Abstract
RNA–protein interactions are vital throughout the HIV-1 life cycle for the successful production of infectious virus particles. One such essential RNA–protein interaction occurs between the full-length genomic viral RNA and the major structural protein of the virus. The initial interaction is between the Gag polyprotein and the viral RNA packaging signal (psi or Ψ), a highly conserved RNA structural element within the 5′-UTR of the HIV-1 genome, which has gained attention as a potential therapeutic target. Here, we report the application of a target-based assay to identify small molecules, which modulate the interaction between Gag and Ψ. We then demonstrate that one such molecule exhibits potent inhibitory activity in a viral replication assay. The mode of binding of the lead molecules to the RNA target was characterized by 1H NMR spectroscopy.
Introduction
Human immunodeficiency virus type 1 (HIV-1) is a positive-sense, diploid RNA retrovirus of the Lentivirus family. HIV-1 is the major cause of acquired immunodeficiency syndrome (AIDS) worldwide. While current antiviral treatments have had a profound impact on the life expectancy of infected individuals, HIV-1 remains a major global health problem with reports estimating 34 million people infected worldwide in 2010 (UNAIDS World AIDS Day Report, 2011; www.unaids.org). New therapeutics are still needed as the virus is highly variable, and drug resistance by mutational escape develops swiftly.1
Most of the traditional and recent antiviral drugs such as emtricitabine, rilpivirine, maraviroc, and raltegravir target the early life cycle of HIV-1, prior to integration of the provirus, the one exception being the protease inhibitor class such as saquinavir and lopinavir.2 One such target of the HIV-1 late life cycle is the interaction between Gag, the major structural protein of HIV-1, and the full-length viral RNA genome3,4 in which the viral polyprotein Gag binds the viral RNA with a high degree of specificity through the Ψ-packaging domain located within the 5′-UTR (Figure 1). Following this initial binding, further Gag protein binding and subsequent trafficking to the host cell membrane allows packaging of the RNA genome into the budding viron.5−7
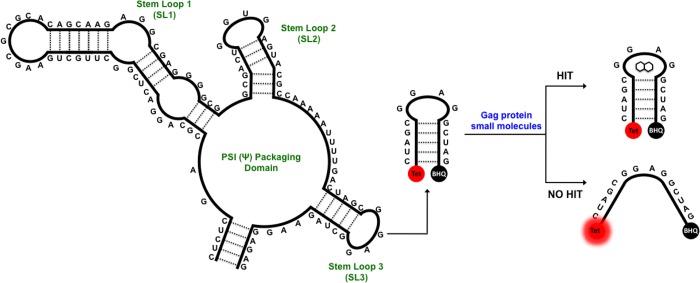
Figure 1.
Structure of the core Ψ-packaging domain of HIV-1. Schematic representation of the in vitro biophysical assay. Attached to the 5′ and 3′ ends of stem loop 3 (SL3) are the TET fluorophore and blackhole quencher (BHQ1), respectively, which form a destabilization assay in the presence of Gag. Small molecules that inhibit the destabilation of SL3 are taken forward, after validation, to a viral replication assay.
Targeting RNA or RNA–protein interactions with small molecules has been a relatively unexplored approach to small molecule intervention compared to targeting protein active sites,8 with few examples, besides targeting the rRNA. Herman et al.9 targeted the internal ribosome entry site (IRES) of the hepatitis C virus (HCV); Butcher et al.10 and al-Hashimi et al.11 targeted the frameshift and transactivation response (TAR) elements from HIV-1, respectively. Cis-acting RNA elements such as the IRES, frameshift sites, TAR and Ψ are far more highly conserved than conventional protein targets, and mutational escape is therefore more difficult.12
The core packaging domain of HIV-1 comprises three stem loops (SL): SL1 is the dimerization initiation site, SL2 contains the major splice donor, and SL3 is known as the critical packaging element (Figure 1).13−16 These regions have been extensively interrogated by mutagenesis to elucidate each SL’s contributions to genomic RNA packaging,5,17 This has led to the concept that a structural switch occurs within the 5′-UTR; associated with the change in function from translation to encapsidation of the full-length RNA genome.17−19 The switch is suggested to be triggered by the binding of one or more Gag proteins to the 5′-UTR.7,20 Gag is the structural polyprotein of HIV-1 consisting of four major domains: the N-terminus matrix (MA-p17), capsid (CA-p24), nucleocapsid (NC-p7), and the C-terminus p6.20 Gag is the effector molecule recognizing the Ψ-packaging domain (and in particular SL3) in vivo. Thus, it is vital to use the authentic RNA ligand, the Gag polyprotein, to identify a compound that will be inhibitory to RNA packaging in vivo, which, until now, has not been done.
Herein we describe the identification of small molecules that are able to disrupt the HIV-1 Gag/RNA interaction in vitro using a destablisation assay based on the interaction of the Gag protein with the 14 base tetraloop of SL3 (Figure 1). We then characterize the identified small molecules using a cell based assay activity and highlight the selective binding of NSC260594 to the tetraloop of SL3 by 1H NMR spectroscopy.
Results and Discussion
Initially we employed a fluorogenic destabilization assay (Figure 1) based on Gag’s ability to affect the structure of a molecular beacon modeled on SL3 of the Ψ-packaging domain.21 This assay was used to screen the LOPAC (1280 molecules, Sigma) and Diversity Set II (1363 molecules, NIH) libraries with positive hits being defined as small molecules showing inhibition greater than three standard deviations (21%) from the mean negative control. On each plate, the positive (SL3) and negative (SL3 and Gag) controls were used to calculate the Z′ score, and any plate that failed to gain a Z′ greater than 0.5 was rejected and the plate was rescreened.22 Confirmation of the primary hits was performed in triplicate under the original screening conditions, and 78 compounds were identified (Figure S1, Supporting Information).
To assess the biological activity, of the compounds shortlisted by the in vitro screen on HIV-1 replication, we employed a viral replication assay based on protocols from Hu et al.23 (Table S2, Supporting Information). The protease inhibitor drugs saquinavir24 and lopinavir25 were used as positive antiviral controls as these FDA-approved drugs affect late stages of the viral life cycle (Figures S3 and S4, Supporting Information). Furthermore, an enzyme-linked immunosorbent assay (ELISA) directed against the CA-p24 domain of the Gag polyprotein was used to quantify inhibition of virus production by the small molecules.26 Because of either high cytotoxicity (≤20 μM) of the small molecule on the 293T cells or low efficacy (≥20 μM) of the small molecule within the viral replication assay 75 of the 78 compounds were discarded (Table S2, Supporting Information) leaving compounds A1895, R0529, and NSC260594 (Table S1, Supporting Information). A1895 was discarded as A1895 has been identified in previous studies looking at the HIV-1 envelope glycoprotein.27−29 R0529 shares structural similarities to the Hoechst series of nucleic acid dyes which are known to interact nonspecifically with DNA and RNA.30,31 NSC260594 is a quinolinium derivative similar in structure to known DNA binding compounds such as SN 6999 and SN 7167.32,33 Quinolinium derivatives are known to bind in the minor groove of DNA, but DNA minor groove binders have previously been documented as having little to no binding toward RNA.22 While R0529 and NSC260594 both showed efficacy, both in the initial screen based on the RNA hairpin SL3 and the viral replication assay, far less is known about the interaction of NSC260594 with RNA.
Furthermore, the effect of NSC260594 on virus production (measured by the CA-p24 ELISA) reflects the effect on viral infectivity (Figure 2), indicating that NSC260594 affects particle production and release. This is consistent with studies involving mutations to Ψ, which show a similar decrease in particle production and infectivity levels. This is in contrast to the protease inhibitors saquinavir and lopinavir, where the effect on p24 production occurs at a far higher concentration than the effect on viral replication (Figures S2 and S3, Supporting Information). For these reasons we decided to focus our attention on NSC260594 and its interaction with the SL3 RNA.
Figure 2.
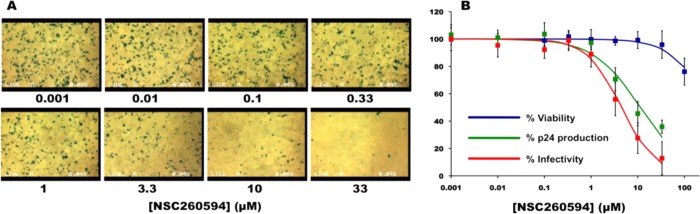
Biological activity of NSC260594. (A) 24 h postinfection the expression of β-galactosidase within the TZM-bl cell were imaged using X-Gal (blue spots in images). (B) Viability of the 293T cells (blue line, 50% inhibition (CC(50) = N/A)), viral production from transfected 293T (green line, 50% inhibition (p24(50) = 11.3 ± 3.4 μM)) and infectivity of harvested viral particles (red line 50% inhibition (IC(50) = 4.5 ± 1.8 μM)) in the presence of different concentrations of NSC260594.
We then employed circular dichroism (CD) (Figure S5, Supporting Information), fluorescence melting (Table S3, Supporting Information) and 1H NMR spectroscopy (Figures 3 and S6) to provide structural insight on the binding of NSC260594 to the RNA SL3 of the HIV-1 Ψ packaging domain. From both the CD and fluorescence melting experiments we determined that NSC260594 was interacting with SL3, with the fluorescence melting data showing that above 30 μM of NSC260594 the RNA hairpin could not be melted at 95 °C (Table S3, Supporting Information).
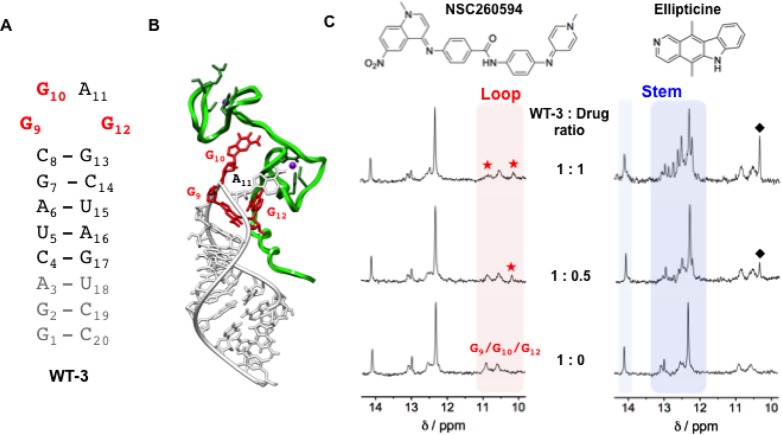
Figure 3.
Interaction between the structural analogue of SL3 (WT-3) and the two small molecules, NSC260594 and ellipticine, as monitored by 1H NMR. (A) Secondary structures of the WT-3 hairpin. In gray are reported the nucleotides added to SL3 in order to have a well-defined secondary structure suitable for NMR spectroscopy and in red the G9, G10, and G12 guanines constituting the tetraloop of SL3. (B) 3-D model of the HIV-1 nucleocapsid–WT-3 complex (PDB 1A1T) depicting the protein and the RNA hairpin with a green and white ribbon, respectively. The G9, G10, and G12 guanines essential for binding to the HIV-1 nucleocapsid are represented in red. (C) WT-3 1H NMR titration experiments with an increasing amount of the compounds NSC260594 and ellipticine. The imino protons between 12 and 14.5 ppm (blue region) are attributed to the Watson–Crick H-bonded base pairs of the stem of the hairpin structure and imino protons between 10 and 11 ppm (red region) are attributed to the WT-3 loop G bases. Red stars (★) highlight the alteration of the G9, G10, and G12 imino signals, and black diamonds (⧫) highlight peaks attributed to ellipticine.
For the 1H NMR spectroscopy Borer et al. had previously characterized and assigned the imino proton shifts of a stabilized SL3 derivative (WT-3).34 The imino proton NMR spectrum for WT-3 at pH 7.0 displayed well-defined peaks between 12 and 14.5 ppm due to Watson–Crick hydrogen bonded base pairs within the stem of the hairpin structure, and by lowering the pH to 5.0, new imino signals appeared in the 10–11 ppm range (Figures 3 and S6).34 These new peaks are attributed to the imino protons of unpaired guanines (G9, G10, and G12) in the loop of SL3. 1H NMR titration experiments were performed at pH 5.0 to monitor the interaction of NSC260594 with WT-3 focusing on the effect on the loop imino protons of the hairpin structure. Titration of an increasing amount of NSC260594 (up to 1.5 mol equiv) caused little change to the imino peaks relative to the stem of WT-3 (peaks between 12 and 14.5 ppm, Figures 3 and S6); by contrast the imino peaks attributed to guanines in the G9-G10-A11-G12 RNA tetraloop (peaks between 10 and 11 ppm) are significantly altered by up to 1 mol equiv of NSC260594 (Figures 3C and S6). A new imino peak at 10.23 ppm appeared upon the addition of 0.5 mol equiv of NSC260594 and further shifted upfield and sharpened following the addition of additional amounts of the compound. The existing peak at 10.90 ppm is broadened upon addition of NSC260594. These results suggest a well-defined recognition of the loop by the small molecules NSC260594. Increasing the molar equivalents of NSC260594 to greater than 1 caused no further changes of imino protons for WT-3 (Figure S6, Supporting Information), consistent with a 1:1 binding stoichiometry of NSC260594 by the RNA WT-3 tetraloop. As a control, similar titration experiments were performed with the known nonspecific intercalator ellipticine. Upon addition of ellipticine, the imino protons of the stem of WT-3 were most perturbed, while no significant changes were observed for the imino protons of unpaired guanines G9-G10-A11-G12 of the loop (Figure 3C). Indeed, the titration gives rise to the appearance of major new sharp resonances around 12.7 ppm, suggesting stabilization of the stem of the hairpin. Furthermore, the imino proton spectra continued to alter at equivalents greater than 1 molar (Figure S6, Supporting Information). These results are consistent with the nonspecific intercalation of ellipticine into the stem of the WT3 hairpin structure.
The alteration of imino protons within the loop of WT-3 by NSC260594 compared to the change of the imino protons within the stem of WT-3 by ellipticine shows that the two small molecules have very different modes of interactions with WT-3, and we hypothesize that NSC260594 recognizes the tetra-nucleotide loop of WT-3 and hinders the binding of the Gag protein. The binding of HIV-1 NC protein to the RNA SL3 has been well characterized by 1H NMR spectroscopy (PDB 1A1T, Figure 3B).35 The SL3/NC structure revealed that tight binding is mediated by specific interactions between the NC protein and the G9-G10-A11-G12 RNA tetraloop.35 In the same way that mutation of the tetraloop inhibits Gag interactions, binding of a small molecule here is likely to have a similar disruptive effect.
In summary, we have developed and applied an assay to identify small molecules that disrupt the Gag/SL3 protein RNA interaction. We then identified a subset of these molecules that showed biological activity in a viral infectivity assay. Compound NSC260594 was found to bind specifically to the tetraloop of SL3 of the HIV-1 Ψ-packaging domain as judged by 1H NMR spectroscopy. These studies represent a proof of concept for the identification of specific small molecule inhibitors of RNA/protein interactions that are critical for the HIV-1 life cycle.
Methods and Materials
Reagents
The RNA oligonucleotides WT (CUA GCG GAG GCU AG) and FQ WT (Tet-CUA GCG GAG GCU AG-BHQ1) and WT-3 (GGA CUA GCG GAG GCU AGU CC) were synthesized and purified by IBA GmbH. Stock solutions (100 μM for WT and FQ WT and 1 mM for WT-3) were made by resuspending the RNA in molecular biology grade water and quantified by A260 at 95 °C, using the ε260 value as provided by the manufacturers, before the RNAs were aliquoted and stored at −80 °C. All samples were freshly prepared prior to each experiment, and the FQ WT was heated at 95 °C for 5 min and snap-cooled on ice for a further 5 min, before the addition of the appropriate buffer. The expression and purification of the GagΔp6 protein have been reported previously with the exception that the purified GagΔp6 was rebuffered in 25 mM NaOAc, pH 6.5, 200 mM NaCl, 1 mM DTT, and 0.25 mM ZnCl2 by gel filtration.21
Small Molecule Screen
The small molecule screen was performed with a combination of the LOPAC 1280 (Sigma) and Diversity Set II (NIH) libraries. The libraries were condensed from their original 10 mM DMSO stocks in a 96-well format to daughter plates containing 2 mM DMSO stocks in a 384-well format, with 320 small molecules and 64 empty wells for controls per plate.
For screening, each small molecule from the daughter plates was first diluted to 100 μM in buffer (25 mM NaOAc, pH 6.5) before 2 μL was transferred to the 384 well assay (low volume flat bottom black NBS treated, Corning 3820). To the small molecules 4 μL of a 0.25 μM solution of FQ WT RNA (25 mM NaOAc, 2.5 mM MgCl2) was added, and the microplate was allowed to incubate at room temperature for 1 h before 4 μL of a 2.5 μM GagΔp6 solution (25 mM NaOAc, pH 6.5, 200 mM NaCl, 1 mM DTT, and 0.25 mM ZnCl2) was added, and the plate was incubated for 45 min at room temperature.
DMSO only (0.5% v/v) wells, which contained no small molecule, were used as a negative control, while positive controls well consisted of either an FQ WT RNA only or WT RNA as a “small molecule” control. Dilution, transfer, and mixing of all solutions were carried out using a Biomek NX liquid handling robot (Beckman Coulter). The fluorescent measurements were taken at 25 °C using a Pherastar+ platereader (BMG LabTech) with an excitation filter of 510 nm and an emission filter of 540 nm.
Tissue Culture
HEK293T (ATCC CRL-11268) and TZM-bl (ATCC) cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin and 100 μg/mL streptomycin, in humidified air with 5% CO2 at 37 °C. Cells were grown in 75 cm2 tissue culture flasks and split 1:10 twice a week. The TZM-bl cell line is a HeLa derivative that has been genetically engineered to express CD4, CCR5, and CXCR4 surface receptors, along with β-galactosidase and firefly luciferase genes under the transcriptional control of the HIV-1 LTR.36
Virus Preparation and Cytotoxicity Assays
Compounds of interest from the small molecules screen were reordered from either Sigma-Aldrich or the NIH and dissolved as 25 mM DMSO stocks. The 293T cells were seeded in 96-well tissue culture plates (Corning) at a density of 1.86 × 105 cells/well in 150 μL of DMEM supplemented with 10% FCS, 24 h prior transfection. To prepare the VSV-G-pseudotyped virus 293T cells were transfected (TransIT-LT1, Mirus) with the Env-defective proviral DNA (pNL-Kp) and VSV-G (pVSV-G) plasmids.37,38 Six hours post transfection, the small molecules were added at the appropriate concentration (0–33 μM) in 25 μL of media, and the cells were cultured for an additional 24 h. Transfection efficiency was measured by the addition of a GFP-expressing vector (pEGFP-N1; Clontech). Viral production was measured by a p24-Capsid ELISA, and cytotoxicity of the small molecules was determined using the CellTiter-Glo assay (Promega) according to the manufacturer’s protocols.
Infectivity Assays
TZM-bl cells were seeded onto 96-well tissue culture plates at a density of 1.86 × 105 cells/well in 150 μL of DMEM supplemented with 10% FCS, 24 h prior to infection. The TZM-bl cells were challenged with 50 μL of supernatant from the transfected 293T cells and incubated for 24 h. Viral infectivity was visualized using 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal, Sigma) and quantified using the 2-nitrophenyl-β-d-galactopyranoside (ONPG, Sigma) measured at 420 nm using a Multiskan microplate reader (Thermo Electron).
P24 Capsid ELISA
Half area, white, high binding 96-well plates (Greiner) were coated overnight with sheep anti-HIV-1 CA-p24 antibody (D7320, Aalto Bio Reagents) diluted in 0.1 M sodium bicarbonate. Prior to use, plates were blocked for 1 h in 5% BSA in TBS followed by four washes in 1× TBS. Samples and standards (recombinant HIV-1 CA-p24: Centre for AIDS Reagents) diluted in 0.05% Empigen in 1× TBS were incubated in the plates for 1.5 h, followed by four washes in 1× TBS and a 1 h incubation with alkaline phosphatase-conjugated anti-HIV-1 CA-p24 mouse monoclonal antibody (BC 1071-AP, Aalto Bio Reagents) diluted 1:8000 in a solution of 1× TBS containing 2% fat-free milk powder (Marvel), 0.05% Tween (BDH) and 20% sheep serum (PAA). Plates were washed four times with 0.1% Tween in PBS, incubated with Lumiphos Plus (Lumigen) detection reagent, and luminescence levels were determined using a Glomax luminometer (Promega).
Fluorescence Melting
Fluorescence melting curves were recorded on a Roche Lightcycler. Melting curves were acquired using FQ WT RNA (25 mM NaOAc, pH 6.5, 50 mM NaCl, 1 mM MgCl2) in the presence of NSC260594 (0–100 μM) with the temperature ramped from 35 to 95 °C at 0.5 °C min–1.
Circular Dichroism
CD spectra were recorded on a Chirascan Plus spectropolarimeter (Applied Biosciences). Spectra were acquired using a WT-3 oligonucleotide sample at 5 μM (25 mM NaOAc, pH 6.5, 50 mM NaCl, 1 mM MgCl2) using an accumulation of two scans from 320 to 190 nm using a 0.1 cm cell, a resolution of 0.1 nm, bandwidth of 1.0 nm, sensitivity of 2 mdeg, response of 2 s, and a scan speed of 50 nm min–1. Titration experiments with an increasing concentration of NSC260594 were performed by adding an increasing amount of a 1 mM aqueous solution.
1H NMR Spectroscopy
NMR spectra were recorded at 278 K using a 500 MHz Bruker Avance TCI spectrometer equipped with a cryogenic TCI ATM probe. Water suppression was achieved using excitation sculpting. Spectra were acquired using a WT-3 oligonucleotide sample at 100 μM strand concentration in NMR buffer: 5 mM PBS (pH 7.0 or 5.0), 25 mM NaCl supplemented with 10% D2O. The samples were heat cycled by equilibrating at 60 °C, then at 85 °C, and again at 60 °C for a few minutes to ensure hairpin formation. The samples were then kept at 4 °C. Titration experiments with an increasing concentration of the small molecules (NSC260594 and ellipticine) were performed by adding an increasing amount of a 1 mM aqueous solution of the small molecules.
Supporting Information Available
Supporting figures and tables mentioned in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by a Medical Research Council Milstein Award (MRC 88493 to S.B. and A.M.L.L.) and the Biomedical Research Centre (Grant RG56162 to A.M.L.L.). The Balasubramanian lab is supported by programme funding from Cancer Research UK.
Supplementary Material
References
- Johnson V. A.; Calvez V.; Gunthard H. F.; Paredes R.; Pillay D.; Shafer R.; Wensing A. M.; Richman D. D. (2011) 2011 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 19, 156–164. [PMC free article] [PubMed] [Google Scholar]
- Pirrone V.; Thakkar N.; Jacobson J. M.; Wigdahl B.; Krebs F. C. (2011) Combinatorial approaches to the prevention and treatment of HIV-1 infection. Antimicrob. Agents Chemother. 55, 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A.; Datta S. A. K.; Jones C. P.; Musier-Forsyth K. (2011) Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 36, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed A. A.; Freed E. O. (2012) HIV Type 1 Gag as a Target for Antiviral Therapy. AIDS Res. Hum. Retroviruses 28, 54–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A. M. (2007) HIV-1 RNA packaging. Adv. Pharmacol. 55, 1–32. [DOI] [PubMed] [Google Scholar]
- Poole E.; Strappe P.; Mok H. P.; Hicks R.; Lever A. M. L. (2005) HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; Analysis by confocal microscopy and FRET. Traffic 6, 741–755. [DOI] [PubMed] [Google Scholar]
- Zeffman A.; Hassard S.; Varani G.; Lever A. (2000) The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the Gag polyprotein. J. Mol. Biol. 297, 877–893 Erratum in J. Mol. Biol. (2000) 301, 1315. [DOI] [PubMed] [Google Scholar]
- Thomas J. R.; Hergenrother P. J. (2008) Targeting RNA with small molecules. Chem. Rev. 108, 1171–1224. [DOI] [PubMed] [Google Scholar]
- Parsons J.; Castaldi M. P.; Dutta S.; Dibrov S. M.; Wyles D. L.; Hermann T. (2009) Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 5, 823–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheschi R. J.; Tonelli M.; Kumar A.; Butcher S. E. (2011) Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication. ACS Chem. Biol. 6, 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer A. C.; Frank A. T.; Kratz J. D.; Swanson M. D.; Gonzalez-Hernandez M. J.; Lee J.; Andricioaei I.; Markovitz D. M.; Al-Hashimi H. M. (2011) Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 7, 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los Alamos HIV database, http://www.hiv.lanl.gov/content/index.
- Huthoff H.; Berkhout B. (2001) Two alternating structures of the HIV-1 leader RNA. RNA 7, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.; Heng X.; Garyu L.; Monti S.; Garcia E. L.; Kharytonchyk S.; Dorjsuren B.; Kulandaivel G.; Jones S.; Hiremath A.; Divakaruni S. S.; LaCotti C.; Barton S.; Tummillo D.; Hosic A.; Edme K.; Albrecht S.; Telesnitsky A.; Summers M. F. (2011) NMR Detection of Structures in the HIV-1 5′-Leader RNA That Regulate Genome Packaging. Science 334, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. M.; Dang K. K.; Gorelick R. J.; Leonard C. W.; Bess J. W. Jr.; Swanstrom R.; Burch C. L.; Weeks K. M. (2009) Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson J. D.; Li H.; Kenyon J. C.; Symmons M.; Klenerman D.; Lever A. M. (2013) Three-dimensional RNA structure of the major HIV-1 packaging signal region. Structure 21, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.; Heng X.; Summers M. F. (2011) Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 410, 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbink T. E.; Berkhout B. (2003) A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278, 11601–11611. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. A.; Gorelick R. J.; Vasa S. M.; Guex N.; Rein A.; Mathews D. H.; Giddings M. C.; Weeks K. M. (2008) High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N. M.; Lever A. M. (2012) HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 21, 136–144. [DOI] [PubMed] [Google Scholar]
- Bell N. M.; Kenyon J. C.; Balasubramanian S.; Lever A. M. (2012) Comparative structural effects of HIV-1 Gag and nucleocapsid proteins in binding to and unwinding of the viral RNA packaging signal. Biochemistry 51, 3162–3169. [DOI] [PubMed] [Google Scholar]
- Zhang J. H.; Chung T. D. Y.; Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Moore M. D.; Fu W.; Soheilian F.; Nagashima K.; Ptak R. G.; Pathak V. K.; Hu W. S. (2008) Suboptimal inhibition of protease activity in human immunodeficiency virus type 1: effects on virion morphogenesis and RNA maturation. Virology 379, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J. C.; Duncan I. B.; Hockley D.; Grief C.; Roberts N. A.; Mills J. S. (1991) Antiviral properties of Ro 31–8959, an inhibitor of human immunodeficiency virus (HIV) proteinase. Antiviral Res. 16, 295–305. [DOI] [PubMed] [Google Scholar]
- Sham H. L.; Kempf D. J.; Molla A.; Marsh K. C.; Kumar G. N.; Chen C. M.; Kati W.; Stewart K.; Lal R.; Hsu A.; Betebenner D.; Korneyeva M.; Vasavanonda S.; McDonald E.; Saldivar A.; Wideburg N.; Chen X.; Niu P.; Park C.; Jayanti V.; Grabowski B.; Granneman G. R.; Sun E.; Japour A. J.; Leonard J. M.; Plattner J. J.; Norbeck D. W. (1998) ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 42, 3218–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedter G.; Pearlman S.; Hofheinz D.; Blakeslee J.; Cockerell G.; Dezzutti C.; Yee J.; Lal R. B.; Lairmore M. (1992) Development of a monoclonal antibody-based p24 capsid antigen detection assay for HTLV-I, HTLV-II, and STLV-I infection. AIDS Res. Hum. Retroviruses 8, 527–532. [DOI] [PubMed] [Google Scholar]
- Blair W. S.; Cao J.; Jackson L.; Jimenez J.; Peng Q.; Wu H.; Isaacson J.; Butler S. L.; Chu A.; Graham J.; Malfait A. M.; Tortorella M.; Patick A. K. (2007) Identification and characterization of UK-201844, a novel inhibitor that interferes with human immunodeficiency virus type 1 gp160 processing. Antimicrob. Agents Chemother. 51, 3554–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath A. K.; Jiang S.; Strick N.; Lin K.; Haberfield P.; Neurath A. R. (1994) Three-dimensional structure-activity analysis of a series of porphyrin derivatives with anti-HIV-1 activity targeted to the V3 loop of the gp120 envelope glycoprotein of the human immunodeficiency virus type 1. J. Med. Chem. 37, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Wang J.; Kang X.; Kuntz I. D.; Kollman P. A. (2005) Hierarchical database screenings for HIV-1 reverse transcriptase using a pharmacophore model, rigid docking, solvation docking, and MM-PB/SA. J. Med. Chem. 48, 2432–2444. [DOI] [PubMed] [Google Scholar]
- Latt S. A.; Stetten G. (1976) Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J. Histochem. Cytochem. 24, 24–33. [DOI] [PubMed] [Google Scholar]
- Latt S. A.; Stetten G.; Juergens L. A.; Willard H. F.; Scher C. D. (1975) Recent developments in the detection of deoxyribonucleic acid synthesis by 33258 Hoechst fluorescence. J. Histochem. Cytochem. 23, 493–505. [DOI] [PubMed] [Google Scholar]
- Rydzewski J. M.; Leupin W.; Chazin W. (1996) The width of the minor groove affects the binding of the bisquaternary heterocycle SN-6999 to duplex DNA. Nucleic Acids Res. 24, 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire C. J.; Clark G. R.; Denny W. A. (1997) Minor groove binding of a bis-quaternary ammonium compound: the crystal structure of SN 7167 bound to d(CGCGAATTCGCG)2. Nucleic Acids Res. 25, 4072–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardo L.; Kerwood D. J.; Pelczer I.; Borer P. N. (1998) Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J. Mol. Biol. 282, 801–818. [DOI] [PubMed] [Google Scholar]
- De Guzman R. N.; Wu Z. R.; Stalling C. C.; Pappalardo L.; Borer P. N.; Summers M. F. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279, 384–388. [DOI] [PubMed] [Google Scholar]
- Wei X. P.; Decker J. M.; Liu H. M.; Zhang Z.; Arani R. B.; Kilby J. M.; Saag M. S.; Wu X. Y.; Shaw G. M.; Kappes J. C. (2002) Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akari H.; Uchiyama T.; Fukumori T.; Iida S.; Koyama A. H.; Adachi A. (1999) Pseudotyping human immunodeficiency virus type 1 by vesicular stomatitis virus G protein does not reduce the cell-dependent requirement of vif for optimal infectivity: functional difference between Vif and Nef. J. Gen. Virol. 80, 2945–2949. [DOI] [PubMed] [Google Scholar]
- Aiken C. (1997) Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71, 5871–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.