Abstract
Kainate receptors (KAR) form a class of glutamate receptors that have been implicated in epilepsy, stroke, Alzheimer’s and neuropathic pain.1 KAR subtypes are known to be segregated to specific locations within neurons and play significant roles in synaptic transmission and plasticity.2 Increasing evidence suggests a the role for ubiqutination in regulating the number of synaptic neurotransmitter receptors.3–5 The ubiquitin pathway consists of activation (E1), conjugation (E2) and ligation (E3). Cullins form the largest family of E3 ligase complexes. We have recently shown that the BTB/Kelch domain proteins, actinfilin and mayven, bind both Cul3 and specific KAR subtypes (GluR6 and GluR5-2b) to target these KARs for ubiquitination and degradation.5 In this chapter we will review how these interactions occur, what they mean for the stability of KARs and their associated proteins and how, in turn, they may affect synaptic functions in the central nervous system.
Introduction
KARs are found pre and postsynaptically and have been implicated in the etiology of epilepsy, as well as stroke-induced neurodegeneration and Huntington’s disease.6,7 Epilepsy occurs when inhibitory adaptation is unable to prevent excess neural activity. The developing cortex is particularly vulnerable and a number of syndromes are associated with epilepsy at an early age.8 Postsynaptic injection of kainic acid causes epileptiform discharges and the death of hippocampal CA3 pyramidal neurons.9,10 Moreover, KARs are subject to developmental and activity-dependent regulation at thalamocortical synapses and are likely to play an important role in the development of hippocampal synaptic circuits.11–13 KARs act as excitatory glutamate- gated ion channels: KAR-mediated excitatory postsynaptic currents were first described at mossy fiber-CA3 pyramidal cell synapses,14,15 while presynaptically, activation of KARs on inhibitory interneurons decreases GABA release which acts to enhance electrical activity, suggesting that presynaptic KARs may be epileptigeneic.16 Notably, the GluR6 subtype of KARs can also increase neuronal excitability via metabotropic regulation of potassium channels.17 To treat pathological conditions it will be crucial to understand the molecular mechanisms that determine localization of specific KARs to specific membrane domains.
Within KARs, there is a considerable diversity of properties, including unitary channel conductance, Ca2+ permeability and rectification, which arise from differences in receptor subunit composition and RNA editing of GluR5 and GluR6.18–20 KARs are tetramers that can be assembled from any one of five receptor subunits encoded by two separate gene families. The first of these includes receptor subunits GluR5, -6 and -7. Each of these subunits can form functional homomeric ion channels or heteromeric mixtures that appear to assemble promiscuously with any available subunit GluR5, -6 or -7.21–23 Alternative splicing of GluR5 yields two isoforms:21 GluR5-1 and GluR5-2, which has three additional splice variants possessing distinct C-terminal sequences. The shortest variant is designated GluR5-2a, while additional exons located in the C-terminal region give rise to GluR5-2b and GluR5-2c; these variants share a C-terminal type 1 PDZ-binding domain that is absent in GluR5-2a.18 The second gene family consists of KA1 and KA2 subunits that are functional only when expressed as heteromeric assemblies with GluR5, -6 or -7.23,24
Alternative splicing and RNA editing of ionotropic glutamate receptors play important roles in receptor assembly and trafficking.25–27 Regulatory steps in the assembly of KA2-containing KARs are governed by at least two trafficking signals located in the cytoplasmic terminal (C-tail) of the KA2 subunit. The first is an arginine-rich motif which operates as an endoplasmic reticulum (ER) retention signal preventing the insertion of homomeric KA2 receptors into the plasma membrane.28 The second is a di-leucine motif which mediates the internalization and subsequent relocalization of surface-expressed KA2 subunits.28 Similarly, GluR5-2b carries a positively charged amino acid motif that acts as a novel ER retention signal.29 In contrast, GluR6, which is highly expressed at the plasma membrane, has a forward trafficking signal in its C-terminal domain critical for ER exit.30,31
These differences in targeting appear to convey specific roles to specific KAR subtypes: In GluR6 knockout mice, mossy fiber long-term potentiation (LTP) was reduced, whereas GluR5 knockout mice exhibited normal LTP.32 The activation of KARs also modulates neurotransmitter release from a number of hippocampal synapses, including GABA release at inhibitory terminals that synapse onto CA1 pyramidal cells.16,33–35 In hippocampal slices, kainate depresses GABA-mediated synaptic inhibition and increases the firing rate of interneurons. These effects are explained by two populations of KARs in CA1 interneurons: GluR6/KA2 located in the somatodendritic compartment and GluR5-GluR6 or GluR5-KA2 at presynaptic terminals.35 It is anticipated that this segregation of KARs will allow us to design drugs that specifically target each function.
Recent evidence suggests that the ubiquitin-proteasome pathway and synaptic activity affects the composition of postsynaptic proteins.36–38 The addition of ubiquitin to proteins leads to a variety of fates for the tagged proteins, one of which is degradation by the 26S proteasome.39 A family of proteins called E3 ligases determines the specificity of ubiquitin addition. E3 ligases frequently consist of a complex of proteins that act together for specific substrate binding and ubiquitin ligation activity. Two major families of E3 ligases have been described: the HECT-domain family that is defined by its homology to the C-terminus of E6-associated protein (E6AP) and the RING family that contains either an intrinsic RING-finger domain or an associated RING-finger protein subunit essential for ubiquitin ligase activity.40 One of the best-characterized subset of the RING E3 ligases is the Skp1/Cul1/F-box protein complex (SCF), in which Cul1 binds an adaptor molecule, Skp1.41,42 Skp1 associates with an F-box protein that in turn binds a phosphorylated substrate. The Cul1 component of the SCF E3 ligase belongs to an evolutionarily conserved family of proteins known as cullins, of which there are six closely related members (Cul1, 2, 3, 4A, 4B and 5) and three distant relatives (Cul7, Parc and APC2).
A major class of Kelch proteins, defined as containing a 6-fold tandem “kelch” element,43 contains an N-terminal BTB/POZ domain and C-terminal kelch repeats and targets different substrates to the Cul3-Roc1 catalytic core.44 For example, the BTB-Kelch protein Keap1, a negative regulator of the transcription factor Nrf2, binds Cul3 and Nrf2 via its BTB and kelch domains, respectively, targeting Nrf2 for ubiquitination and subsequent degradation by the proteasome.45 The BTB-Kelch family also includes the closely related protein mayven, an actin-binding protein and gigaxonin, which is mutated in a human autosomal recessive neurodegenerative disorder named giant axonal neuropathy.46 Mutations in E3 ubiquitin ligases have also been associated with Parkinson’s disease and breast cancer.47
KAR Regulation by the Ubiquitin-Proteasome Pathway
To search for proteins involved in the regulation of KARs, we performed a yeast two-hybrid screen of an adult rat brain cDNA library using the C-terminus of GluR6 as bait. Strong interactions were detected between GluR6 and actinfilin. Actinfilin (AF) is a novel BTB/Kelch protein that was identified as a brain-specific actin-binding protein in postsynaptic densities (PSDs).48 Co-immunoprecipitation studies performed using HEK293 cell and rat brain extracts show that actinfilin binds GluR6 and GluR5-2b, but not with other glutamate receptors and ion channels.5 Because actinfilin is highly similar to another brain BTB/Kelch protein member, mayven49 (55% amino acid identity), we cloned this cDNA and, upon expression, found that it also co-immuno-precipitated with GluR6.
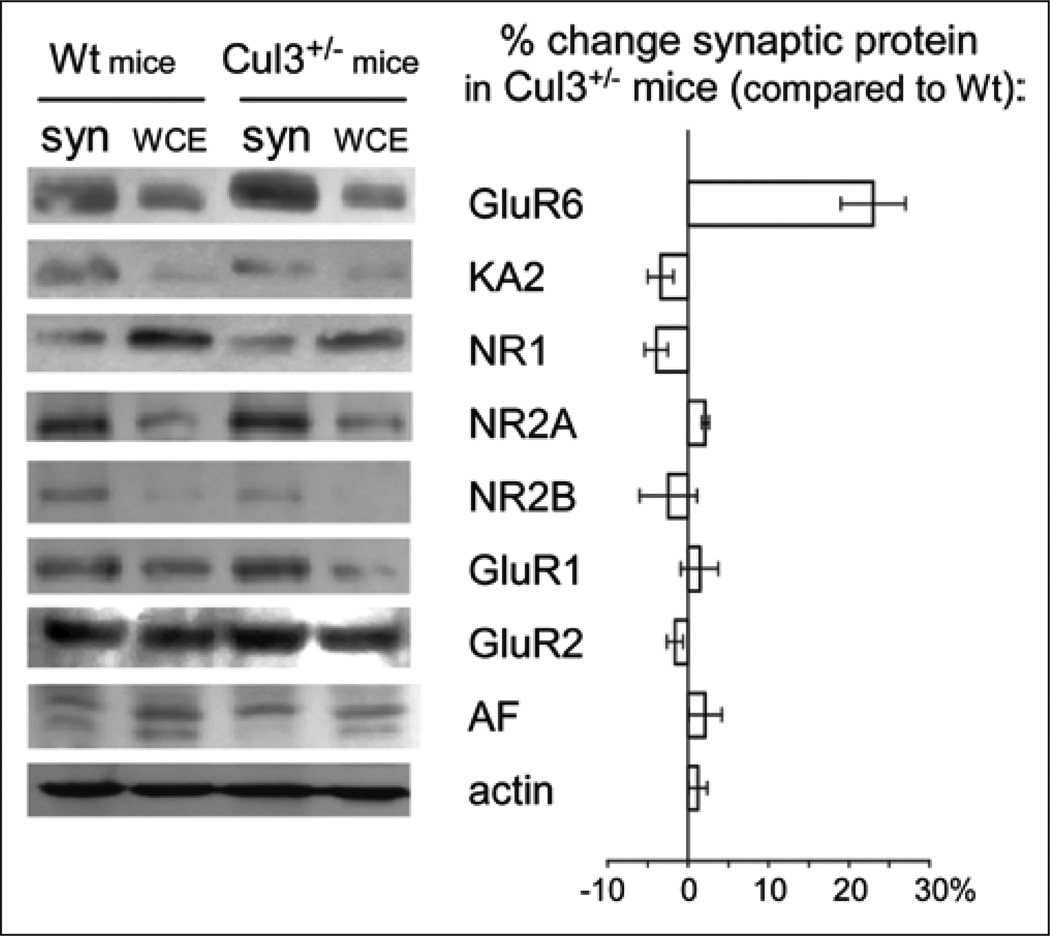
Actinfilin was found to interact with Cul3 to promote proteasomal degradation of GluR6 in vitro and in vivo.5 Expression of GluR6 with an HA-tagged ubiquitin (Ub-HA) in HEK293 cells showed a characteristic ladder indicating that GluR6 was ubiquitinated. Conversely, treatment with the 26S proteasomal inhibitor, MG132, greatly enhanced ubiquitination and stabilized GluR6 expression, demonstrating that GluR6 protein is fairly short-lived. Furthermore, co-immunoprecipitation studies verified that actinfilin interacts with Cul3, but not Cul1, in brain. The interactions of cullins with their adaptors o%en cause mutual degradation. Importantly, Cul3 appears to specifically regulate KAR levels in vivo (Fig. 1): In synaptosomes prepared from heterozygous Cul3 knockout mice, GluR6 levels are substantially increased, a small effect is observed on KA2 levels, but significantly, no effect on AMPA or NMDA receptors can be detected. These data suggest that Cul3 promotes degradation of KARs.
Figure 1.
Synaptic GluR6 levels are much higher in synaptosomes (syn) from +/− Cul3 mice than in wild-type (Wt) mice and KA2 show a small increase. Significantly, neither GluR2, NR1 nor NR2B detectably changed. WCE, whole-cell extract.
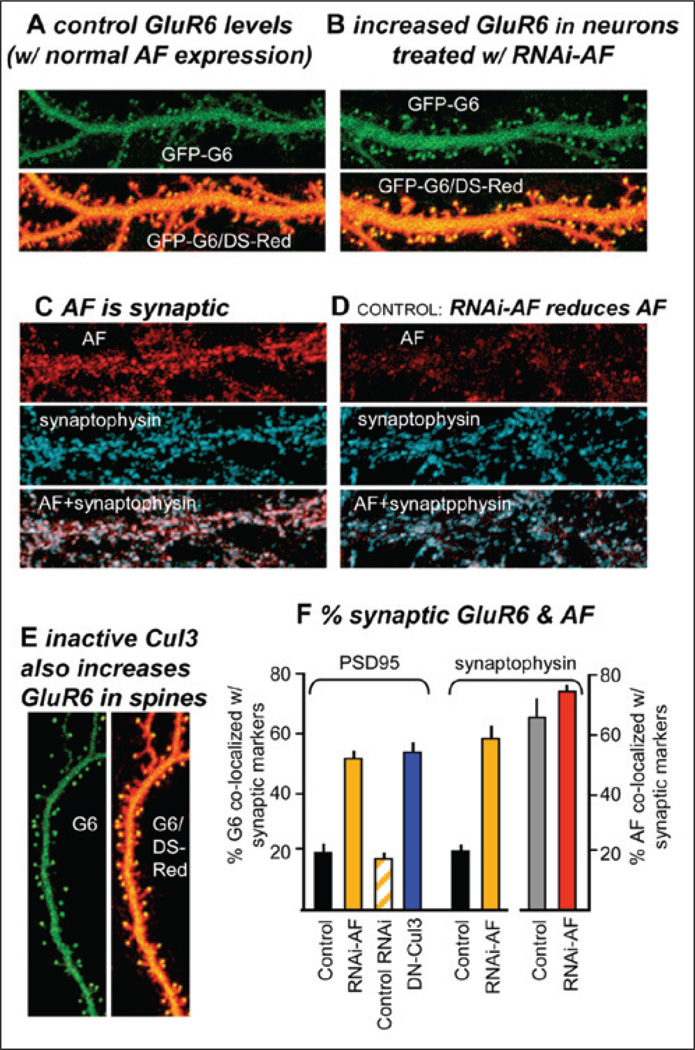
We also found that actinfilin was localized synaptically in hippocampal and cortical neurons and that it negatively regulates KAR expression (Fig. 2). A high degree of colocalization of AF and GluR6 was observed in dendritic spines. To establish tools to determine the role of actinifilin in the trafficking and/or synaptic localization of GluR6, we have developed a short hairpin inhibitory RNA (RNAi) to actinfilin that eliminates actinifilin expression (Fig. 2D). Specifically, we found that decreasing actinfilin levels via RNAi and overexpressing an inactive Cul3 both induced increased surface GluR6 expression at synapses, suggesting that actinfilin-Cul3-mediated degradation may provide an important mechanism for regulating neuronal GluR6.
Figure 2.
Actinfilin (AF) and Cul3 negatively regulate surface expression of GluR6. A–B) Hippocampal neurons were transfected with GluR6 tagged extracellularly with GFP (GFP-G6) and, to identify cellular morphology, DS-Red. Neurons (A) with normal AF levels exhibit relatively low GluR6 levels, while reducing AF levels with RNAi-AF strongly increases the amount of GluR6 found in dendritic spines (B). C–D) Actinfilin largely colocalizes with synaptic markers (here, synaptophysin), but the RNAi-AF strongly reduces AF levels (see also F). E) Overexpressing a dominant negative Cul3 mutant results in high surface levels of endogenous GluR6. F) Reducing AF increases GluR6 colocalization with synaptic markers: The % colocalization of GluR6 with synaptic markers (PSD-95 or synaptophysin) was determined (means ± SEMs).
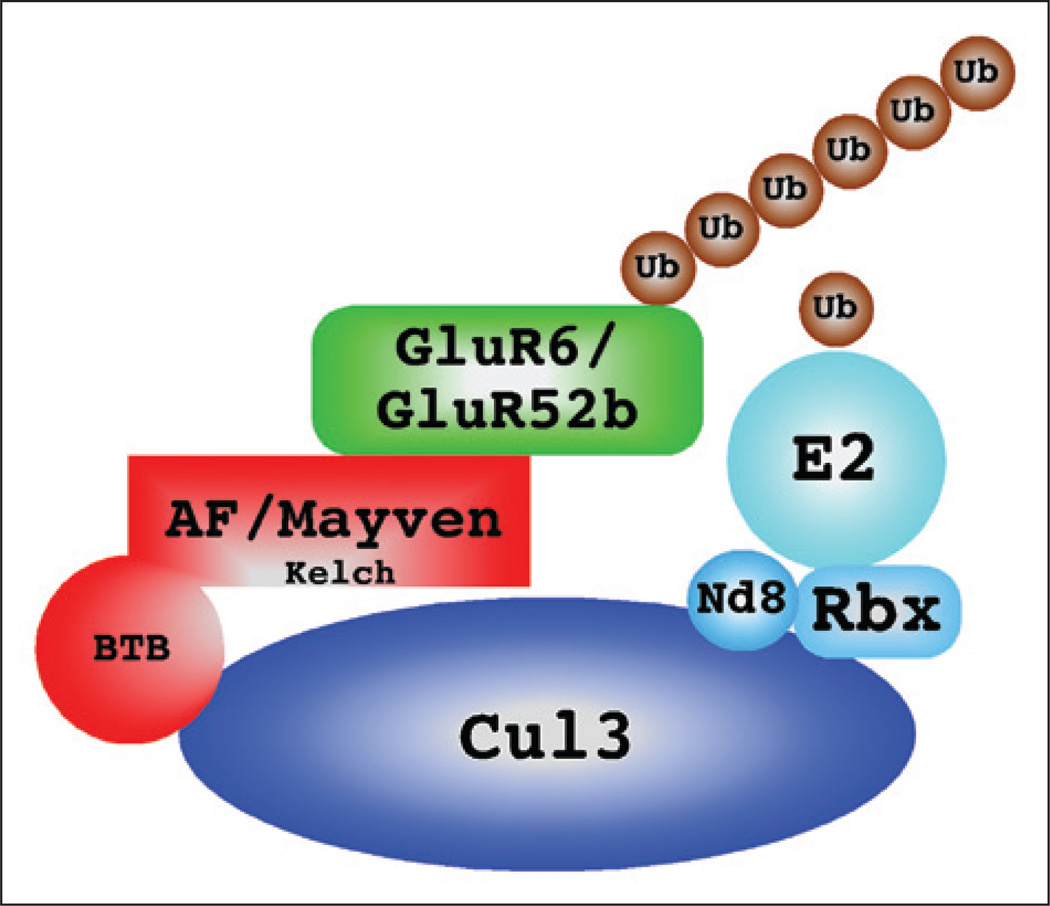
A model of how actinfilin may link GluR6 to Cul3 and the E3 ubiquitin ligase complex is presented in Figure 3. It should be noted that Cul3 is a component of an E3 ubiquitin ligase complex, also composed of the proteins Nedd8 (N8), Rbx1 and a ubiquitin conjugating enzyme or E2. Actinfilin or mayven would then act as adaptors to link GluR6 or GluR5-2b to this complex, binding the receptors through their kelch domains and Cul3 through their BTB domains and enabling ubiquitination of the receptor.
Figure 3.
Proposed regulation of KARs by AF and Cul3. The Cul3-based E3 ligase consists of Cul3, which acts as a scaffold to bring several essential proteins in close proximity. These proteins include Nedd 8 (Nd8), Rbx1 and a ubiquitin conjugating enzyme (E2). Substrate specificity is mediated by proteins containing a BTB domain that bind Cul3 in its amino-terminal end. Other domains on the BTB domain-containing proteins are involved in recognition of substrates; in the cases of actinfilin and mayven, these are kelch domains.
AF Regulation of Shank, a Parallel Path to Regulate Surface GluR Expression
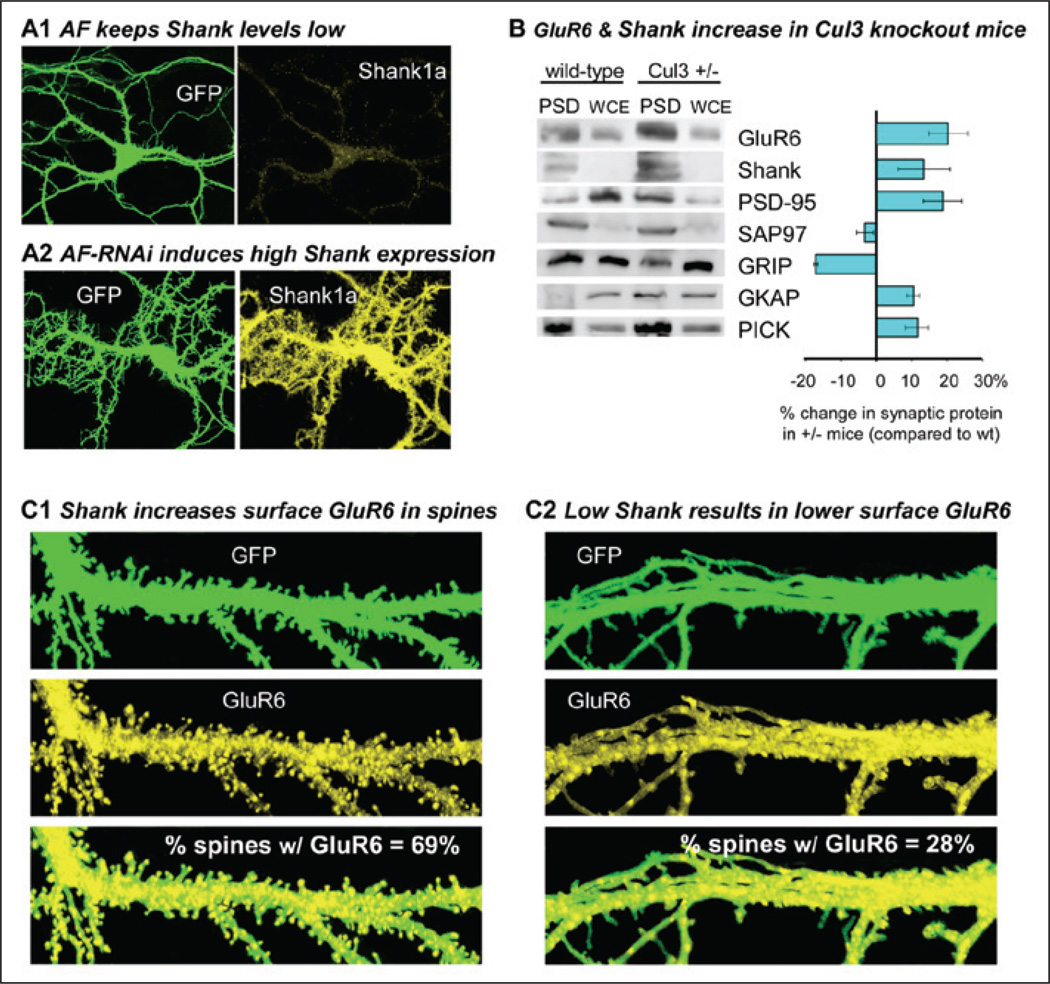
AF appears to negatively regulate the formation of dendritic spines in cortical neurons (Fig. 4). Potentially, this effect is not directly via regulating glutamate receptors, but could involve ubiquitination of scaffolding proteins.50 Shank proteins (Shanks1–3, also known as ProSAPs) constitute a group of postsynaptic, multidomain proteins that link glutamate receptors to intracellular calcium stores51 and are involved in the enlargement and maturation of dendritic spines.52 Importantly, Shank is known to be ubiquitinated.38 Specifically, we found that in synaptosomes prepared from heterozygous Cul3 knockout mice or by decreasing actinfilin levels via RNAi, Shank levels are substantially increased (Fig. 4). Elevating Shank levels by overexpressing Shank3 in cortical neurons increases not only the number of spines, but the likelihood of detecting surface GluR6 in a spine (Fig. 4). These results suggest that actinfilin-Cul3-mediated Shank regulation may provide an important mechanism for regulating spine development and synaptic KAR localization.
Figure 4.
Shank/ProSAP promotes surface GluR6 in dendritic spines and is down-regulated by actinfilin (AF) and Cul3, both of which decrease GluR6. A) AF keeps Shank levels low, but RNAi to AF strongly promotes expression of Shank in hippocampal neurons. B) Reduction of Cul3 (Cul3+/− mice) results in increased GluR6 and Shank in postsynaptic densities (PSD). WCE, whole cell extract. Also shown is the % change in each synaptic protein in Cul3+/− mice. C) Elevating Shank levels by overexpressing Shank3 in cortical neurons increases not only the number of dendritic spines, but the likelihood of detecting surface GluR6 in a spine (C1). For comparison, control neurons with lower Shank levels are also shown (C2). The % values indicate the % of spines that have detectable surface GluR6.
Development of Novel Peptidomimetic Drugs for the Treatment of Neurological Disorders
In neurons, actinfilin is preferentially localized to the dendritic spine, a structure rich in actin and critical for synapse formation.48 Moreover, synaptic activity is known to promote the redistribution of proteasomes from dendritic sha%s to spines via an association with actin filaments.53 Actinfilin and other BTB-Kelch proteins, such as Keap1, have been shown to associate with the actin cytoskeleton and interactions with actin are necessary for Keap1 to regulate Nrf2 levels.54 Similarly, the actin cytoskeleton and/or other scaffolding proteins may regulate actinfilin function, suggesting possible roles for these proteins in regulating KAR-actinfilin binding or the traf- ficking of actinfilin-bound GluR6 to the degradation machinery. We find that down-regulating actinfilin in cortical neurons increases both the synaptic localization and the size of synaptic GluR6-containing KAR clusters (Fig. 4). Because GluR6 has been implicated in excitotoxic neuronal death, in particular with damage associated with cerebral ischemia, stroke and epileptic seizures, our data imply that actinfilin may provide an important means for ensuring the correct regulation of GluR6 surface expression.
Recently, using NMR to determine the structure of the binding sites regulating GluR interactions with scaffolding and regulatory proteins, we have begun developing compounds that specifically target individual pathways that modulate synaptic GluR levels. One such reagent, CN2180, is a cyclized peptidomimetic compound that targets PSD-95 to inhibit GluR6 clustering. 55 This compound and related analogues, were designed to be membrane permeable, highly selective and resistant to protease digestion. In an in vivo retinal toxicity model,56 they are taken up rapidly by retinal neurons and attenuate the KA-induced PARP-1 hyperactivation associated with retinal neuron death (Marshall and Goebel, unpublished data). Based on our studies, we conclude that these compounds target the ‘main circuit breaker’ of the KAR-mediated cell death pathway. The newly developed compounds are expected to improve the tolerance for treatment because they do not affect the ability of glutamate receptors to perform normal neuronal signaling, but they do prevent the disease-related damage.
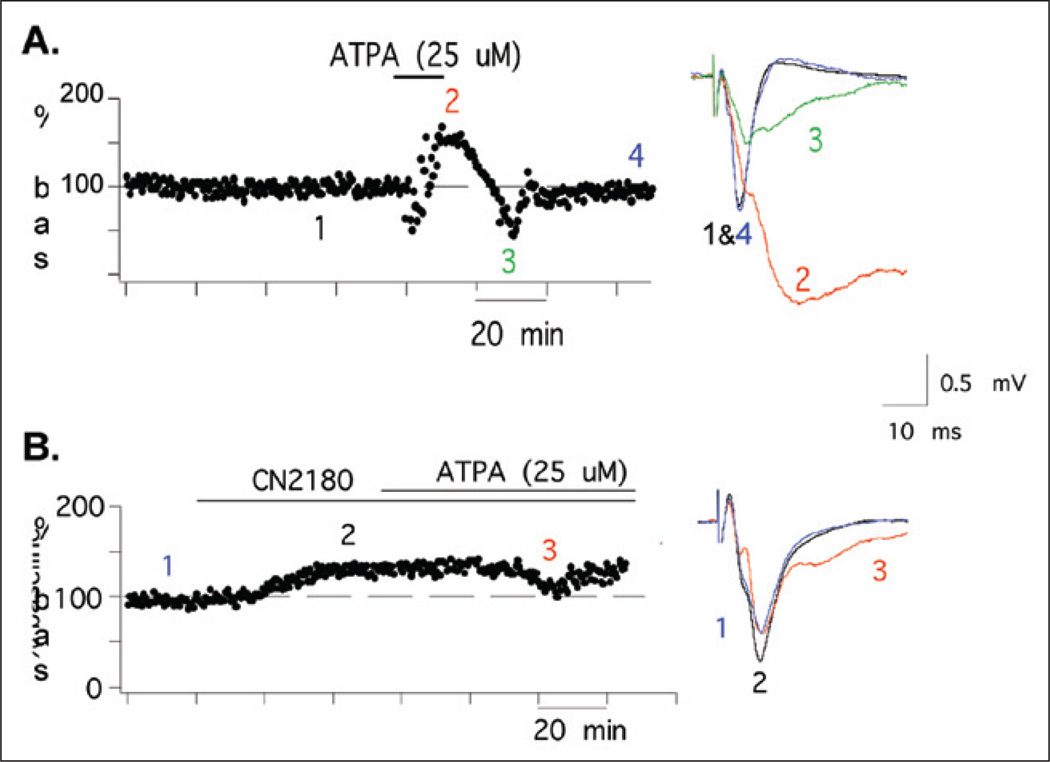
The effects of these compounds on motor cortical slices are also being examined. ATPA, a GluR5—GluR6/KA2 agonist, causes seizure-like activity, inducing a large, transient increase and widening in field potentials (Fig. 5A). However, we find that CN2180 largely prevents the seizure-like effect (Fig. 5B), without affecting either AMPAR or NMDAR activities. Overall, these studies indicate the applicability of this approach to design drugs specific for KAR-regulation and suggest that actinfilin may also prove a useful therapeutic target to control endogenous synaptic GluR6.
Figure 5.
CN2180, a reagent that reduces GluR6 surface expression, strongly reduces seizure-like activity in the motor cortex. A) ATPA, a KAR agonist, induces seizure-like in cortical slices. Time course of field potential amplitudes showing that APTA induces a large, transient increase in field potentials (1,2) followed by loss of evokable potentials (2,3). B) CN2180 pretreatment almost completely blocks the ATPA effects.
Conclusion
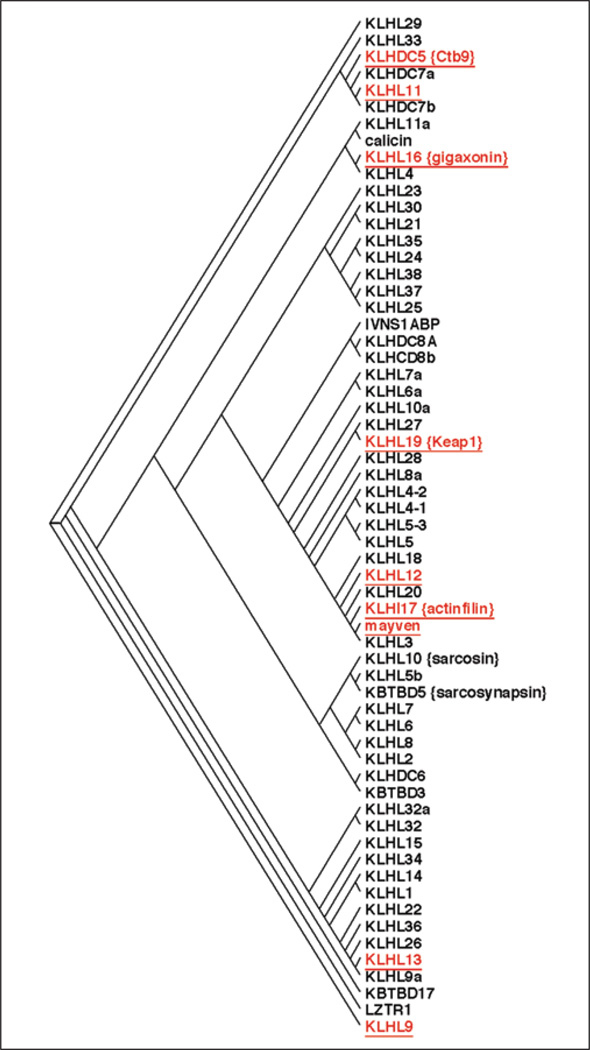
How general is this degradation mechanism for the regulation of kainate receptors? As is shown in Figure 6, there are 61 proteins predicted in the human protein database that contain both a BTB domain and kelch domains. Many of these have been shown to be adaptors for the Cul3-based E3 ligase (red, underlined) in mammals. In addition, KLHL8 which is similar to Kel-8 from C. elegans is involved in regulation of the GluR-1 receptor.57 There are a number of proteins that are very closely related to both mayven and actinfilin which are good candidates for Cul3 substrate adaptors for other related receptors. We think it is very likely that these proteins will serve such a role.
Figure 6.
Dendogram showing the 61 human proteins predicted to have both a BTB domain and kelch repeats. The red (underlined) proteins have been shown to act as substrate adaptors for the Cul3-based E3 ligase. The length of the lines connecting the proteins is related to the degree of homology, with shorter distances meaning more closely related (higher percentage of homology). A color version of this figure is available online at www.landesbioscience.com/curie.
Almost all the other mammalian BTB/Kelch proteins shown to be Cul3 adaptors appear to regulate either receptors or the cytoskeleton. These include KLHL9/KLHL13 which regulate Aurora B kinase,58 KLHL12 regulates the dopamine D4 receptor59 and KLHDC5 which regulates KATNA1, a katanin that is involved in microtubule remodeling.60 Other BTB/Kelch proteins, such as gigaxonin which is involved in peripheral axon development61 and sarcosin which is involved in cell motility,62 may also regulate neuronal function through protein ubiquitination. Taken together, Cul3 and its associated BTB/Kelch domain containing substrate adaptors play a critical role in both the development and maintanance of mammalian neurons.
Acknowledgements
We would like to thank Dr. Dennis Goebel (Wayne State University) for conducting the studies on retinal toxicity. We would also like to thank Drs. Greg Salinas and Mengia Rioult-Pedotti for their contributions. This work was supported by NIH R21.2970803 to JM and NIH 1R01GM082940-01A1 to JS.
References
- 1.Dingledine R, Borges K, Bowie D, et al. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Lerma J. Roles and rules of kainate receptors in synaptic transmission. Nat Rev Neurosci. 2003;4:481–495. doi: 10.1038/nrn1118. [DOI] [PubMed] [Google Scholar]
- 3.Burbea M, Dreier L, Dittman JS, et al. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002;35:107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 4.Colledge M, Snyder EM, Crozier RA, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salinas GD, Blair LA, Needleman LA, et al. Actinfilin is a Cul3 substrate adaptor, linking GluR6 kainate receptor subunits to the ubiquitin-proteasome pathway. J Biol Chem. 2006;281:40164–40173. doi: 10.1074/jbc.M608194200. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Ari Y, Cossart R. Kainate, a double agent that generates seizures: two decades of progress. Trends Neurosci. 2000;23:580–587. doi: 10.1016/s0166-2236(00)01659-3. [DOI] [PubMed] [Google Scholar]
- 7.Savinainen A, Garcia EP, Dorow D, et al. Kainate receptor activation induces mixed lineage kinase-mediated cellular signaling cascades via postsynaptic density protein 95. J Biol Chem. 2001;276:11382–11386. doi: 10.1074/jbc.M100190200. [DOI] [PubMed] [Google Scholar]
- 8.Guerrini R, Andermann E, Avoli M, et al. Cortical dysplasias, genetics and epileptogenesis. Adv Neurol. 1999;79:95–121. [PubMed] [Google Scholar]
- 9.Lerma J. Kainate reveals its targets. Neuron. 1997;19:1155–1158. doi: 10.1016/s0896-6273(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 10.Frerking M, Malenka RC, Nicoll RA. Synaptic activation of kainate receptors on hippocampal inter-neurons. Nat Neurosci. 1998;1:479–486. doi: 10.1038/2194. [DOI] [PubMed] [Google Scholar]
- 11.Kidd FL, Isaac JT. Developmental and activity-dependent regulation of kainate receptors at thalamo-cortical synapses. Nature. 1999;400:569–573. doi: 10.1038/23040. [DOI] [PubMed] [Google Scholar]
- 12.Lauri SE, Bortolotto ZA, Bleakman D, et al. A critical role of a facilitatory presynaptic kainate receptor in mossy fiber LTP. Neuron. 2001;32:697–709. doi: 10.1016/s0896-6273(01)00511-6. [DOI] [PubMed] [Google Scholar]
- 13.Lauri SE, Segerstrale M, Vesikansa A, et al. Endogenous activation of kainate receptors regulates glutamate release and network activity in the developing hippocampus. J Neurosci. 2005;25:4473–4484. doi: 10.1523/JNEUROSCI.4050-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–186. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- 15.Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388:179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Moreno A, Lerma J. Kainate receptor modulation of GABA release involves a metabotropic function. Neuron. 1998;20:1211–1218. doi: 10.1016/s0896-6273(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 17.Fisahn A, Heinemann SF, McBain CJ. The kainate receptor subunit GluR6 mediates metabotropic regulation of the slow and medium AHP currents in mouse hippocampal neurones. J Physiol. 2005;562(Pt 1):199–203. doi: 10.1113/jphysiol.2004.077412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sommer B, Burnashev N, Verdoorn TA, et al. A glutamate receptor channel with high affinity for domoate and kainate. Embo J. 1992;11:1651–1656. doi: 10.1002/j.1460-2075.1992.tb05211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herb A, Burnashev N, Werner P, et al. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron. 1992;8:775–785. doi: 10.1016/0896-6273(92)90098-x. [DOI] [PubMed] [Google Scholar]
- 20.Isaac JT, Mellor J, Hurtado D, et al. Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacol Ther. 2004;104:163–172. doi: 10.1016/j.pharmthera.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Bettler B, Boulter J, Hermans-Borgmeyer I, et al. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- 22.Cui C, Mayer ML. Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6 and GluR7. J Neurosci. 1999;19:8281–8291. doi: 10.1523/JNEUROSCI.19-19-08281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiffer HH, Swanson GT, Heinemann SF. Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron. 1997;19:1141–1146. doi: 10.1016/s0896-6273(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 24.Wenthold RJ, Trumpy VA, Zhu WS, et al. Biochemical and assembly properties of GluR6 and KA2, two members of the kainate receptor family, determined with subunit-specific antibodies. J Biol Chem. 1994;269:1332–1339. [PubMed] [Google Scholar]
- 25.Standley S, Roche KW, McCallum J, et al. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 26.Scott DB, Blanpied TA, Swanson GT, et al. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greger IH, Khatri L, Kong X, et al. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- 28.Ren Z, Riley NJ, Garcia EP, et al. Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J Neurosci. 2003;23:6608–6616. doi: 10.1523/JNEUROSCI.23-16-06608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Z, Riley NJ, Needleman LA, et al. Cell surface expression of GluR5 kainate receptors is regulated by an endoplasmic reticulum retention signal. J Biol Chem. 2003;278:52700–52709. doi: 10.1074/jbc.M309585200. [DOI] [PubMed] [Google Scholar]
- 30.Yan S, Sanders JM, Xu J, et al. A C-terminal determinant of GluR6 kainate receptor trafficking. J Neurosci. 2004;24:679–691. doi: 10.1523/JNEUROSCI.4985-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaskolski F, Coussen F, Nagarajan N, et al. Subunit composition and alternative splicing regulate membrane delivery of kainate receptors. J Neurosci. 2004;24:2506–2515. doi: 10.1523/JNEUROSCI.5116-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contractor A, Swanson G, Heinemann SF. Kainate receptors are involved in short- and long-term plasticity at mossy fiber synapses in the hippocampus. Neuron. 2001;29:209–216. doi: 10.1016/s0896-6273(01)00191-x. [DOI] [PubMed] [Google Scholar]
- 33.Clarke VR, Collingridge GL. Characterisation of the effects of ATPA, a GLU(K5) kainate receptor agonist, on GABAergic synaptic transmission in the CA1 region of rat hippocampal slices. Neuropharmacology. 2004;47:363–372. doi: 10.1016/j.neuropharm.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Bureau I, Dieudonne S, Coussen F, et al. Kainate receptor-mediated synaptic currents in cerebellar Golgi cells are not shaped by diffusion of glutamate. Proc Natl Acad Sci USA. 2000;97:6838–6843. doi: 10.1073/pnas.97.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen JK, Paternain AV, Selak S, et al. A mosaic of functional kainate receptors in hippocampal interneurons. J Neurosci. 2004;24:8986–8993. doi: 10.1523/JNEUROSCI.2156-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegde AN, DiAntonio A. Ubiquitin and the synapse. Nat Rev Neurosci. 2002;3:854–861. doi: 10.1038/nrn961. [DOI] [PubMed] [Google Scholar]
- 37.Murphey RK, Godenschwege TA. New roles for ubiquitin in the assembly and function of neuronal circuits. Neuron. 2002;36:5–8. doi: 10.1016/s0896-6273(02)00943-1. [DOI] [PubMed] [Google Scholar]
- 38.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 39.Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 40.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 41.Furukawa M, Ohta T, Xiong Y. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J Biol Chem. 2002;277:15758–15765. doi: 10.1074/jbc.M108565200. [DOI] [PubMed] [Google Scholar]
- 42.Furukawa M, He YJ, Borchers C, et al. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 43.Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 44.Geyer R, Wee S, Anderson S, et al. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 45.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bomont P, Cavalier L, Blondeau F, et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26:370–374. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto A, Friedlein A, Imai Y, et al. Parkin phosphorylation and modulation of its E3 ubiquitin ligase activity. J Biol Chem. 2005;280:3390–3399. doi: 10.1074/jbc.M407724200. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Derin R, Petralia RS, et al. Actinfilin, a brain-specific actin-binding protein in postsynaptic density. J Biol Chem. 2002;277:30495–30501. doi: 10.1074/jbc.M202076200. [DOI] [PubMed] [Google Scholar]
- 49.Soltysik-Espanola M, Rogers RA, Jiang S, et al. Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol Biol Cell. 1999;10:2361–2375. doi: 10.1091/mbc.10.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng M, Pak DT. Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu Rev Physiol. 2000;62:755–778. doi: 10.1146/annurev.physiol.62.1.755. [DOI] [PubMed] [Google Scholar]
- 51.Gao L, Blair LA, Salinas GD, et al. Insulin-like growth factor-1 modulation of CaV1.3 calcium channels depends on Ca2+ release from IP3-sensitive stores and calcium/calmodulin kinase II phosphorylation of the alpha1 subunit EF hand. J Neurosci. 2006;26:6259–6268. doi: 10.1523/JNEUROSCI.0481-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sala C, Piech V, Wilson NR, et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 53.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 54.Kang MI, Kobayashi A, Wakabayashi N, et al. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci USA. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piserchio A, Salinas GD, Li T, et al. Targeting specific PDZ domains of PSD-95; structural basis for enhanced affinity and enzymatic stability of a cyclic peptide. Chem Biol. 2004;11:469–473. doi: 10.1016/j.chembiol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Goebel DJ, Winkler BS. Blockade of PARP activity attenuates poly(ADP-ribosyl)ation but offers only partial neuroprotection against NMDA-induced cell death in the rat retina. J Neurochem. 2006;98:1732–1745. doi: 10.1111/j.1471-4159.2006.04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaefer H, Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell. 2006;17:1250–1260. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sumara I, Peter M. A Cul3-based E3 ligase regulates mitosis and is required to maintain the spindle assembly checkpoint in human cells. Cell Cycle. 2007;6:3004–3010. doi: 10.4161/cc.6.24.5068. [DOI] [PubMed] [Google Scholar]
- 59.Rondou P, Haegeman G, Vanhoenacker P, et al. BTB Protein KLHL12 targets the dopamine D4 receptor for ubiquitination by a Cul3-based E3 ligase. J Biol Chem. 2008;283:11083–11096. doi: 10.1074/jbc.M708473200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mesrobian CM, Bentley CA, Perdue SA, et al. The Cul3/KLHL5 E3 Ligase regulates P60/Katanin and is required for normal mitosis in mammalian cells. doi: 10.1074/jbc.M809374200. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dequen F, Bomont P, Gowing G, et al. Modest loss of peripheral axons, muscle atrophy and formation of brain inclusions in mice with targeted deletion of gigaxonin exon 1. J Neurochem. 2008;107:253–264. doi: 10.1111/j.1471-4159.2008.05601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greenberg CC, Connelly PS, Daniels MP, et al. Krp1 (Sarcosin) promotes lateral fusion of myofibril assembly intermediates in cultured mouse cardiomyocytes. Exp Cell Res. 2008;314:1177–1191. doi: 10.1016/j.yexcr.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]