Abstract
Condensins are conserved multi-subunit protein complexes that participate in eukaryotic genome organization. Well known for their role in mitotic chromosome condensation, condensins have recently emerged as integral components of diverse interphase processes. Recent evidence shows that condensins are involved in chromatin organization, gene expression, and DNA repair and indicates similarities between the interphase and mitotic functions of condensin. Recent work has enhanced our knowledge of how chromatin architecture is dynamically regulated by condensin to impact essential cellular processes.
Keywords: Condensin, Chromosome condensation, SMC proteins, Chromatin, Nuclear organization, Interphase chromosome
Introduction
It is a generally accepted view that genetic information encoded in DNA exists and functions within the context of chromatin. Chromatin organization is dynamic, and changes in chromatin structure can either facilitate or inhibit DNA accessibility. The packaging of DNA into chromatin presents a significant challenge to essential cellular processes such as transcription, DNA replication and repair, and chromosome segregation. These processes rely on enzymatic activities that require access to the DNA molecule and are therefore coordinated with precise modulation of chromatin structure [1–4]. Furthermore, compelling evidence suggests that the nucleus is organized into functional compartments containing different types of chromatin [5, 6]. The question of how chromatin is organized in three-dimensional space within the eukaryotic nucleus has been a long-standing interest of geneticists and cell biologists, and is critical for understanding the regulation of these essential cellular processes.
That an important connection exists between the function of genetic material and its spatial organization has been supported by the identification of numerous defects in nuclear morphology and chromatin organization in a variety of human pathologies. What has remained a largely open question is that of causation: Are defects in spatial organization of chromatin a cause of human disease and cellular dysfunction, or are these morphological defects simply a result of one or more defunct pathways? Perhaps this question is only relevant if we still view biological pathways as linear. Given the interconnected nature of biological networks, it is likely that morphological defects are both a cause and a result of cellular processes gone awry. A particular challenge to the advancement of this field has been the lack of testable mechanistic models where specific chromatin and nuclear organizational states can be experimentally manipulated so as to ask how changes in organization may cause defects in essential processes.
Condensins are conserved protein complexes that are best known for their function in chromosome condensation during mitosis. However, emerging evidence has uncovered numerous non-mitotic functions suggesting that condensin complexes are key players in eukaryotic chromosome organization. This review will focus specifically on the emerging role of condensin in interphase genome organization; therefore, discussion of the mitotic and meiotic roles of condensin is outside the scope of this review. For recent reviews on these topics, see Hudson et al. [7] and Thadani et al. [3].
Condensin Complexes
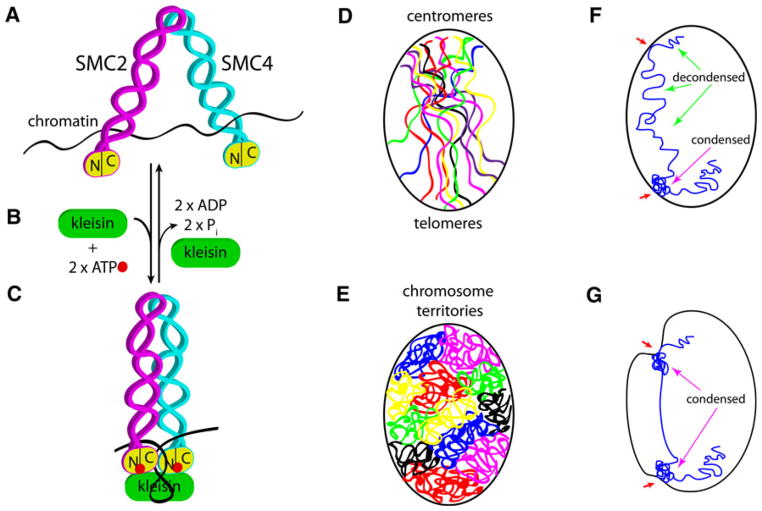
Eukaryotes have two different condensin complexes, condensin I and condensin II, which are highly conserved across eukaryotic species. Both are five-subunit complexes that share a core heterodimeric SMC2–SMC4 subunit belonging to the structural maintenance of chromosome (SMC) family [8]. The two complexes each have three unique non-SMC subunits (Fig. 1); condensin I contains CAP-D2, CAP-G, and CAP-H, while condensin II contains CAP-D3, CAP-G2, and CAP-H2. CAP-D2, CAP-D3, CAP-G, and CAP-G2 contain HEAT repeats, which are thought to be important for protein-protein interactions [9]; CAP-H and CAP-H2 belong to the kleisin family of proteins [10]. The SMC heterodimer forms a V-shaped structure, characteristic of all SMC proteins, with an ATP-binding catalytic “head” domain and a “hinge” domain required for dimerization (Fig. 1a–c) [11, 12].
Fig. 1.
Condensins drive changes in chromosome organization and nuclear shape in interphase. a Eukaryotic condensin complexes consist of a heterodimer of two SMC proteins, SMC2 and SMC4. Each SMC subunit has half its ATPase on its N-terminus (N) and the other half on the C-terminus (C), which come together to form a functional ATPase “head,” shown in yellow. SMC proteins form a coiled-coil domain and a hinge domain, where dimerization occurs. SMC dimers can interact directly with DNA or chromatin (black line). Condensin not bound to ATP is thought to have an “open” conformation. b The SMC dimer can bind one ATP molecule (red) in each of the two ATPase heads, and ATP-bound head domains can then recruit a kleisin subunit (green). Barren/Cap-H is a condensin I-specific kleisin; Cap-H2 is a condensin II-specific kleisin. Other chromosome associated proteins (Cap) can also be recruited to the complex (not shown) in a condensn I- or II-specific manner. Additional Cap subunits are thought to mediate specific protein-protein interactions. See Table 1 for a complete list of SMC and Cap subunit genes. c ATP-bound SMC2/4 dimer induces a conformational change to a “closed” state. This conformational change is thought to drive axial shortening of chromosomes by inducing compaction of chromatin. Kleisin binding inhibits ATP hydrolysis and may serve to stabilize the closed SMC conformational state. High levels of kleisin favor the closed conformation. Dissociation of kleisin and ATP hydrolysis (as in b) reestablishes the open conformation and allows decondensation of chromatin. d Chromosomes (colored lines) are contained within the nuclear envelope (black oval) and exist in the Rabl conformation, where centromeres and telomeres are at opposite ends of the nucleus. Centromeres, telomeres, and other regions are thought to be tethered to the inner nuclear membrane through chromatin interactions with envelope-associated proteins. e Interphase chromosomes can adopt territories where each chromosome occupies a discrete position in the three-dimensional space of the nucleus. The position of each chromosome relative to the nuclear envelope and to other chromosomes is important for the expression of genes. It has been proposed that condensin II compaction forces in interphase are required for organizing chromosomes into territories. Chromatin tethers to the envelope are speculated to serve as anchors of condensin-driven gathering of chromatin as it condenses. f An interphase nucleus is shown with one chromosome (blue) for simplicity. Chromosomes can have regions that are relatively decondensed (green arrows) and condensed (magenta arrow) that reflect tissue-specific chromatin and gene expression states. Chromatin can be tethered to the inner nuclear membrane (red arrows). g A speculative model where local condensation states can be modulated, likely by local condensin activation, and envelope-tethered chromatin anchors may be drawn toward the interior of the nucleus. Invaginations and distortions of the envelope (red arrows) may result from local chromatin condensation or defects in nuclear envelope structure (see text for further details)
Condensin I and II have distinct spatial and temporal localization patterns, with condensin II localizing to the nucleus throughout the cell cycle while condensin I is localized to the cytoplasm, only accessing the chromosomes following nuclear envelope breakdown in prometaphase [13–15]. This differential localization suggests that the condensin complexes may have distinct roles in chromosome organization. For example, condensin I facilitates lateral compaction of mitotic chromosomes while condensin II facilitates axial compaction [16•]. Recent studies suggest that the non-redundant roles of the eukaryotic condensin complexes are not limited to mitosis, but are important for a wide range of processes during interphase.
Condensin Functions in Interphase
Homolog Pairing and Polytene Disassembly
One of the most dramatic chromatin organizations is the pairing of homologous chromosomes. This entails the physical juxtaposition of maternal and paternal DNA sequences along the entire lengths of chromosomes, specifically between homologs. Homologous chromosome pairing is a key event in meiosis, and is required for recombination and chromosome segregation (reviewed in [17]). Homolog pairing also occurs in somatic cells, first described in Dipteran insects [18, 19], and has been observed in a number of organisms [20–22]. A unique feature of somatic homolog pairing that distinguishes it from meiosis is that homology-dependent pairing of sequences does not require DNA breaks, recombination machinery, or synaptonemal complex proteins that typically mediate meiotic pairing [23, 24]. Somatic homolog pairing has been studied most extensively in Drosophila, where pairing initiates in mitotic division 13 during embryogenesis and persists throughout development [25–29]. In tissues that are developmentally programed for polyploidy, pairing also occurs to form giant polytene chromosomes, where thousands of DNA strands are aligned along the length of the chromosome [30]. Polytene chromosomes occur in tissues of many species, including the giant trophoblast cells of the mammalian placenta [31].
Pairing of homologous chromosomes in diploid somatic cells has been implicated in regulation of gene expression through transvection as well as DNA damage repair [32, 33], but the mechanisms underlying pairing remain poorly understood. Transvection refers to the physical interactions of enhancers on one chromosome with promoters of another chromosome, resulting in activation or repression of transcription [32]. Suppressor of Hairy-wing [34] and topoisomerase II [35] were the first factors identified that function to promote somatic pairing. Conversely, the CAP-H2 subunit of condensin II promotes disassembly of polytene chromosomes into unpaired homologs and chromatid fibers during mid-oogenesis in Drosophila ovarian nurse cells and antagonizes transvection in diploid nuclei [36]. Because other condensin II subunits were also required for this unpairing activity, condensin II was proposed to function as an anti-pairing factor. The activity of condensin II is regulated by the SCFSlimb ubiquitin ligase, which targets CAP-H2 for degradation, thus resulting in homolog pairing and inhibition of interphase chromosome compaction [37, 38•]. Lending further support to the anti-pairing role of condensin II, a recent study utilizing high-throughput FISH combined with RNAi identified Cap-H2, Cap-D3, and SMC2 as factors that antagonize homolog pairing [38•]. Additionally, this and another recent study identified a number of novel genes that promote or antagonize somatic pairing, further suggesting that homolog pairing is a dynamic process that is regulated by both pairing and anti-pairing factors [38•, 39]. Among the anti-pairing factors that have been identified are chromatin proteins such as HP1, ORC1, and the Drosophila homolog of the human Mortality Factor 4 (Morf4), Mrg15 [38•, 40]. Identification of factors that both promote and disrupt pairing is important because it suggests that pairing and unpairing are both actively regulated. The traditional view has been that pairing is promoted by specific factors, while unpairing is the default state driven by entropy. Moreover, this brings up the exciting possibility that pairing/unpairing may be actively modulated at the local gene or region-specific level, thus regulating important trans-chromosomal interactions. It has been speculated that compaction activity on interphase chromatin disrupts pairing by driving intrachromosomal accordion folds that exclude interchromosomal interactions [41, 42•]. That condensin II can promote interphase chromatin compaction has been demonstrated in Drosophila and mouse [40, 42•, 43]; however, it remains unclear how this higher-order chromatin folding contributes to disruption of trans-chromosomal interactions.
Chromosome Territories
Interphase chromosomes in higher eukaryotes are organized into distinct sub-nuclear regions referred to as chromosome territories (CTs) (Fig. 1e) [44–48]. Formation of CTs results in the partitioning of the genome into functional domains, facilitating separation of actively transcribed genes from inactive genes and repetitive DNA sequences [49, 50]. In Drosophila ovarian nurse cell disruption of polytene pairing in mid-oogenesis coincides with formation of globular territories reminiscent of mammalian CTs [51]. Condensin II is required for disassembly of polytene chromosomes at this transition and also required for the proper formation of CTs [42•]. FISH in ovarian nurse cells showed that Cap-H2 mutants are unable to alter their orientation from the Rabl configuration (Fig. 1d), in which the centromeres are localized at one pole of the nucleus and the telomeres are positioned near the opposite pole. The finding that CAP-H2 promotes axial compaction and CT formation in both nurse cells and salivary glands, along with the observation that CT formation in meiotic spermatocytes is dependent on condensin II [41], suggests that condensin II has a role in regulating CT formation in multiple tissue types. Furthermore, these findings suggest that the interphase function of condensin II is similar to its role in axial compaction of meiotic chromosomes [13, 16•, 52–54]. An interesting implication arising from this work is that there is competition between cis (intrachromosomal) and trans (interchromosomal) interactions, and that condensin II may regulate chromatin organization by tipping the scales in favor of cis interactions.
How chromosomes transition from Rabl to CT configuration is not clearly understood. Because condensin II compaction forces have been implicated in CT formation [42•], it has been proposed that the intrinsic self-gathering properties of chromosome condensation can lead to discrete and non-overlapping CT formation. Moreover, chromatin tethers to the nuclear envelope act as anchoring points to reel in specific chromosomal regions to at each tethering point [42•]. Although chromatin tethers to the nuclear matrix can also exist, only tethers at or near the envelope can serve to draw CTs away from the center of the nucleus and form an inner nuclear space depleted of chromatin.
Maintenance of rDNA Stability
The most abundant genes in the eukaryotic genome are those encoding ribosomal RNA (rRNA), an integral component of ribosomes. rRNA genes are arranged in clusters of repeats, allowing cells to produce sufficient amounts of rRNA when demand for ribogenesis is high. In Saccharomyces cerevisiae, ~200 copies of rRNA genes (rDNA) are arranged on chromosome XII in tandem arrays consisting of a coding sequence for 35S rRNA that is transcribed by Pol I, 5S rRNA that is transcribed by Pol III, and two non-transcribed spacers (NTS1 and NTS2) [55–57]. The highly repetitive nature of rDNA, however, causes it to be intrinsically unstable as it is prone to losing copies of the repeats through homologous recombination. Copy number maintenance is achieved through regulation of recombination by the protein FOB1 [58]. Condensin, previously found to be required for proper mitotic condensation and segregation of rDNA regions [59], was identified in a genetic screen as an additional factor required for rDNA maintenance. Condensin is recruited by FOB1 to the replication fork barrier (RFB) sequence in early S-phase [60], a finding that points to a role for condensin in interphase cells and suggests a link between condensin loading and replication termination.
The role of condensin in rDNA maintenance is further supported by a report that condensin is loaded onto chromatin to promote condensation of rDNA in response to transcriptional repression during nutrient starvation [61]. Conditions such as nutrient starvation inhibit Target of rapamycin complex 1 (TORC1), resulting in rDNA transcription inhibition, nucleolar contraction, and condensin-mediated rDNA condensation [61–64]. Under these conditions, condensin acts antagonistically to Rad52, a component of the homologous recombination (HR) machinery, by preventing its localization to the nucleolus where it can mediate inappropriate HR among the rDNA repeats [65]. Consequently, inactivation of condensin leads to rDNA repeat instability [61]. The idea that condensin-mediated compaction generally restricts access to DNA by excluding binding proteins like Rad52 from DNA surfaces is also consistent with recent findings in human cells where condensin II compaction quenches checkpoint signaling of double-strand breaks (DSBs) ([66] and see section below on DNA damage repair). Furthermore, it has been hypothesized that repression of recombination within rDNA repeats might be due, in part, to the ability of condensin to mediate both cis- and trans- chromatin interactions [67]. For example, interactions between condensin-bound RFB sites scattered throughout the genome could promote the formation of chromatin folding, which would limit interactions among the rDNA repeats, thereby repressing recombination.
Pol III Gene Clustering
The budding yeast S. cerevisiae contains 274 tRNA genes dispersed throughout the genome. Throughout the cell cycle, however, these widely distributed genes are clustered in the nucleolus [68, 69]. While nucleolar positioning of tRNA genes is a microtubule-dependent process, tRNA gene clustering occurs by a separate condensin-dependent mechanism. Mutations in the five budding yeast condensin mutants smc2-8, smc4-1, ycg1-2, ysc4-1, and brn1-9 cause varying degrees of tRNA gene positioning defects and partially inhibit tRNA gene-mediated silencing, in which transcribed tRNA genes suppress RNA polymerase II-dependent transcription of nearby genes [69]. Yeast condensin was shown to bind to tRNA genes as well as genomic regions bound by the Pol III transcription factor TFIIIC in both small-scale and genome-wide ChIP experiments [69, 70]. These observations suggest that condensin is recruited to tRNA genes by TFIIIC where it might facilitate nucleolar clustering by participating in numerous interactions with multiple condensin complexes throughout the genome.
A more recent study has shown that condensin associates with Pol III genes at the c417 locus in S. pombe in both mitosis and interphase [71]. Pol III-bound tRNA and 5S rRNA genes that are dispersed throughout the linear genome are clustered in centromeric regions near the nuclear periphery [71, 72]. Centromeric localization of Pol III genes is mediated by condensin and can be counteracted by active Pol III transcription, which is thought to result in dissociation of TFIIIC and condensin from these regions [71]. Condensin-mediated localization of Pol III genes within the nucleus contributes to the three-dimensional organization of the genome in both budding and fission yeast; however, it remains to be determined whether this organization is conserved in higher eukaryotes. Interestingly, TFIIIC binding sites corresponding to tRNA genes have recently been shown by 4C to cluster in the nucleus of human cells [73]. It is tempting to speculate that the nuclear organization of Pol III genes mediated by TFIIIC is conserved among eukaryotes and that condensins might be important effectors of this process.
Gene Regulation
Increasing lines of evidence suggest that condensins play important roles in the regulation of gene expression, and this process is thought to be intimately linked to the role of condensins in regulation of chromosome architecture. For example, condensin has been proposed to maintain the silenced state of homeotic genes by regulating chromosome topology [74]. In budding yeast, loss of condensin binding to rDNA in Smc2p mutants results in relocation of the silencing protein Sir2p from telomeres to rDNA [75], suggesting that condensin helps to ensure the correct balance of nucleolar and telomeric Sir2p and that condensin might act as a barrier to prevent the spread of silent chromatin into active regions.
Condensins have been implicated in Drosophila position effect variegation (PEV). Condensin subunits have been linked not only to suppression, but also enhancement of PEV [74, 76–78], a finding that is surprising considering its well-established role in chromosome condensation. It is interesting to note that reporter genes located in close proximity to rDNA arrays are most sensitive to the effects of condensin mutations [78]. This raises the possibility that, as in S. cerevisiae [59, 79, 80], condensins may be enriched at rDNA in Drosophila, although to date the potential involvement of condensins at these loci has not been explored further.
Dosage compensation in C. elegans represents a well-established model system for studying condensin function in regulation of gene expression. The dosage compensation complex (DCC) achieves a two-fold downregulation of each of two X chromosomes in hermaphrodites and ensures a level of X-linked gene expression equal to that from the single male X chromosome [81]. The DCC consists of ten proteins, five of which are homologous to condensin complex subunits (Table 1), [82–86] and binds to two distinct classes of sites. The rex (recruitment element on X) sites recruit the DCC in an autonomous, sequence-dependent manner via a 12-base-pair sequence motif called MEX (motif enriched on X) [87, 88], whereas dox (dependent on X) sites are only able to recruit the DCC when located on the X chromosome [88].
Table 1.
Eukaryotic condensin proteins
| Species | Subunits | Interphase function | |
|---|---|---|---|
| S. cerevisiae | Core SMC | Smc2 | Nuclear organization [69] |
| Smc4 | Nuclear organization [69] | ||
| Non-SMC | Ycs4 | Nuclear organization [69] | |
| Ycs5/Ycg1 | Nuclear organization [69] | ||
| Brn1 | Nuclear organization [69] | ||
| S. pombe | Core SMC | Cut14 | DNA repair [95], nuclear organization [71] |
| Cut3 | DNA repair [94], nuclear organization [71] | ||
| Non-SMC | Cnd1 | ||
| Cnd3 | |||
| Cnd2 | DNA repair [93] | ||
| A. thaliana | Core SMC | CAP-E1 and CAP-E2 | |
| CAP-C | |||
| Non-SMC | CAB72176 (I) | ||
| BAB08309 (I) | |||
| AAC25941 (I) | |||
| At4g15890.1 (II) | |||
| CAP-G2/HEB1 (II) | DNA repair [101] | ||
| CAP-H2/HEB2 (II) | DNA repair [101] | ||
| C. elegans | Core SMC | MIX-1 | Gene expression [85] |
| SMC-4 | |||
| DPY-27 (IDC) | Gene expression [84] | ||
| Non-SMC | DPY-28 (I) | Gene expression [82] | |
| CAPG-1 (I) | Gene expression [82] | ||
| DPY-26 (I) | Gene expression [82] | ||
| HCP-6 (II) | |||
| CAP-G2 (II) | |||
| KLE-2 (II) | |||
| D. melanogaster | Core SMC | SMC2 | Nuclear organization [37, 38•] |
| SMC4/Gluon | Gene expression [78], nuclear organization[42•] | ||
| Non-SMC | CAP-D2 (I) | ||
| CAP-G (I) | Gene expression [78] | ||
| CAP-H/Barren (I) | Gene expression [78] | ||
| CAP-D3 (II) | Gene expression [117], nuclear organization [38•] | ||
| – | |||
| CAP-H2 (II) | Gene expression [38•], nuclear organization [38•] | ||
| Vertebrates | Core SMC | CAP-E/SMC2 | DNA repair [66] |
| CAP-C/SMC4 | |||
| Non-SMC | CAP-D2 (I) | ||
| CAP-G (I) | DNA repair [99] | ||
| CAP-H(I) | |||
| CAP-D3 (II) | DNA repair [100] | ||
| CAP-G2 (II) | Gene expression [43], nuclear organization [43, 104] | ||
| CAP-H2/nessy (II) | Gene expression [43], nuclear organization [43, 104] |
Since rex and dox sites are separated by distances up to 90 kb, long-range communication is essential to facilitate DCC binding on the X. The similarity between the DCC and condensin complexes suggests that the DCC could facilitate chromatin looping, bringing rex and dox sites into close proximity to one another. This looping model is reminiscent of the role of condensin complexes in promoting clustering of yeast tRNA genes [69], and ChIP-chip experiments have shown that DCC is located at sites near a majority of C. elegans tRNA genes [88]. Direct DCC binding to the promoter or coding sequence of a gene is not the determining factor in whether that gene will be compensated, an observation that lends further support to the idea that the reduction of gene expression over long distances occurs as a result of DCC-mediated changes in chromatin structure.
The finding that mutations in DCC components result in increased RNA Pol II binding to the X chromosome provided the first evidence that dosage compensation occurs at the transcriptional level [89]. Using a strategy for mapping transcription start sites (TSSs), it was shown that dosage compensation in C. elegans occurs by reduction of Pol II recruitment to X-linked gene promoters [90]. These observations suggest that the condensin-like DCC prevents Pol II recruitment to promoters and may disrupt long-range interactions between enhancers and promoters or reduce accessibility of Pol II to promoter regions [90]. The proposed function of the DCC in altering chromosome architecture to limit or facilitate interactions between distant regulatory elements is analogous to the function of boundary elements. Boundary activity is consistent with the role of condensin in rDNA locus organization as well as the clustering of tRNA genes, which are known to act as chromatin boundaries. The recent finding that the C. elegans DCC promotes X chromosome enrichment of H4K20me1 [91] supports the idea that inhibition of Pol II recruitment is a result of increased X chromosome compaction, which might function to reduce promoter accessibility of Pol II. Condensin II components N-CAPD3 and N-CAPG2 have recently been found to bind directly to methylated histone H4 on lysine-20 (H4K20me1), raising the possibility that the DCC might bind directly to H4K20me1 to affect compaction of chromatin [92].
DNA Damage Repair
Emerging evidence has linked condensin function to DNA repair processes in S. pombe [93]. In addition to defects in condensation, mutants for Cnd2, a non-SMC condensin subunit homologous to the Drosophila Barren protein, exhibited hypersensitivity to UV, hydroxyurea (HU), and methylmethane sulphonate. Recovery from HU-induced S phase arrest and activation of the checkpoint kinase Cds1 (a Chk2 homolog) requires Cnd2, and other condensin subunits, indicating the entire condensin complex is required for Cds1 activation. The hypersensitivity of cnd2-1 mutants is suppressed by overexpression of Cti1, an interactor of Cut3/SMC4 [94]. Cti1 is more abundant on chromatin after HU-induced DNA damage, and Cti1 likely recruits condensin to DNA damage sites. A mutation in the S. pombe Cut14/SMC2 exhibited sensitivity to DNA damaging agents similar to those observed in Cnd2 mutants, and these defects were suppressed by mutation in the replication protein A (RPA)-endcoding gene ssb1 [95]. Thus, condensins act antagonistically to the ssDNA-binding RPA by promoting its removal from DNA, suggesting condensin may function to remove repair proteins from DNA in preparation for mitosis.
In higher eukaryotes, both condensin I and condensin II are involved in DNA damage repair. Condensin I plays a role in single-strand break (SSB) repair through its interaction with PARP1, a DNA nick-sensor that is thought to play a role in organizing chromatin at the site of DNA damage and in the recruitment of repair proteins [96–98]. Condensin I interacts with PARP-1 specifically in inter-phase and forms a complex with PARP1 and its binding partner, the base excision repair (BER) factor XRCC11, in response to SSB damage [99]. The repair function of condensin I is specific for SSBs, as hCAP-D2 depletion causes no defects in DSB repair. Furthermore, condensin I was found to be recruited directly to sites of DNA damage where it is stabilized by its interaction with PARP1 [100].
Condensin II is also thought to function in DNA DSB repair. In A. thaliana, heb1-1 and heb2-1 encode the CAP-G2 and CAP-H2 subunits of condensin II, and mutations in these genes render these plants hyper-sensitive to boron-induced DNA breaks [101]. The mechanism by which condensins confer boron resistance remains unclear. In human cells and A. thaliana, condensin II is involved in HR-mediated repair [101, 102], and condensins also promote HR-mediated repair of DSBs at rDNA loci in yeast [103]. It has been proposed, however, that condensin might instead play a role in prevention of DNA damage induced by genotoxic stress, as evidenced by its role organizing and stabilizing the genome in response to nutrient starvation [61]. Conversely, in human cells the bromodomain protein Brd4 promotes inhibition of DNA damage signaling and DSB repair through condensin II-mediated chromatin compaction inhibition of DNA damage signaling and DSB repair [66], pointing to species-specific functions of condensin II in DNA damage repair.
Cell-Type-Specific Roles of Condensins
Recently, condensin subunits have been identified as key players in epigenetic regulation of cell-type specific gene expression. For example, murine CAP-G2 promotes chromatin condensation and transcriptional repression during erythroid cell differentiation [104]. Similarly, naïve T lymphocytes remain in a quiescent state until they undergo T cell receptor inducted T-cell activation. This transition involves changes in condensin-mediated higher order chromatin structure that allow the expression of proliferation-specific genes [43]. Mutations in the kleisin β (CAP-H2) subunit of condensin II lead to chromatin condensation defects as well as misregulation of genes that are normally silenced in naïve T-cells, indicating that condensin maintains chromatin in a condensed state during the quiescent period to suppress proliferation.
A recent study has shown that YY1, a PcG protein, functions in B-cell development. YY1 physically interacts with condensin subunit SMC4 through its REPO domain, which is necessary for Ig VJ segment rearrangement. YY1 also co-localizes with SMC4, SMC2, and BRRN1, the human CAP-H homolog, within the Ig [105]. Since the Ig loci are thought to be organized into loops that form rosette-like structures, it has been proposed that condensin might promote long-range interactions between YY1 binding sites that would facilitate rearrangement of Ig locus genes.
Conclusion
It has become evident that in addition to their well-established role in proper condensation and segregation of mitotic chromosomes, condensin complexes function in diverse interphase processes. Condensin proteins have therefore emerged as important regulators of chromatin organization throughout the cell cycle. It remains unknown, however, whether the mitotic and interphase functions of condensin complexes share similar molecular mechanisms. One proposed model for condensation is that condensin promotes positive supercoiling [106–108]. Perhaps a similar mechanism underlies the diverse interphase functions of condensin. Indeed, evidence suggests that supercoiling of DNA can facilitate long-range interactions [109]. Furthermore, dynamic supercoiling influences transcription [110], recombination [111, 112], and homolog pairing [35, 113]. Future work will be important to elucidate the molecular mechanisms underlying condensin-mediated regulation of interphase chromatin organization as well as to determine commonalities and differences among various condensin-mediated processes.
Understanding the molecular mechanisms by which condensins function may also provide insights into human disease. The first link between chromosome condensation and disease came from the observations that condensin II contributes to premature chromosome condensation in autosomal recessive primary microcephaly and that MCPH1 inhibits condensin II activity [114]. Condensin complexes function in numerous processes that are important to preserve genomic stability; therefore, it is not surprising that mutations in condensin subunits have been linked to tumorigenesis. For example, loss of heterozygosity in the chromosomal region containing the Cap-D3 gene is often associated with breast cancer, and mutations in SMC2 and SMC4 have been identified in several cell lines and tumor samples from patients with pyothorax-associated lymphoma [115]. Furthermore, the Drosophila retinoblastoma family protein Rfb1 is required for targeting CAP-D3 to chromatin, suggesting that aneuploidy in Rb mutants might result from loss of condensin II function [116]. Additionally, both CAP-D3 and RBF1 regulate genes involved in development and cell fate determination [117]. Finally, a mutation in Drosophila CAP-G results in cell cycle delays and increased apoptosis in retinal cells, linking condensin I to genome instability [118].
Higher-order chromatin organization has been implicated in a number of other human diseases. Cornelia de Lange syndrome (CdLS) is a developmental disorder that results from mutations in NIPBL, which acts with the cohesin complex to promote long-range chromatin interactions that are important for regulating gene expression. Interestingly, cells from CdLS patients exhibit extensive chromatin decondensation [119]; however, it is unclear whether condensin function is altered in these patients. Laminopathies, such as Hutchinson-Gilford progeria syndrome, are caused by mutations in nuclear lamins that are thought to affect levels of gene expression. Cells from patients with laminopathies exhibit abnormal nuclear envelope morphologies and changes in chromatin organization [120]. Similar defects in nuclear envelope morphology were observed upon inappropriate activation of condensin II by depletion of the SCFSlimb ubiquitin ligase [37]. Modulation of dynamic changes in nuclear organization might transduce mechanical forces to the nuclear envelope, resulting in aberrant nuclear envelope distortions in cells lacking normal lamin function [121]. It is tempting to speculate that condensin may play a role in this process (Fig. 1f, g). Certainly further research will be necessary in order to explore the potential function of condensins in diseases involving defects in higher-order chromatin organization.
Footnotes
Conflict of Interest: HA Wallace declares no conflicts of interest. G Bosco declares no conflicts of interest.
Human and Animal Rights and Informed Consent: This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Heather A. Wallace, Email: Heather.Wallace@dartmouth.edu.
Giovanni Bosco, Email: Giovanni.Bosco@dartmouth.edu.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
- 1.Groth A, et al. Chromatin challenges during DNA replication and repair. Cell. 2007;128(4):721–33. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 2.Macalpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harb Perspect Biol. 2013 doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thadani R, Uhlmann F, Heeger S. Condensin, chromatin crossbarring and chromosome condensation. Curr Biol. 2012;22(23):R1012–21. doi: 10.1016/j.cub.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet. 2013;14(1):62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol. 2011;192(5):711–21. doi: 10.1083/jcb.201010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson DF, Marshall KM, Earnshaw WC. Condensin: architect of mitotic chromosomes. Chromosome Res. 2009;17(2):131–44. doi: 10.1007/s10577-008-9009-7. [DOI] [PubMed] [Google Scholar]
- 8.Hirano T. Condensins: organizing and segregating the genome. Curr Biol. 2005;15(7):R265–75. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 9.Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10(10):1445–52. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schleiffer A, et al. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11(3):571–5. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 11.Anderson DE, et al. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156(3):419–24. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melby TE, et al. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142(6):1595–604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirota T, et al. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci. 2004;117(Pt 26):6435–45. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- 14.Gerlich D, et al. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16(4):333–44. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 15.Ono T, et al. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15(7):3296–308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011;25(14):1464–9. doi: 10.1101/gad.2060311. This paper describes the establishment of a cell-free system used to determine the contributions of each condensin complex to chromosome condensation. This study concluded that the ratio of condensin I to condensin II is important for determining chromosome shape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai JH, McKee BD. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci. 2011;124(Pt 12):1955–63. doi: 10.1242/jcs.006387. [DOI] [PubMed] [Google Scholar]
- 18.Stevens NM. A study of the germ cells of certain diptera, with reference to the heterochromosomes and the phenomena of synapsis. J Exp Zool. 1907;5(1–3):359–74. [Google Scholar]
- 19.Metz CW. Chromosome studies on the Diptera II The paired association of chromosomes in the Diptera, and its significance. J Exp Zool. 1916;21(2):213–79. [Google Scholar]
- 20.Apte MS, Meller VH. Homologue pairing in flies and mammals: gene regulation when two are involved. Genet Res Int. 2012;2012:430587. doi: 10.1155/2012/430587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant-Downton RT, Dickinson HG. Plants, pairing and phenotypes—two’s company? Trends Genet. 2004;20(4):188–95. doi: 10.1016/j.tig.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13(12):1627–41. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krueger C, et al. Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PLoS One. 2012;7(7):e38983. doi: 10.1371/journal.pone.0038983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busch H, Rothblum L. The cell nucleus. New York: Academic Press; 1974. pp. 1–12. [Google Scholar]
- 25.Fung JC, et al. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141(1):5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiraoka Y, et al. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J Cell Biol. 1993;120(3):591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta. 2004;1677(1–3):165–80. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Csink AK, Henikoff S. Large-scale chromosomal movements during interphase progression in Drosophila. J Cell Biol. 1998;143(1):13–22. doi: 10.1083/jcb.143.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gemkow MJ, Verveer PJ, Arndt-Jovin DJ. Homologous association of the Bithorax-Complex during embryogenesis: consequences for transvection in Drosophila melanogaster. Development. 1998;125(22):4541–52. doi: 10.1242/dev.125.22.4541. [DOI] [PubMed] [Google Scholar]
- 30.Painter TS. A new method for the study of chromosome rearrangements and the plotting of chromosome maps. Science. 1933;78(2034):585–6. doi: 10.1126/science.78.2034.585. [DOI] [PubMed] [Google Scholar]
- 31.Zybina EV, Zybina TG. Polytene chromosomes in mammalian cells. Int Rev Cytol. 1996;165:53–119. doi: 10.1016/s0074-7696(08)62220-2. [DOI] [PubMed] [Google Scholar]
- 32.Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila-melano-gaster. Am Nat. 1954;88(841):225–39. [Google Scholar]
- 33.Rong YS, Golic KG. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 2003;165(4):1831–42. doi: 10.1093/genetics/165.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fritsch C, Ploeger G, Arndt-Jovin DJ. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 2006;14(4):451–64. doi: 10.1007/s10577-006-1068-z. [DOI] [PubMed] [Google Scholar]
- 35.Williams BR, et al. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics. 2007;177(1):31–46. doi: 10.1534/genetics.107.076356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322(5906):1384–7. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]
- 37.Buster DW, et al. SCFSlimb ubiquitin ligase suppresses condensin II-mediated nuclear reorganization by degrading Cap-H2. J Cell Biol. 2013;201(1):49–63. doi: 10.1083/jcb.201207183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Joyce EF, et al. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 2012;8(5):e1002667. doi: 10.1371/journal.pgen.1002667. This paper identified factors that both promote and disrupt somatic homolog pairing and provided evidence that pairing and unpairing are both actively regulated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bateman JR, et al. A genome-wide screen identifies genes that affect somatic homolog pairing in Drosophila. G3 (Bethesda) 2012;2(7):731–40. doi: 10.1534/g3.112.002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith HF, et al. Maintenance of interphase chromosome compaction and homolog pairing in Drosophila is regulated by the condensin Cap-H2 and its partner Mrg15. Genetics. 2013 doi: 10.1534/genetics.113.153544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartl TA, et al. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 2008;4(10):e1000228. doi: 10.1371/journal.pgen.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Bauer CR, Hartl TA, Bosco G. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 2012;8(8):e1002873. doi: 10.1371/journal.pgen.1002873. This paper demonstrates that condensin II contributes to the compartmentalization of the interphase nucleus into chromosome territories and points to common mechanisms among mitotic and interphase activities of condensin II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rawlings JS, et al. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 2011;30(2):263–76. doi: 10.1038/emboj.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 45.Santos AP, et al. The architecture of interphase chromosomes and gene positioning are altered by changes in DNA methylation and histone acetylation. J Cell Sci. 2002;115(Pt 23):4597–605. doi: 10.1242/jcs.00160. [DOI] [PubMed] [Google Scholar]
- 46.Pecinka A, et al. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113(5):258–69. doi: 10.1007/s00412-004-0316-2. [DOI] [PubMed] [Google Scholar]
- 47.Berr A, et al. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 2006;48(5):771–83. doi: 10.1111/j.1365-313X.2006.02912.x. [DOI] [PubMed] [Google Scholar]
- 48.Shaw PJ, et al. The architecture of interphase chromosomes and nucleolar transcription sites in plants. J Struct Biol. 2002;140(1–3):31–8. doi: 10.1016/s1047-8477(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 49.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148(3):458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126(2):293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- 52.Green LC, et al. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci. 2012;125(Pt 6):1591–604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139(1):73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu HG, Koshland DE. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J Cell Biol. 2003;163(5):937–47. doi: 10.1083/jcb.200308027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci USA. 1979;76(1):410–4. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell GI, et al. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977;252(22):8118–25. [PubMed] [Google Scholar]
- 57.Planta RJ, Raue HA. Control of ribosome biogenesis in yeast. Trends Genet. 1988;4(3):64–8. doi: 10.1016/0168-9525(88)90042-x. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi T, et al. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12(24):3821–30. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149(4):811–24. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johzuka K, et al. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26(6):2226–36. doi: 10.1128/MCB.26.6.2226-2236.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26(2):448–58. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10(4):987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsang CK, et al. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22(22):6045–56. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaragoza D, et al. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18(8):4463–70. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsang CK, Zheng XF. Opposing role of condensin and radiation-sensitive gene RAD52 in ribosomal DNA stability regulation. J Biol Chem. 2009;284(33):21908–19. doi: 10.1074/jbc.M109.031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Floyd SR, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498(7453):246–50. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johzuka K, Horiuchi T. The cis element and factors required for condensin recruitment to chromosomes. Mol Cell. 2009;34(1):26–35. doi: 10.1016/j.molcel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 68.Thompson M, et al. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302(5649):1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haeusler RA, et al. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22(16):2204–14. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D’Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22(16):2215–27. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwasaki O, et al. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell. 2010;21(2):254–65. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noma K, et al. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125(5):859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 73.Raab JR, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31(2):330–50. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lupo R, et al. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol Cell. 2001;7(1):127–36. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- 75.Machin F, et al. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr Biol. 2004;14(2):125–30. [PubMed] [Google Scholar]
- 76.Dej KJ, Ahn C, Orr-Weaver TL. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168(2):895–906. doi: 10.1534/genetics.104.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savvidou E, et al. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci. 2005;118(Pt 11):2529–43. doi: 10.1242/jcs.02392. [DOI] [PubMed] [Google Scholar]
- 78.Cobbe N, Savvidou E, Heck MM. Diverse mitotic and inter-phase functions of condensins in Drosophila. Genetics. 2006;172(2):991–1008. doi: 10.1534/genetics.105.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang BD, et al. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol. 2005;25(16):7216–25. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell. 2002;13(2):632–45. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer BJ. X-chromosome dosage compensation. WormBook: the online review of C. elegans biology. 2005:1–14. doi: 10.1895/wormbook.1.8.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Csankovszki G, et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol. 2009;19(1):9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7(5):311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 84.Chuang PT, Albertson DG, Meyer BJ. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79(3):459–74. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 85.Lieb JD, et al. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92(2):265–77. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 86.Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 2002;16(4):399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- 87.McDonel P, et al. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444(7119):614–8. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jans J, et al. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009;23(5):602–18. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25(5):499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kruesi WS, et al. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vielle A, et al. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet. 2012;8(9):e1002933. doi: 10.1371/journal.pgen.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466(7305):508–12. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aono N, et al. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417(6885):197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- 94.Chen ES, Sutani T, Yanagida M. Cti1/C1D interacts with condensin SMC hinge and supports the DNA repair function of condensin. Proc Natl Acad Sci USA. 2004;101(21):8078–83. doi: 10.1073/pnas.0307976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akai Y, et al. Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol. 2011;1(4):110023. doi: 10.1098/rsob.110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31(6):446–54. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 97.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst) 2008;7(7):1077–86. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 99.Heale JT, et al. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21(6):837–48. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kong X, et al. Condensin I recruitment to base damage-enriched DNA lesions is modulated by PARP1. PLoS One. 2011;6(8):e23548. doi: 10.1371/journal.pone.0023548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakamoto T, et al. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell. 2011;23(9):3533–46. doi: 10.1105/tpc.111.086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wood JL, et al. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283(43):29586–92. doi: 10.1074/jbc.M804080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ide S, et al. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327(5966):693–6. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 104.Xu Y, et al. MTB, the murine homolog of condensin II subunit CAP-G2, represses transcription and promotes erythroid cell differentiation. Leukemia. 2006;20(7):1261–9. doi: 10.1038/sj.leu.2404252. [DOI] [PubMed] [Google Scholar]
- 105.Pan X, et al. YY1 controls Igkappa repertoire and B-cell development, and localizes with condensin on the Igkappa locus. EMBO J. 2013;32(8):1168–82. doi: 10.1038/emboj.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cuylen S, Metz J, Haering CH. Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol. 2011;18(8):894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- 107.Baxter J, Aragon L. A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet. 2012;28(3):110–7. doi: 10.1016/j.tig.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 108.Baxter J, et al. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331(6022):1328–32. doi: 10.1126/science.1201538. [DOI] [PubMed] [Google Scholar]
- 109.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70(6):2548–56. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sinden RR. The helix turns at 60: writhing free in chromosomes. Nat Struct Mol Biol. 2013;20(3):251–3. doi: 10.1038/nsmb.2533. [DOI] [PubMed] [Google Scholar]
- 111.Trigueros S, Roca J. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomer-ase-deficient yeast cells. J Biol Chem. 2002;277(40):37207–11. doi: 10.1074/jbc.M206663200. [DOI] [PubMed] [Google Scholar]
- 112.Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388(6644):798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- 113.Strick TR, Croquette V, Bensimon D. Homologous pairing in stretched supercoiled DNA. Proc Natl Acad Sci USA. 1998;95(18):10579–83. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamashita D, et al. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol. 2011;194(6):841–54. doi: 10.1083/jcb.201106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ham MF, et al. Condensin mutations and abnormal chromosomal structures in pyothorax-associated lymphoma. Cancer Sci. 2007;98(7):1041–7. doi: 10.1111/j.1349-7006.2007.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Longworth MS, et al. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22(8):1011–24. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Longworth MS, et al. A shared role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet. 2012;8(4):e1002618. doi: 10.1371/journal.pgen.1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Seipold S, et al. Non-SMC condensin I complex proteins control chromosome segregation and survival of proliferating cells in the zebrafish neural retina. BMC Dev Biol. 2009;9:40. doi: 10.1186/1471-213X-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nolen LD, et al. Regional chromatin decompaction in Cornelia de Lange syndrome associated with NIPBL disruption can be uncoupled from cohesin and CTCF. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dauer WT, Worman HJ. The nuclear envelope as a signaling node in development and disease. Dev Cell. 2009;17(5):626–38. doi: 10.1016/j.devcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 121.Shimi T, Butin-Israeli V, Goldman RD. The functions of the nuclear envelope in mediating the molecular crosstalk between the nucleus and the cytoplasm. Curr Opin Cell Biol. 2012;24(1):71–8. doi: 10.1016/j.ceb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]