Abstract
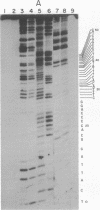
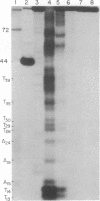
We have investigated the sites of DNA damage by the antitumor antibiotics neocarzinostatin and bleomycin by using a 5'-end-labeled DNA fragment of defined sequence as a substrate. At the high drug concentrations used here, neocarzinostatin creates single-strand breaks in DNA at positions of adenine and thymine in the presence of 2-mercaptoethanol, and bleomycin cleaves DNA at GC and GT sequences and to a lesser extent at TA sequences with its degradative activity enhanced by 2-mercaptoethanol. In the presence of ferrous ions, bleomycin cleaves DNA at TT, AT, and TA, as well as at GC and GT sequences. Both antibiotics make double-strand breaks in DNA at specific sites and it is likely that these result from two independent single-strand breaks at nearby sites on opposite strands of the DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arditti R. R., Scaife J. G., Beckwith J. R. The nature of mutants in the lac promoter region. J Mol Biol. 1968 Dec;38(3):421–426. doi: 10.1016/0022-2836(68)90396-3. [DOI] [PubMed] [Google Scholar]
- Bearden J. C., Jr, Lloyd R. S., Haidle C. W. Bleomycin-induced breakage of closed-circular DNA. Biochem Biophys Res Commun. 1977 Mar 21;75(2):442–448. doi: 10.1016/0006-291x(77)91062-2. [DOI] [PubMed] [Google Scholar]
- Beerman T. A., Goldberg I. H. The relationship between DNA strand-scission and DNA synthesis inhibition in HeLa cells treated with neocarzinostatin. Biochim Biophys Acta. 1977 Mar 18;475(2):281–293. doi: 10.1016/0005-2787(77)90019-3. [DOI] [PubMed] [Google Scholar]
- Crooke S. T., Bradner W. T. Bleomycin, a review. J Med. 1976;7(5):333–428. [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidle C. W. Fragmentation of deoxyribonucleic acid by bleomycin. Mol Pharmacol. 1971 Nov;7(6):645–652. [PubMed] [Google Scholar]
- Haidle C. W., Weiss K. K., Kuo M. T. Release of free bases from deoxyribonucleic acid after reaction with bleomycin. Mol Pharmacol. 1972 Sep;8(5):531–537. [PubMed] [Google Scholar]
- Haseltine W. A., Kleid D. G., Panet A., Rothenberg E., Baltimore D. Ordered transcription of RNA tumor virus genomes. J Mol Biol. 1976 Sep 5;106(1):109–131. doi: 10.1016/0022-2836(76)90303-x. [DOI] [PubMed] [Google Scholar]
- Lown J. W., Sim S. K. The mechanism of the bleomycin-induced cleavage of DNA. Biochem Biophys Res Commun. 1977 Aug 22;77(4):1150–1157. doi: 10.1016/s0006-291x(77)80099-5. [DOI] [PubMed] [Google Scholar]
- Maizels N. M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meienhofer J., Maeda H., Glaser C. B., Czombos J., Kuromizu K. Primary structure of neocarzinostatin, an antitumor protein. Science. 1972 Nov 24;178(4063):875–876. doi: 10.1126/science.178.4063.875. [DOI] [PubMed] [Google Scholar]
- Ohtsuki K., Ishida N. Neocarzinostatin-induced breakdown of deoxyribonucleic acid in HeLa-S3 cells. J Antibiot (Tokyo) 1975 Feb;28(2):143–148. doi: 10.7164/antibiotics.28.143. [DOI] [PubMed] [Google Scholar]
- Poon R., Beerman T. A., Goldberg I. H. Characterization of DNA strand breakage in vitro by the antitumor protein neocarzinostatin. Biochemistry. 1977 Feb 8;16(3):486–493. doi: 10.1021/bi00622a023. [DOI] [PubMed] [Google Scholar]
- Rhaese H. J., Freese E. Chemical analysis of DNA alterations. I. Base liberation and backbone breakage of DNA and oligodeoxyadenylic acid induced by hydrogen peroxide and hydroxylamine. Biochim Biophys Acta. 1968 Feb 26;155(2):476–490. [PubMed] [Google Scholar]
- Rhaese H. J., Freese E., Melzer M. S. Chemical analysis of DNA alterations. II. Alteration and liberation of bases of deoxynucleotides and deoxynucleosides induced by hydrogen peroxide and hydroxylamine. Biochim Biophys Acta. 1968 Feb 26;155(2):491–504. [PubMed] [Google Scholar]
- Samy T. S., Hu J. M., Meienhofer J., Lazarus H., Johnson R. K. A facile method of purification of neocarzinostatin, an antitumor protein. J Natl Cancer Inst. 1977 Jun;58(6):1765–1768. doi: 10.1093/jnci/58.6.1765. [DOI] [PubMed] [Google Scholar]
- Sausville E. A., Peisach J., Horwitz S. B. A role for ferrous ion and oxygen in the degradation of DNA by bleomycin. Biochem Biophys Res Commun. 1976 Dec 6;73(3):814–822. doi: 10.1016/0006-291x(76)90882-2. [DOI] [PubMed] [Google Scholar]
- Sim S. K., Lown J. W. The mechanism of the neocarzinostatin-induced cleavage of DNA. Biochem Biophys Res Commun. 1978 Mar 15;81(1):99–105. doi: 10.1016/0006-291x(78)91635-2. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa H., Maeda K., Takeuchi T., Okami Y. New antibiotics, bleomycin A and B. J Antibiot (Tokyo) 1966 Sep;19(5):200–209. [PubMed] [Google Scholar]