Abstract
Seasonal and pandemic influenza viruses continue to be a leading global health concern. Emerging resistance to the current drugs and the variable efficacy of vaccines underscore the need for developing new flu drugs that will be broadly effective against wild-type and drug-resistant influenza strains. Here, we report the discovery and development of a class of inhibitors targeting the cap-snatching endonuclease activity of the viral polymerase. A high-resolution crystal form of pandemic 2009 H1N1 influenza polymerase acidic protein N-terminal endonuclease domain (PAN) was engineered and used for fragment screening leading to the identification of new chemical scaffolds binding to the PAN active site cleft. During the course of screening, binding of a third metal ion that is potentially relevant to endonuclease activity, was detected in the active site cleft of PAN in the presence of a fragment. Using structure-based optimization, we developed a highly potent hydroxypyridinone series of compounds from a fragment hit that defines a new mode of chelation to the active site metal ions. A compound from the series demonstrating promising enzymatic inhibition in a fluorescence-based enzyme assay with an IC50 value of 11 nM was found to have an antiviral activity (EC50) of 11 μM against PR8 H1N1 influenza A in MDCK cells.
Influenza pandemics of 1918 H1N1 (Spanish flu), 1957 H2N2 (Asian flu), 1968 H3N2 (Hong Kong flu), and 2009 H1N1 (swine flu) signify the need for wider vaccination and therapeutic options for a future outbreak. Emergence of drug-resistant variants of circulating influenza A strains, adaptation of H5N1 for human-to-human transmission, and the very recent outbreak of an unusual H7N9 strain are among the central concerns. 1 Vaccination in the form of either inactivated or live attenuated viruses is the primary defense against influenza. However, at times including the year 2012–13, the vaccines are only partially effective2 and timely production of vaccines in the event of a pandemic influenza outbreak would be challenging. Currently, antivirals used for prophylactic and therapeutic treatment of influenza infection target either the neuraminidase or the M2 ion channel protein.3-5 There continues to be a need for novel, more efficacious drugs due to drug resistance by seasonal influenza viruses6 as well as emerging potential pandemic strains H5N17 and H7N98.
Influenza virus contains eight negative-stranded RNA genomic segments. The three largest RNA segments encode for the polymerase acidic protein (PA), polymerase basic protein 1 (PB1), and polymerase basic protein 2 (PB2) that assemble to form a heterotrimeric viral RNA-dependent RNA polymerase (RdRP).9-11 The influenza RdRP is highly conserved among all influenza strains and subtypes, and is essential for viral transcription and replication. Transcription is initiated by hijacking a 5’ cap from the host cell pre-mRNAs via a ‘cap snatching’ mechanism.12,13 This entails the binding of PB2 to the 5’-cap (m7G) of the host cell’s pre-mRNA, which is then cleaved 10–13 nucleotides downstream by the endonuclease activity of influenza RdRP.13 The cleaved 5’-capped RNA segment is used as a primer to initiate transcription of the viral mRNA.
The cap-snatching event is not a normal cellular process; however, it is an essential step in the influenza virus lifecycle. Additionally, recent discovery of an alternative encoded protein, PA-X, which has an overlapping sequence and similar enzymatic activity as PAN, may contribute to the host protein shutdown observed during influenza A infection.14 Thus, inhibition of the endonuclease activity of influenza RdRP is an attractive target for novel antiviral therapy.
Biochemical and structural studies have elucidated that the endonuclease active site resides in the N-terminal domain of the PA subunit (PAN).15-17 Crystal structures of H5N116 and H3N217 PAN revealed the architecture of the enzymatic domain. The domain fold and active site arrangement of PAN were found to be similar to those of the PD-(D/E)XK family of nucleases. Mutagenesis of catalytic residues within the RdRP showed that endonuclease-dependent transcription was abolished while the endonuclease-independent RNA replication was maintained.16 Endonuclease activity of PAN was further validated by the hydrolysis of both single-stranded RNA and single-stranded DNA substrates.17 Crystal structures of 2009 pandemic H1N118 and an internally truncated variant of H5N119 PAN in complexes with endonuclease inhibitors have been reported recently.
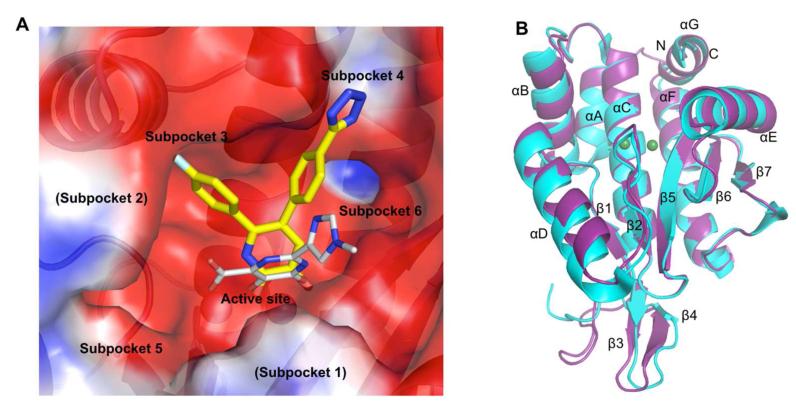
Early endonuclease inhibitors, which include diketo acids,20,21 flutimide,22 N-hydroxamic acids,23 N-hydroxyimides,23 tetramic acids,24 and the green tea catechins,25,26 were identified through pharmacological screening. However, lack of critical information regarding the location of the endonuclease site and its structure did not permit pursuit of structure-based drug design (SBDD). Structures of PAN have revealed a deep active site cleft containing multiple subpockets that can be exploited for drug design (Figure 1A). Crystal structures of PAN in complexes with inhibitors have revealed a common chelation mode with the two active site metal ions (M1 and M2) despite extending into different subpockets.18,19 The structural characterization of PAN has renewed interest in the development of endonuclease inhibitors. Crystal structures of pH1N1 endonuclease were determined in complexes with nucleotides and known inhibitors.18 Baughman et al. used a fluorescence polarization assay to establish structure-activity relationships (SAR) for a carboxamide-based series of inhibitors reported to have activity in the micromolar to nanomolar range.27 Antiviral activity for one of these compounds was determined to be 12.7 μM (see Supplemental Table 1 for antiviral activity comparisons of known influenza endonuclease inhibitors). Crystal structures for a subset of the compounds in complex with H5N1 PAN helped to rationalize the observed potency for the series.19
Figure 1.
Description of PAN structure. (A) APBS51 calculated electrostatic potential surface of the active site cleft of PAN. The binding of a ligand described in this study (7) is shown with yellow carbon atoms and a carboxamide derivative with three chelating groups crystallized by Dubois et al. is shown in gray (PDB code: 4E5I).19 The subpockets are labeled as previously described.19 (B) Superposition of the pH1N1 2009 PAN structures from this study and one previously described (PDB code: 4AVQ).18
Here, we report results from a small molecule fragment screening effort against 2009 pandemic H1N1 (pH1N1) PAN. SBDD for a promising fragment was pursued to develop a series of compounds that demonstrated antiviral activity. The compounds exhibit a mode of chelation to the active site metal ions that is different from the chelating mode observed for typical endonuclease inhibitors, such as diketo acid compounds.
Results and Discussion
Crystal Engineering of PAN for SBDD
The initial crystal forms for influenza endonuclease were not suitable for SBDD due to occlusion of the active site15 and difficulty of reproducing the crystals.19 To search for an appropriate crystal form, the H5N1 endonuclease construct (obtained from Zhiyong Lou and Zihe Rao16) was mutagenized and verified to be identical to the sequence of the 2009 pH1N1 PAN (A/California/07/2009) and various C-terminal truncations were made to obtain proteolytically stable pH1N1 PAN fragments for crystallization trials. A construct with an HRV14 3C protease cleavable N-terminal dual-hexahistidine tag28 truncated at residue 204, was found to give the maximum yield among the tested constructs (Supplemental Table 2). This construct could be crystallized in multiple conditions. A condition with PEG 8000 as precipitant in the presence of 5 mM Mg2+ and 1 mM Mn2+ produced crystals that reproducibly diffracted X-rays to 1.7 Å resolution. These crystals could be routinely soaked in solutions containing 10 mM small molecule fragments29 in 10% (v/v) DMSO. This crystal form has one molecule per asymmetric unit and the endonuclease active site was not occluded by crystal contacts, thus allowing for binding of small molecule inhibitors to PAN. Although the crystal form is distinct from the recently described 2009 pH1N1 PAN18 structure, the superposition of both structures revealed a backbone root-mean-square deviation (RMSD) of only 0.24 Å (Figure 1B).
Fragment Screening
Fragment screening by X-ray crystallography was conducted using a previously assembled diverse compound library30 by soaking fragments into preformed crystals. Although the fragment library was not biased to contain metal chelators, 159 compounds were found to contain potentially chelating moieties. Of the 159 potential metal chelators, eight fragments inhibited PAN (Supplemental Table 3). Initial screening of 775 fragments was done using cocktails of four to eight compounds designed to ensure maximum structural diversity and minimum chemical reactivity.30 Fragment hits identified from cocktail soaking were verified by individual compound soaking experiments. Screening revealed eight compounds binding to the endonuclease domain yielding a hit rate of 1.0% (Supplemental Table 4). All fragments were also subjected to a secondary screen using a high-throughput fluorescence resonance energy transfer based inhibition assay,28 analogous to those developed by Kowalinski et al.17 as well as Noble et al.31 The measured IC50 for each compound was then used to calculate the apparent ligand efficiency (LE), which is the binding contribution per non-hydrogen atom (NHA) of a compound.32,33 LE, a widely used metric in fragment screening experiments to rank small molecules based on their average af?nity contribution per atom, was used as a guide for hit selection and lead optimization.
Third Metal Binding at the Active Site
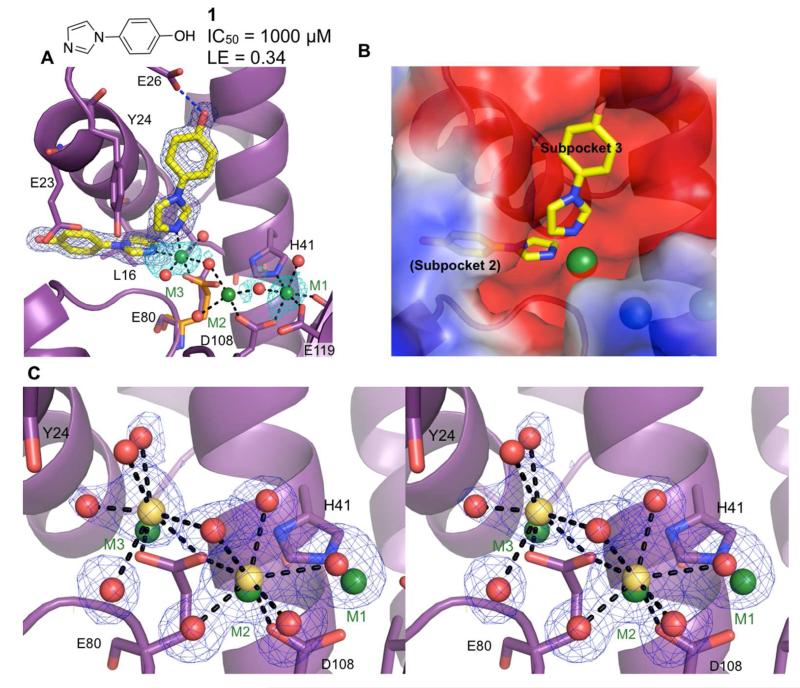
Of the eight fragments identified during the course of our fragment screening, four fragments chelated to the two catalytic metal ions, M1 and M2, in a mode similar to the known endonuclease inhibitors. Surprisingly, binding of compound 1 revealed the presence of a previously undetected third metal ion (M3) in the active site cleft, at a distance ~4 Å from M2 (Figure 2A). Compound 1 binds to two sites: subpocket 2, which consists of Leu16, Lys19, Ala20, Glu23, Gly81, and Asp83, as well as subpocket 3 surrounded by Tyr24, Glu26, Lys34, and Ile38 (Figure 2A). The imidazole moieties of both molecules coordinate with M3. The side chain of Glu80 can no longer coordinate with M2 due to steric clash with M3, which is located 1.0 Å from the Glu80 of the apo structure. Coordination to M3 requires the side chain of Glu80 to shift such that the carboxylate rotates 90° and no longer coordinates with M2. As a result, weak electron density is detected for M2 relative to M1 and M3. Although strong electron density was observed for 1, the inhibitory activity (IC50) was found to be only 1 mM.
Figure 2.
Discovery of a third active site metal. Metal-coordinating bonds are depicted as black dashed lines and hydrogen bonds as blue dashed lines. Residues with significant structural changes upon binding of a ligand are displayed with the apo structure shown in orange. (A) Structure of 1 bound to subpockets 2 and 3. Compound 1 coordinates to a third metal (M3), which coordinates to Glu80. The electron density (dark blue mesh) is calculated from a Fo-Fc omit map and contoured at 5σ. Electron density calculated from anomalous data (cyan) is contoured at 3σ. (B) Structure of 1 calculated electrostatic potential surface (APBS51) and subpocket labels. (C) Stereoview of the active site cleft showing electron density for the metal ions and coordinating waters in the apo PAN structure from a 100 mM CaCl2 soak. The locations of the metal ions from the soak of 1 are shown in green whereas the locations of the calcium cations from a 100 mM soak are shown in yellow. The electron density for the metal and coordinating waters is shown (omit map contour of 2.5σ).
Calorimetric results from the titration of endonuclease with Mn2+ or Mg2+ in the absence of substrate support a two-metal binding model for both Mn2+ and Mg2+, with Mn2+ having higher binding affinity than Mg2+.34 It is possible that the third metal observed in the active site due to binding of 1 is stabilized by coordination to the substrate. In order to determine if M3 is an artifact of fragment binding, we titrated metals (specifically manganese and calcium) into EDTA-treated crystals of PAN, which was necessary to remove residual metal ions from the crystallization condition. Due to difficulties in determining a crystal structure of PAN in complex with RNA, soaking of high concentrations of various metals into crystals of PAN without substrate present was used to compensate for the lack of substrate coordination to M3. Soaks of magnesium, manganese, zinc, and nickel salts did not give conclusive results due to loss of X-ray diffraction resolution. A calcium soak, however, gave high-resolution X-ray diffraction with clear electron density for the metal ions with their coordinating residues and water molecules.
It was previously determined that calcium thermally stabilized PAN without enhancing endonuclease activity.17,35 Surprisingly, a 100 mM calcium soak revealed metal binding at the M2 and M3 sites but not at the M1 site (Figure 2B); binding of a Mg2+ ion only at the M2 site and not the M1 site was earlier observed in the H5N1 PAN structure.16 Soaking revealed that the position of M3, 2.5 Å away from Glu80, allowed Glu80 to coordinate with both M3 and M2, which was not seen in PAN in complex with 1. Bipyramidal pentavalent coordination with the protein and water molecules confirmed the bound metal ions to be calcium and not residual manganese or magnesium in our structure.
The above experiment confirmed the existence of a third metal-binding site, suggesting that M3 is not an artifact of fragment binding, but may have functional relevance. A three-metal ion coordination mode has also been reported for other endonucleases, including T5 flap endonuclease, endonuclease IV, and EcoRV.36-38 A three-metal catalysis model for EcoRV has been developed using a combination of structural and molecular modeling data. Briefly, in this model, metal at site I (analogous to M1 in PAN) is directly involved in catalysis whereas metal at site II (analogous to M2 in PAN) provides structural support and metal at site III (analogous to M3 in PAN) is believed to stabilize the negative charge of the transition state and recruit a water to protonate the leaving group.39-41 Although this may not necessarily be the case with respect to influenza endonuclease, the presence of the third metal binding near the active site suggests the need for further studies particularly in the presence of ssRNA substrates. Indeed, a third metal near the active site maybe relevant for other enzymes in the PD-(D/E)XK family of nucleases.42
Hit-to-lead Development
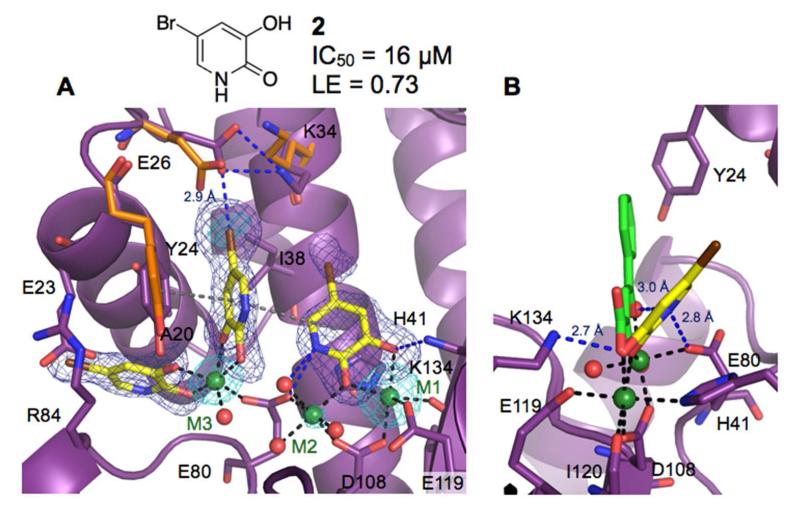
Fragment screening subsequently identified 5-chloro-3-hydroxypyridin-2(1H)-one as an active site chelating ligand in the presence of the third metal. Stronger electron density was observed for a bromine derivative of this initial hit (2). Subsequent refinement of the structure revealed three copies of 2 bound to PAN. Two molecules of 2 were bound to the subpockets 2 and 3 and chelated to M3, analogous to the binding of 1, while the third molecule of 2 coordinated with M1 and M2 at the endonuclease active site (Figures 3A-B). An IC50 value of 16 μM was determined for 2 using the enzymatic assay. The ligand efficiency of 0.73 kcal/mol·NHA was calculated assuming the inhibition was solely due to binding of the compound to the active site.
Figure 3.
Structure of 2 bound to PAN. Metal-coordinating bonds are depicted as black dashed lines, hydrophobic and cation-π interactions as grey, and hydrogen bonds as blue dashed lines. Residues with significant structural changes upon binding of a ligand are depicted with the apo structure shown in orange. Electron density calculated from anomalous scattering data (cyan) is contoured at 3σ. (A) Compound 2 bound at subpockets 2 and 3, and the active site (omit map contoured at 4σ). (B) Side view of 2 bound at the active site superposed with 2,4-dioxo-4-phenylbutanoic acid (PDB code: 4E5G).19
Binding of 2 to subpockets 2 and 3 creates an interaction network between the hydroxyls of 2, M3, and a coordinating water molecule. M3 is shifted 1.8 Å from its location in the structure in complex with 1, which allows Glu80 to coordinate with both M2 and M3. A non-optimal hydrophobic interaction network is created upon binding of 2 to subpockets 2 and 3. This network consists of π-stacking interaction between the side chains of Arg84 and Glu23, which, in turn, weakly interacts with the bromine of 2 bound to subpocket 2. The side chains of Ala20 and Tyr24 help bridge the network by participating in weak hydrophobic interactions with 2 bound to both subpockets 2 and 3. Compound 2 binding to subpocket 3 forms hydrophobic interactions with Lys34 and Ile38. Additionally, the bromine of 2 bound to subpocket 3 forms a halogen bond with a carboxylate oxygen of Glu26 at a distance of 2.9 Å.
Compound 2 bound at the active site also creates an interaction network of its own by coordinating with M1 and M2 and forming a hydrogen bond with Lys134 at a distance of 2.7 Å. The typical binding motif for influenza endonuclease inhibitors entails three chelating groups forming four coordinating interactions with M1 and M2 (43 for review). However, 2 forms three chelating interactions through two chelating groups. This type of coordination is common for magnesium and manganese chelating inhibitors of other targets (44 for review). The position analogous to the third metal chelating group from reported endonuclease inhibitors18,19 is occupied by a water molecule that chelates to M2 and forms a hydrogen bond with the pyridinone nitrogen of 2 at a distance of 3.0 Å (Figure 3B). The pyridinone nitrogen is also within hydrogen-bonding distance (2.8 Å) from a side-chain oxygen of Glu80. A striking difference between 2 and known three group chelating inhibitors is that the pyridinone ring of 2 is tilted by an angle of ~50° towards His41 (Figure 3B). This orientation of 2 allows for favorable π-stacking interactions with the aromatic side chain of His41, an invariant metal-chelating residue in PAN of all influenza viruses. Moreover, the distinct binding mode and chelation of 2 at the active site provided opportunities for designing new chemical classes of endonuclease inhibitors, and hit-to-lead optimization was conducted in this case.
Synthetically feasible chemical modifications were designed using structural information and molecular docking of the designed compounds was undertaken using AutoDock Vina.45 The most promising substitutions/modifications predicted to improve the binding affinity were considered for chemical synthesis. Each of the synthesized pyridinone derivatives was evaluated for endonuclease inhibitory activity using the described enzymatic assay. The binding of each compound was ascertained by determining the crystal structure in complex with PAN. SAR was developed for this series by iterative cycles of molecular docking, chemical synthesis, enzymatic assay, and structure determination. Substitutions at the 4, 5, and 6-positions of the pyridinone were explored. Detailed chemical synthesis and SAR for the compounds in this series have been described elsewhere.46
Substitution at the 4-Position
Computational docking using the structure of 2 complexed to endonuclease identified possible substitutions at the 4-position. Surprisingly, even small chemical substitutions at the 4-position of the pyridinone were found to be deleterious for inhibition of endonuclease activity. A plausible explanation is that substitutions at the 4-position may perturb the electronic arrangements of the chelating oxygens of the pyridinone ring.
Substitution at the 5-Position
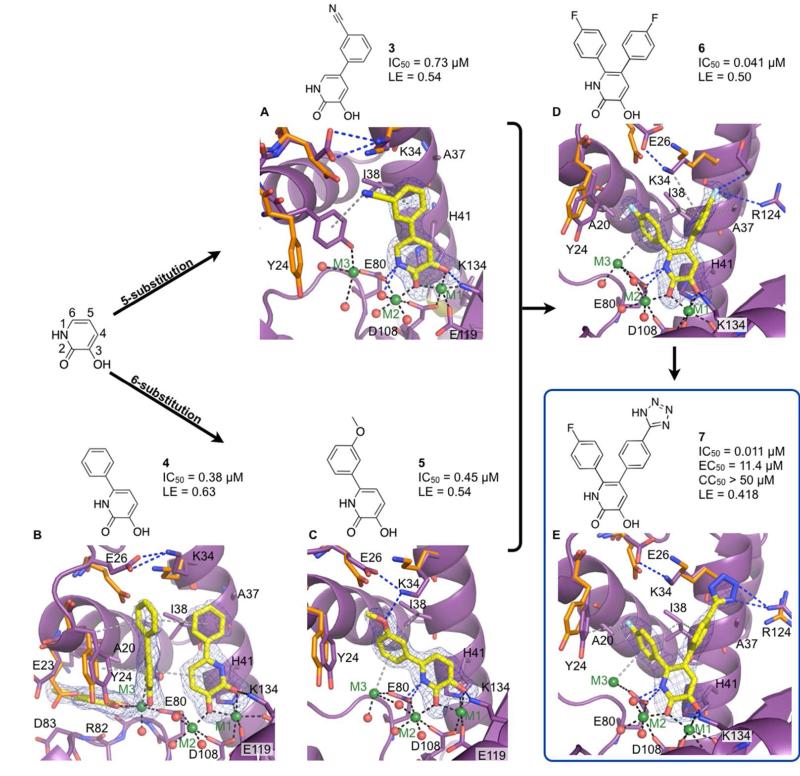
The structure of 2 bound to PAN revealed room for extension at the 5-position into subpocket 4. Compared to 2, a 20-fold improvement in inhibitory activity was seen by adding a 3-benzonitrile at the 5-position (3). Unlike 2, only one molecule of 3 is bound to the active site of PAN, and the benzonitrile group is extended into subpocket 4 (Figure 4A). The phenyl ring forms hydrophobic contacts with side chains of Ala24 and Ile38. Hydrophobic interactions are also seen between the nitrile of 3 and side chains of Lys34 and Ile38. Binding of 3 induces several changes in the active site of PAN compared to the apo structure. Expansion of subpocket 3 occurs due to rearrangements of side chains of Glu26 and Lys34, which now form a new salt bridge (Figure 4A). Stabilization of Glu23, which is a part of subpocket 2, was also observed via a hydrogen-bonding interaction with a water molecule that coordinates with M3. Even though M3 is present in the structure, 3 does not directly interact with M3.
Figure 4.
Development of the pyridinone series of compounds. Metal coordinating bonds are depicted as black dashes, hydrophobic and cation-π interactions are grey, and hydrogen bonds as blue dashes. Residues with significant structural changes upon binding of a ligand are shown with the apo structure colored orange. (A) 3 bound at the active site (omit map contoured at 3σ). (B) Compound 4 bound at subpockets 2 and 3, and the active site (omit map 4σ contours). (C) 5 bound at the active site (omit map contour of 4.5σ). (D) 6 bound at the active site (omit map with 4σ contours). (E) 7 bound at the active site (omit map contoured at 4σ).
The most significant change in the cleft involved Tyr24, a residue that has been structurally shown to be a highly adaptive and involved in nucleotide binding.18 Compared to the apo structure, binding of 3 caused a ~1 Å backbone shift of Tyr24. A 45° rotation of the side-chain torsion angle χ1 downwards allows the hydroxyl of Tyr24 to chelate to M3, which maintains an octahedral coordination with Tyr24, Glu80, and three surrounding water molecules. Suboptimal weak π interaction between the nitrile moiety of 3 and the aromatic ring of Tyr24 was also observed.
Substitution at the 6-Position
Relative to 2, a 40-fold increase in inhibitory potency was observed upon addition of a phenyl ring at the 6-position of the pyridinone (4). The structure of PAN complexed with 4 revealed a binding pattern analogous to 2. Three molecules of 4 bound to the active site and subpockets 2 and 3 (Figure 4B). Molecule 4 occupying subpocket 3 forms a favorable π stacking interaction with Tyr24 while maintaining an edge-to-face stacking interaction with 4 bound at the active site. Additional hydrophobic interactions were gained between the phenyl of 4 bound to subpocket 3 and the side chains of Glu26, Lys34, and Ile38.
Compared to the binding modes of 2 and 3, a new mode of binding was observed for 4 at the active site. The chelation environment remained unaltered despite the pyridinone ring being flipped 180°. This flipped orientation causes loss of a hydrogen bond that is typically observed between the nitrogen of the pyridinone ring and a coordinating water of M2. However, the C4 atom maintains the 3.0 Å distance to the coordinating water molecule indicating that a C-H…O hydrogen bond may partially compensate for the loss of the N-H…O hydrogen bond. This orientation of the pyridinone ring positions the phenyl ring of 4 in subpocket 4, thereby permitting simultaneous binding of another 4 molecule to subpocket 3.
Substitutions on the phenyl ring of 4 were explored to improve the potency and binding affinity. Of the compounds tested, the strongest electron density was seen for 5, which has a m-methoxyphenyl group at the 6-position of the pyridinone ring (Figure 4C). The crystal structure of 5 in complex with PAN revealed the binding of only one molecule to the active site with all three metal ions present. Compound 5 chelated to M1 and M2 at the active site and developed a cation-π interaction between the m-methoxyphenyl and M3. Substitution at the 6-position with the m-methoxyphenyl did not allow for the flipped orientation of the pyridinone ring that was seen in the binding of 4 to the active site. The oxygen from the m-methoxyphenyl group forms a hydrogen bond with the side chain of Lys34 (3.1 Å), which forms a salt bridge with the carboxylate of Glu26. Hydrophobic interactions between the m-methoxyphenyl and side chains of Ala20, Tyr24, and Ile38 were also observed. The IC50 value of 5 was determined to be 0.45 μM in the enzymatic assay with an LE of 0.54 kcal/(mol·NHA). Although 5 is slightly less active than 4 (0.38 μM), the inhibition by one molecule of 5, which maintained key interactions with all three metal ions as well as residues comprising the active site and subpocket 3 of PAN, was considered to be a remarkable improvement.
Di-substitution at the 5- and 6-Positions
The combination of SAR, molecular docking, and a single, highly superimposable binding mode for the mono-substituted pyridinone ring within the crystal structures indicated that di-substitution at the 5- and 6-positions could be advantageous. This hypothesis was confirmed by a significant improvement in the inhibitory activity seen for the di-substituted compounds compared to 2. Of the di-substituted compounds synthesized and tested, 6, which has a p-fluorophenyl group at both the 5- and the 6-positions, had a 400-fold improvement in inhibitory activity compared to 2.
The structure of 6 revealed that the p-fluorophenyl at the 6-position maintained the interactions of the analogous m-methoxyphenyl moiety of 5 with subpocket 3, including the cation-π interaction with M3 (Figure 4D). The crystal structure indicates weak hydrophobic interaction between the p-fluorophenyl and Tyr24. The weak electron density observed for the Tyr24 side chain indicates that the residue is flexible, thus requiring further optimization of our inhibitors to strengthen this interaction. The p-fluorophenyl at the 5-position extending towards subpocket 4 forms hydrophobic interactions with the side chains of Lys34, Ala37, and Ile38. The fluorine at the para position of the phenyl ring of 6 forms an electrostatic interaction with Arg124, which causes the crystallographic ethylene glycol seen in many of our crystal structures to be shifted upwards by 2 Å. This structural finding suggests room for additional expansion from the para position of the phenyl ring to enhance interactions with Arg124 and other neighboring residues.
Modification at the para position of the phenyl ring extending off the 5-position of the pyridinone with a p-tetrazole (7) enhanced the potency by 4-fold compared to 6 to give an IC50 value of 11 nM and LE of 0.42 kcal/(mol·NHA) (Figure 4E). The structure of 7 in complex with PAN revealed that the tetrazole completely displaces the ethylene glycol to interact with Arg124. Additionally, the tetrazole forms hydrophobic interactions with the side chain of Lys34. The potency of 7 in the enzymatic assay warranted investigation of this compound in an antiviral assay.
Multicycle Antiviral Activity and Cytotoxicity of Compound 7
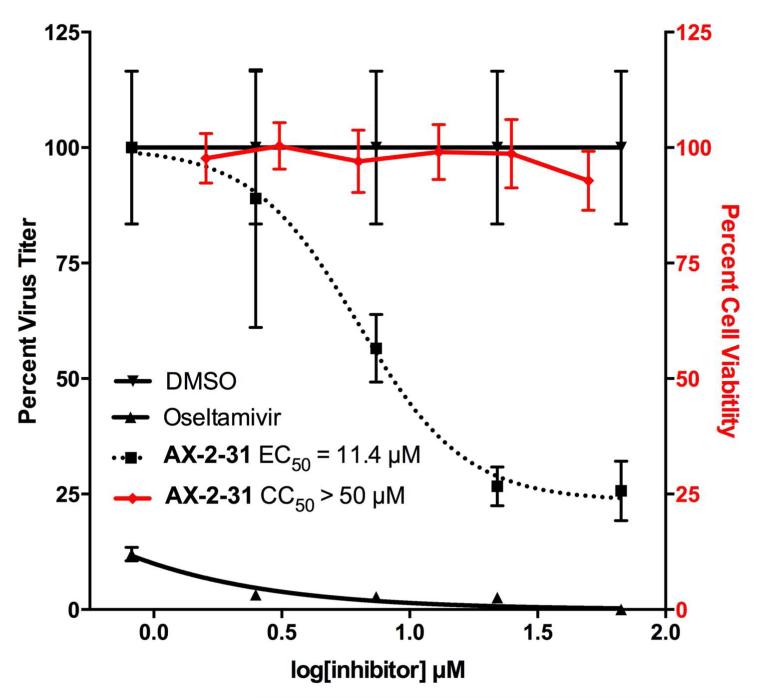
A virus yield assay was used to determine the inhibition of influenza A/Puerto Rico/8/1934 (H1N1; PR8) in MDCK cells by 7. Virus titers in supernatants were determined at 24 hours post infection at the indicated concentrations of 7 by fluorescent forming unit (FFU) assays (Figure 5).47-49 DMSO and oseltamivir were included as negative and positive controls, respectively. The EC50 value for 7 was determined to be 11 μM with no cytotoxicity detected (CC50 > 50 μM) as measured by MTT assay at 24 hours. As expected, DMSO did not have any effect on viral replication while oseltamivir had an EC50 value of 0.015 μM, similar to that previously shown for PR8.50 These results demonstrate the antiviral properties of 7 against a homologous H1N1 subtype virus and the lack of cytotoxicity at the tested concentrations.
Figure 5.
Viral yield inhibition assay and cytotoxicity assay. Virus yield assay using MDCK cells and the A/Puerto Rico/8/1934 (H1N1) influenza strain was used to determine the antiviral activity of 7; oseltamivir and 0.1% (v/v) DMSO (only) were used as the positive and negative controls. Cytotoxicity for 7 (shown in red) was determined from MDCK cells after 24 hours using the MTT assay.
Conclusions
Protein engineering was successful in obtaining a high-resolution crystal form of 2009 pH1N1 PAN that allowed for fragment screening by X-ray crystallography and high-throughput SBDD. Unexpectedly, fragment binding revealed the presence of a third metal ion in the active site cleft and subsequent analyses indicated that the third metal ion may have a functional role. Fragment screening identified a novel pharmacophore, hydroxypyridinone, which was optimized through multiple rounds of enzyme assays, X-ray crystallography, molecular modeling, and medicinal chemistry to a compound, 7, that has an antiviral EC50 activity of 11 μM.
Methods
Details about protein engineering, expression, purification, crystallization are given in Supporting Information. Methodologies for fragment screening and the inhibition assay are specified in Supporting Information. Full descriptions of the chemical synthesis of the described compounds are also included in Supporting Information as well as the materials and methods for the influenza virus yield assay. Statistics for data collection and refinement of the X-ray crystal structures are listed in Supplemental Table 5.
Supplementary Material
Acknowledgements
We thank Zhiyong Lou and Zihe Rao for providing the plasmid of H5N1 PAN, and the laboratories of Ann Stock and Gaetano Montelione for access to equipment used in this study. Data collection was largely conducted at the Cornell High Energy Synchrotron Source (CHESS) and the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-0225180, and the MacCHESS resource is supported by NIH/NCRR award RR-01646. Use of NSLS is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. Research in LM-S’s laboratory is funded by the NIH grants R01 AI077719, R21NS075611-01, R03AI099681-01A, The University of Rochester Center for Biodefense Immune Modeling (CBIM) contract HHSN272201000055C, and by the NIH/NIAID network of Centers of Excellence in Influenza Research and Surveillance (CEIRS) contract HHSN266200700008C. This study was supported by a research agreement between Prodaptics Pharmaceuticals, Inc., and Rutgers, The State University of New Jersey (E.A. and E.J.L.). We are grateful to Michael Pazzani for support of this project.
References
- 1.Parry J. H7N9 avian flu infects humans for the first time. British Med. J. 2013;346:f2151. doi: 10.1136/bmj.f2151. [DOI] [PubMed] [Google Scholar]
- 2.CDC Interim adjusted estimates of seasonal Influenza vaccine effectiveness – United States February 2013. Centers for disease control and prevention. 2013 [Online] Available: http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6207a2.htm?s_cid=mm6207a2_w. [PMC free article] [PubMed]
- 3.Das K, Aramini JM, Ma LC, Krug RM, Arnold E. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 2010;17:530–538. doi: 10.1038/nsmb.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Clercq E, Neyts J. Avian influenza A (H5N1) infection: targets and strategies for chemotherapeutic intervention. Trends Pharmacol. Sci. 2007;28:280–285. doi: 10.1016/j.tips.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govorkova EA, Webster RG. Combination chemotherapy for influenza. Viruses. 2010;2:1510–1529. doi: 10.3390/v2081510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Global Influenza Programme World Health Organization. 2013 [Online] Available: http://www.who.int/influenza/en. [Google Scholar]
- 7.Govorkova EA, Baranovich T, Seiler P, Armstrong J, Burnham A, Guan Y, Peiris M, Webby RJ, Webster RG. Antiviral resistance among highly pathogenic influenza A (H5N1) viruses isolated worldwide in 2002-2012 shows need for continued monitoring. Antiviral Res. 2013;98:297–304. doi: 10.1016/j.antiviral.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y, Lu S, Song Z, Wang. W, Hao P, Li J, Zhang X, Yen H, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Zhang X, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet Epub. 2013 doi: 10.1016/S0140-6736(13)61125-3. May 29, 2013. DOI:10.1016/S0140-6736(08)61345-8. [DOI] [PubMed] [Google Scholar]
- 9.Krug RM, Alonso-Caplen FV, Julkunen I, Katze MG. Expression and replication of the influenza virus genome. In: Krug RM, editor. In The Influenza Viruses. Plenum Publishing Corp.; New York: 1989. [Google Scholar]
- 10.Huang TS, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J.Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda A, Uéda K, Nagata K, Ishihama A. Identification of the RNA polymerase-binding site on genome RNA of influenza virus. J. Biochem. 1987;102:1241–1249. doi: 10.1093/oxfordjournals.jbchem.a122163. [DOI] [PubMed] [Google Scholar]
- 12.Beaton AR, Krug RM. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res. 1981;9:4423–4436. doi: 10.1093/nar/9.17.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)- dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 14.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Firth JK, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara K, Schmidt FI, Crow M, Brownlee GG. Amino acid residues in the N-terminal region of the PA subunit of Influenza A virus RNA polymerase play a crictical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 2006;80:7789–7798. doi: 10.1128/JVI.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 17.Dias A, Bouvier D, Creṕin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 18.Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar OHJ, Ruigrok RWH, Cusack S. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 2012;8:e10002831. doi: 10.1371/journal.ppat.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois RM, Slavish PJ, Baughman BM, Yun MK, Bao J, Webby RJ, Webb TR, White SW. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 2012;8:e1002830. doi: 10.1371/journal.ppat.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomassini J, Selnick H, Davies ME, Armstrong ME, Baldwin J, Bourgeois M, Hastings J, Hazuda D, Lewis J, McClements W, Ponticello G, Radzilowski E, Smith G, Tebben A, Wolfe A. Inhibition of Cap (m7GpppXm)-dependent endonucelase of influenza-virus by 4-subsituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother. 1994;38:2827–2837. doi: 10.1128/aac.38.12.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastings JC, Selnick H, Wolanski B, Tomassini JE. Anti-influenza virus activities of 4-substituted 2,4-dioxobutanoic acid inhibitors. Antimicrob. Agents Chemother. 1996;40:1304–1307. doi: 10.1128/aac.40.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomassini JE, Davies ME, Hastings JC, Lingham R, Mojena M, Raghoobar SL, Singh SB, Tkacz JS, Goetz MA. A novel antiviral agent which inhibits the endonuclease of influenza viruses. Antimicrob. Agents Chemother. 1996;40:1189–1193. doi: 10.1128/aac.40.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cianci C, Chung TDY, Meanwell N, Putz H, Hagen M, Colonno RJ, Krystal M. Identification of N-hydroxamic acid and N-hydroxyimide compounds that inhibit the influenza virus polymerase. Antivir. Chem. Chemother. 1996;7:353–360. [Google Scholar]
- 24.Parkes KEB, Ermert P, Fassler J, Ives J, Martin JA, Merrett JH, Obrecht D, Williams G, Klumpp K. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem. 2003;46:1153–1164. doi: 10.1021/jm020334u. [DOI] [PubMed] [Google Scholar]
- 25.Kuzuhara T, Iwai Y, Takahashi H, Hatakeyama D, Echigo N. Green tea catechins inhibit the endonuclease activity of influenza A virus RNA polymerase. PLoS Curr. 2009;1:RRN1052. doi: 10.1371/currents.RRN1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song JM, Lee KH, Seong BL. Antiviral effect of catechins in green tea on influenza virus. Antiviral Res. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Baughman BM, Slavish PJ, DuBois RM, Boyd VA, White SW, Webb TR. Identification of Influenza Endonuclease Inhibitors Using a Novel Fluorescence Polarization Assay. ACS Chem. Biol. 2012;7:526–534. doi: 10.1021/cb200439z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan F, He M, Taussig MJ. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on ni-nitrilotriacetic acid surfaces. Anal. Chem. 2006;78:3072–3079. doi: 10.1021/ac060184l. [DOI] [PubMed] [Google Scholar]
- 29.Sagong HY, Parhi A, Bauman JD, Patel D, Vijayan RSK, Arnold E, LaVoie EJ. 3-Hydroxylquinolin-2(1H)-ones as inhibitors of Influenza A endonuclease. ACS Med. Chem. Lett. 2013 May 7; doi: 10.1021/ml4001112. DOI:10.1021/ml4001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauman JD, Patel D, Dharia C, Fromer MW, Ahmed S, Frenkel Y, Vijayan RSK, Eck TJ, Ho WC, Das K, Shatkin AJ, Arnold E. Detecting allosteric sites of HIV-1 reverse transcriptase by X-ray crystallographic fragment screening. J. Med. Chem. 2013;56:2738–2746. doi: 10.1021/jm301271j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble E, Cox A, Deval J, Kim B. Endonuclease substrate selectivity characterized with full-length PA of influenza A virus polymerase. Virology. 2012;433:27–34. doi: 10.1016/j.virol.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Disc. Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 34.Creṕin T, Dias A, Palencia A, Swale C, Cusack S, Ruigrok RW. Mutational and metal binding analysis of the endonuclease domain of the influenza virus polymerase PA subunit. J. Virol. 2010;84:9096–9104. doi: 10.1128/JVI.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doan L, Handa B, Roberts NA, Klumpp K. Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry. 1996;38:5612–5619. doi: 10.1021/bi9828932. [DOI] [PubMed] [Google Scholar]
- 36.Syson K, Tomlinson C, Chapados BR, Sayers JR, Tainer JA, Williams NH, Grasby JA. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J. Biol. Chem. 2008;283:28741–28746. doi: 10.1074/jbc.M801264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov I, Tainer JA, McCammon JA. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1465–1470. doi: 10.1073/pnas.0603468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sam MD, Perona JJ. Catalytic roles of divalent metal ions in phosphoryl transfer by EcoRV endonuclease. Biochemistry. 1999;38:6576–6586. doi: 10.1021/bi9901580. [DOI] [PubMed] [Google Scholar]
- 39.Kovall RA, Matthews BW. Type II restriction endonucleases: structural, functional, and evolutionary relationships. Curr. Op. Chem. Biol. 3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 40.Horton NC, Newberry KJ, Perona JJ. Metal ion-mediated substrate assisted catalysis in type II restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13489–13494. doi: 10.1073/pnas.95.23.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horton NC, Perona JJ. DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites. Biochemistry. 2004;43:6841–6857. doi: 10.1021/bi0499056. [DOI] [PubMed] [Google Scholar]
- 42.Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das K. Antivirals targeting influenza A virus. J. Med. Chem. 2012;55:6263–6277. doi: 10.1021/jm300455c. [DOI] [PubMed] [Google Scholar]
- 44.Rouffet M, Cohen SM. Emerging trends in metalloprotein inhibition. Dalton Trans. 2011;40:3445–3454. doi: 10.1039/c0dt01743d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parhi AK, Xiang A, Bauman JD, Patel D, Vijayan RSK, Das K, Arnold E, LaVoie EJ. Submitted. 2013 doi: 10.1016/j.bmc.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 47.Schickli JH, Flandorfer A, Nakaya T, Martinez-Sobrido L, Garcia-Sastre A, Palese P. Plasmid-only rescue of influenza A virus vaccine candidates. Phil. Trans. R. Soc. B. 2001;356:1965–1973. doi: 10.1098/rstb.2001.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, Garcia-Sastre A. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 2010;84:2157–2163. doi: 10.1128/JVI.01433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker SF, Guo H, Albrecht RA, Garcia-Sastre A, Topham DJ, Martinez-Sobrido L. Protection against lethal influenza with a viral mimic. J. Virol. 2013 Epub May 29, 2013. DOI:10.11128/JVI.01081-13. [Google Scholar]
- 50.McSharry JJ, McDonough AC, Olson BA, Drusano GL. Phenotypic drug susceptibility assay for influenza virus neuraminidase inhibitors. Clin. Diagn. Lab. Immunol. 2004;11:21–28. doi: 10.1128/CDLI.11.1.21-28.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker NA, Sept. D, Joseph S, Hoist MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.