. 2006;81(8):1196. doi: 10.1097/01.tp.0000205788.05322.42. [DOI] [PubMed] [Google Scholar]
- 20.Del Galdo F, Jimnez S. T cells expressing allograft inflammatory factor 1 display increased chemotaxis and induce a profibrotic phenotype in normal fibroblasts in vitro. Arthritis rheumatism. 2007;56(10):3478. doi: 10.1002/art.22877. [DOI] [PubMed] [Google Scholar]
- 21.Wuyts A, Van Osselaer N, Haelens A, et al. Characterization of synthetic human granulocyte chemotactic protein 2: usage of chemokine receptors CXCR1 and CXCR2 and in vivo inflammatory properties. Biochemistry. 1997;36(9):2716. doi: 10.1021/bi961999z. [DOI] [PubMed] [Google Scholar]
- 22.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab.Invest. 2004;84(3):353. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 23.Kotsch K, Kunert K, Merk V, et al. Novel markers in zero-hour kidney biopsies indicate graft quality and clinical outcome. Transplantation. 2010;90(9):958. doi: 10.1097/TP.0b013e3181f546e8. [DOI] [PubMed] [Google Scholar]
- 24.Einecke G, Reeve J, Sis B, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J.Clin.Invest. 2010;120(6):1862. doi: 10.1172/JCI41789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matas AJ, Gillingham KJ, Humar A, Dunn DL, Sutherland DE, Najarian JS. Immunologic and nonimmunologic factors: different risks for cadaver and living donor transplantation. Transplantation. 2000;69(1):54. doi: 10.1097/00007890-200001150-00011. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan B, Schold J. Transplantation: neural networks for predicting graft survival. Nature reviews.Nephrology. 2009;5(4):190. doi: 10.1038/nrneph.2009.24. [DOI] [PubMed] [Google Scholar]
- 27.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann.Intern.Med. 1999;130(6):461. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 28.Archer KJ, Reese SE. Detection call algorithms for high-throughput gene expression microarray data. Brief Bioinform. 2010;11(2):244. doi: 10.1093/bib/bbp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini YaH Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc B. 1995;57(1):289. [Google Scholar]
- 30.Liaw A, Wiener M. Classification and Regression by randomForest. Glass. 2002;2(December):18. [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat.Appl.Genet.Mol.Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.