Abstract
Expansion of CAG/CTG trinucleotide repeats causes numerous inherited neurological disorders, including Huntington's disease (HD), several spinocerebellar ataxias and myotonic dystrophy type 1. Expanded repeats are genetically unstable with a propensity to further expand when transmitted from parents to offspring. For many alleles with expanded repeats, extensive somatic mosaicism has been documented. For CAG repeat diseases, dramatic instability has been documented in the striatum, with larger expansions noted with advancing age. In contrast, only modest instability occurs in the cerebellum. Using microarray expression analysis, we sought to identify the genetic basis of these regional instability differences by comparing gene expression in the striatum and cerebellum of aged wild-type C57BL/6J mice. We identified eight candidate genes enriched in cerebellum, and validated four—Pcna, Rpa1, Msh6 and Fen1—along with a highly associated interactor, Lig1. We also explored whether expression levels of mismatch repair (MMR) proteins are altered in a line of HD transgenic mice, R6/2, that is known to show pronounced regional repeat instability. Compared with wild-type littermates, MMR expression levels were not significantly altered in R6/2 mice regardless of age. Interestingly, expression levels of these candidates were significantly increased in the cerebellum of control and HD human samples in comparison to striatum. Together, our data suggest that elevated expression levels of DNA replication and repair proteins in cerebellum may act as a safeguard against repeat instability, and may account for the dramatically reduced somatic instability present in this brain region, compared with the marked instability observed in the striatum.
INTRODUCTION
Expansions of CAG/CTG trinucleotide repeat sequences (TNRs) cause numerous inherited neurological disorders, including Huntington's disease (HD), myotonic dystrophy (DM1) and several spinocerebellar ataxias (SCAs) (1,2). Longer CAG repeat sequences inversely correlate with worsening disease severity and earlier age of onset. TNR tracts <35 units are generally stable, while tracts >35 units, referred to as expanded trinucleotide repeats, become unstable and have the propensity to expand further. The molecular events that underlie trinucleotide repeat instability are still poorly understood (3). For some expanded trinucleotide repeat alleles, extensive ‘somatic mosaicism’ has been documented, wherein variation in repeat length (both between and within tissues) exists (4,5). Somatic mosaicism has been shown to be age-dependent, highly tissue-specific and associated with disease progression (2,6).
The CAG repeat diseases display significant somatic mosaicism, with HD being the first CAG repeat disease documented to show this phenomenon (7). One striking feature of repeat mosaicism in HD CNS is the non-random differential pattern of expanded repeats present between different central nervous system (CNS) regions. Analysis of autopsied HD patient brains and transgenic mice indicates that the striatum displays the largest range of CAG repeats, while the cerebellum contains CAG repeats that are much smaller (8). Thus, instability is greater in the striatum than in the cerebellum in HD. Studies of dentatorubral-pallidoluysian atrophy (DRPLA), DM1, SBMA, SCA1 and SCA3 have also demonstrated that the smallest expansions occur in the cerebellum (1,9–14). These profiles parallel the patterns observed in CNS tissues in HD, despite the fact that different populations of neurons degenerate in each of these six diseases. This suggests that CNS somatic mosaicism patterns may be independent of the disease process or repeat locus and that instead, the cellular characteristics of a tissue type dictate differences in the degree of repeat instability.
Mouse models of CAG/CTG repeat instability accurately recapitulate somatic instability patterns documented in human patients, with the extent of mosaicism observed in different somatic tissues and brain regions mimicking that seen in human patients, although they do not accurately model the dramatic intergenerational expansions seen in human disease (15–19). PCR analysis of repeat length variation in striata from HD and SCA1 mouse models revealed dramatic instability - with rare changes of >150 CAG repeats, whereas cerebellar repeat variation was modest by comparison (19,20). In these disease models and a SCA7 mouse model, age correlates with somatic instability, with extensive variation in repeat length size only clearly emerging once the mice are well into adulthood, with prominent repeat instability noted both in the CNS (cortex, striatum) and periphery (kidney, liver) (15,19,20). It is well known that CAG repeat-expanded RNAs and polyglutamine-expanded proteins result in altered splicing and expression of many genes (21–24). Yet the presence of cell-type and age-specific repeat instability in the cerebellum and striatum in multiple different diseases suggests that tissue-specific differences in the expression of trans-acting protein factors may account for differences in somatic instability between these two brain regions. In this work, we considered repeat instability mechanisms operating in different regions of the brain, as most CAG/CTG repeat diseases are characterized by progressive neurodegeneration accompanied by differences in expanded repeat lengths in various CNS regions. The fact that somatic mosaicism of expanded repeats appears invariant in certain brain regions led us to ask whether comparison of a highly unstable brain region (striatum) with a highly stable brain region (cerebellum) might yield insights into the trans-acting factors that either promote instability or prevent it.
RESULTS
Microarray expression analysis of cerebellum and striatum
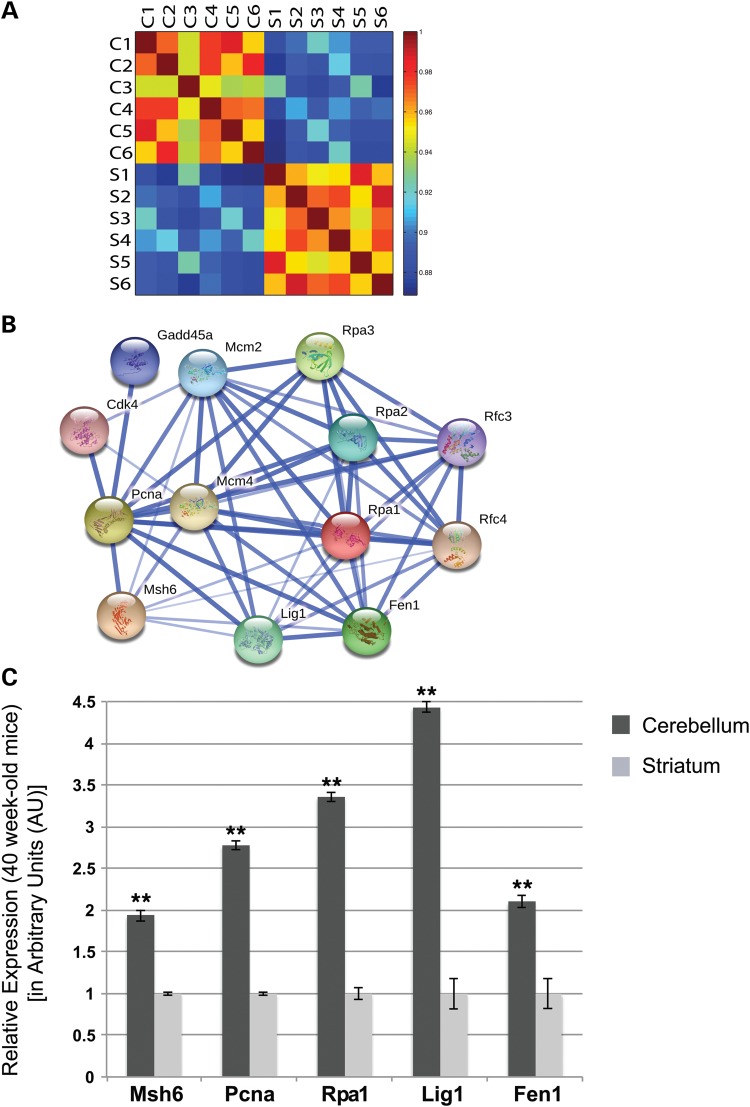
To identify differentially regulated transcripts that might underlie somatic instability differences between the cerebellum and striatum, we performed a microarray expression analysis. Somatic instability increases dramatically with aging; thus, we studied wild-type C57BL/6J mice at 40 weeks of age, as marked regional instability differences emerge in most repeat disease mouse models by this time. For this study, we dissected out the striatum and cerebellum of six mice, three males and three females, and isolated total RNA. We then analyzed these samples using the Affymetrix Murine 430A GeneChip array, containing ∼14 000 annotated mouse genes. To assess the validity of this data set, we generated a microarray correlation heat map of the transformed sample intensities. Importantly, we found that the transcriptional signatures of the six individual cerebellar samples were highly correlative and the transcriptional signatures of the six striatal samples were also highly correlative, but they were not correlative with each other (Fig. 1A). In addition, we found that there was no profile clustering based on mouse gender (Fig. 1A). This indicates that the samples represent reliable transcriptional profile replicates within each tissue, while there are marked differences between the two tissue types, as expected. By referencing a C57BL/6J in situ hybridization expression library (www.alleninstitute.org; http://mouse.brain-map.org, last accessed on 5 November 2013.) (25), we further validated the reliability of our microarray findings by examining the genes with the highest expression in each tissue from our microarray, and observing that these genes were respectively highly expressed cerebellar genes (Supplementary Material, Fig. S1A and B and Table S1), and highly expressed striatal genes (Supplementary Material, Fig. S1C and D and Table S2). Analysis of the microarray expression array data set was performed using Affymetrix expression console software. By applying Robust Multichip Average (RMA) analysis (26–28), we obtained intensity values for each probe. Probe values for the cerebellar data set were then compared with probe values for the striatal data set, using a paired t-test to identify probes that are differentially expressed among the tissue types. In this way, we identified 2655 differentially expressed genes with a P-value of <0.002.
Figure 1.
DNA replication and repair pathway genes are differentially expressed in cerebellum and striatum. (A) Microarray correlation heat map of transformed intensities from the six different cerebellar samples (C1–C6) and six striatal samples (S1–S6). Strong correlations are seen between samples from the same tissue type, while there is a weak correlation between the different tissue types. (B) STRING protein interaction network centered on PCNA, RPA1 and MSH6, which are differentially expressed in cerebellum in comparison to the striatum, highlights key players in DNA replication and repair. (C) Real-time RT–PCR confirmation of microarray expression differences between Rpa1, Pcna, Msh6, Lig1 and Fen1 in 40-week-old mice (n = 6; mean ±s.e.m., three independent experiments; **P < 0.01; t-test).
Identification of differentially regulated pathways
To investigate which pathways are down regulated in the striatum in comparison to cerebellum, we used DAVID (Database for Annotation, Visualization and Integrated Discovery) to analyze all significantly regulated genes (29,30), with a threshold significance of P < 0.002 and a fold difference of at least 1.4, which yielded 929 genes (Supplementary Material, Table S3). KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathway analysis output (31,32) highlighted a number of pathways (Supplementary Material, Table S4), with two pathways emerging as enriched in our data set: (i) DNA replication (Prim1, Mcm7, Rpa1, Pcna, Rfc4, Fen1 and Pold2) genes and (ii) mismatch repair (MMR) (Msh6, Pold2, Pcna, Rpa1 and RFfc4) genes (Table 1). These pathways have previously been implicated as key modulators of repeat instability (1,2,33–35). Of the genes listed in Table 1, we selected a subset (Pcna, Rpa1 and Msh6) with well-established roles in repeat instability (3,36) to study in detail.
Table 1.
DNA replication and mismatch repair gene expression alterations
| Gene name | Symbol | Probe | Fold change (Cb/Str) | P-value |
|---|---|---|---|---|
| Replication protein A1 | Rpa1 | 1437309_a_at | ↑1.99 | 6.6E-5 |
| MutS homolog 6 | Msh6 | 1416915_at | ↑1.84 | 0.0017 |
| Proliferating cell nuclear antigen | PCNA | 1417947_at | ↑1.50 | 1.9E-5 |
| Minichromosome maintenance deficient 7 (S. cerevisiae) | Mcm7 | 1438320_s_at | ↑1.53 | 2.2E-5 |
| Replication factor C (activator 1) 4 | Rfc4 | 1438161_s_at | ↑1.55 | 0.0001 |
| Flap structure specific endonuclease 1 | Fen1 | 1421731_a_at | ↑1.68 | 0.0005 |
| Polymerase (DNA directed), delta 2, regulatory subunit | Pold2 | 1448277_at | ↑1.42 | 0.0001 |
| DNA primase, p49 subunit | Prim1 | 1418369_at | ↑1.42 | 6.4E-5 |
Cb, cerebellum; Str, striatum.
Pathway analysis underscores importance of DNA replication and DNA repair in somatic instability differences
Using the ‘Search Tool for the Retrieval of INteracting Genes/proteins’ (STRING v9.0) (37) analysis to interrogate the protein interaction network centered at PCNA, RPA1 and MSH6, we noted that the resulting network highlights essential components of DNA replication and repair (Fig. 1B). Our STRING analysis yielded a network of predicted functional partners of PCNA, RPA1 and MSH6, including LIG1 and FEN1. The most highly associated interacting genes/proteins all exhibited higher expression levels in the cerebellum in comparison to the striatum in our data set (Supplementary Material, Table S5). The majority of the genes revealed by STRING analysis belong to the KEGG Pathway for the ‘DNA replication complex’. The DNA replication complex is an enriched pathway in the cerebellum compared with the striatum in the microarray data, revealing that a substantial number of key players in the pathway are differentially regulated between the two tissue types. Proteins involved in DNA replication are critically important for maintaining DNA integrity, when DNA damage occurs (1). Our data thus suggest that increased expression of DNA metabolism genes in the cerebellum may contribute to the stability of CAG repeat expansions in this brain region.
Confirmation of RNA expression level changes for Pcna, Rpa1, Msh6, Lig1 and Fen1
To confirm that the DNA replication complex is enriched in the cerebellum in comparison to the striatum, we performed real-time RT–PCR analysis on Pcna, Rpa1, Msh6, Lig1 and Fen1 using the TaqMan approach (38). Significant enrichment (P < 0.01) was detected for Pcna, Rpa1, Msh6, Lig1 and Fen1 in the cerebellum compared with the striatum in 40-week-old C57BL/6J mice (Fig. 1C). These results confirm the alterations reported for the microarray expression comparisons, and further validate the findings of this microarray study.
PCNA, RPA1 and MMR proteins are markedly enriched in cerebellum
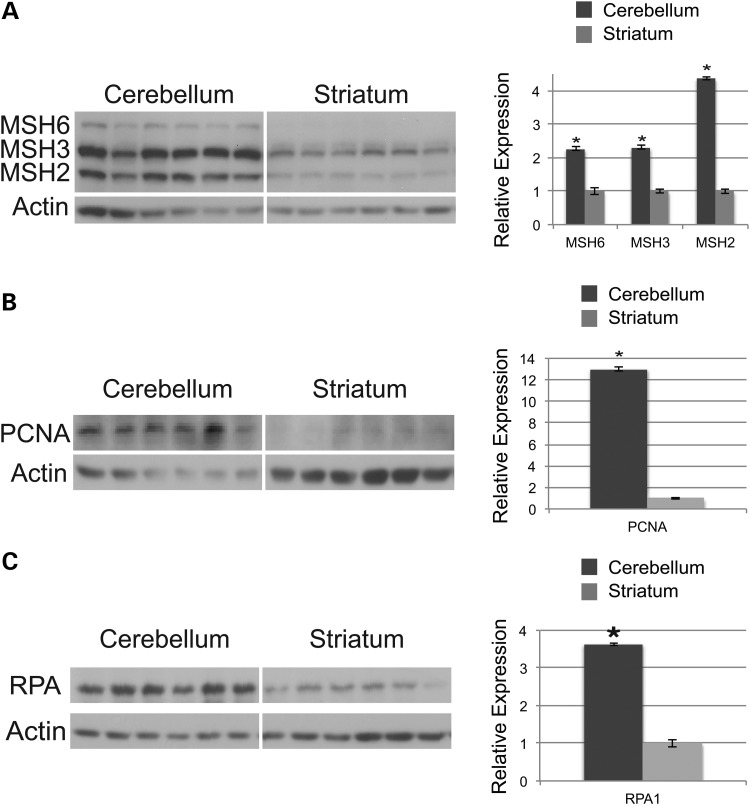
As gene transcript expression levels may not correlate with protein expression levels (39), we chose to analyze cerebellar and striatal protein levels of PCNA, RPA1 and the three principal MMR proteins, MSH2, MSH3 and MSH6, as they form a set of multimeric complexes: MutSα (MSH2-MSH6) and MutSβ (MSH2-MSH3). Protein lysates were obtained from the cerebellum and striatum of six 40-week-old wild-type C57BL/6J mice, and MMR protein levels were measured by simultaneous western blot analysis, an established and previously published method for sensitive detection of MMR expression level variation (39–42). Using this approach, we found that all three MMR proteins were markedly enriched in the cerebellum compared with the striatum (Fig. 2A). MSH6 protein expression was enriched by 2.3-fold, MSH3 protein expression was enriched by 2.3-fold and MSH2 protein expression was enriched by 4.4-fold in the cerebellum versus the striatum (Fig. 2A). Western blot analysis of RPA1 and PCNA also revealed higher protein expression levels in the cerebellum (Fig. 2B and C). Cerebellar RPA1 protein expression was enriched by 3.6-fold and cerebellar PCNA protein expression was enriched by ∼13-fold (Fig. 2B and C).
Figure 2.
DNA replication and repair protein expression levels are significantly increased in the cerebellum. (A) Mismatch repair (MMR) protein expression levels in the cerebellum and the striatum from six age-matched C57BL/6 mice were analyzed by the simo-blot technique. MMR protein levels were quantified by Image J, normalized to actin and presented to the right in arbitrary units, with striatal expression level for each MMR protein arbitrarily set to 1. Cerebellar MMR protein expression is significantly higher for each MMR protein in comparison to striatum MMR protein expression (mean ± s.e.m., three independent experiments; *P < 0.05; t-test). (B) Proliferating cell nuclear antigen (PCNA) protein expression levels in the cerebellum and the striatum from six age-matched C57BL/6 mice were analyzed by western blot analysis. PCNA protein levels were quantified by Image J, normalized to actin, and presented to the right in arbitrary units, with striatal expression level arbitrarily set to 1. Cerebellar PCNA protein expression is significantly higher in comparison to striatum PCNA protein expression (mean ± s.e.m., three independent experiments; *P < 0.05; t-test). (C) Replication protein A1 (RPA1) protein expression levels in the cerebellum and the striatum from six age-matched C57BL/6 mice were analyzed by western blot analysis. RPA protein levels were quantified by Image J, normalized to actin and presented to the right in arbitrary units, with striatal expression level arbitrarily set to 1. Cerebellar RPA protein expression is significantly higher in comparison to striatum RPA protein expression (mean ± s.e.m., three independent experiments; *P < 0.05; t-test).
Brain region-specific differences in Pcna, Rpa1, Msh6, Lig1 and Fen1 are age-independent
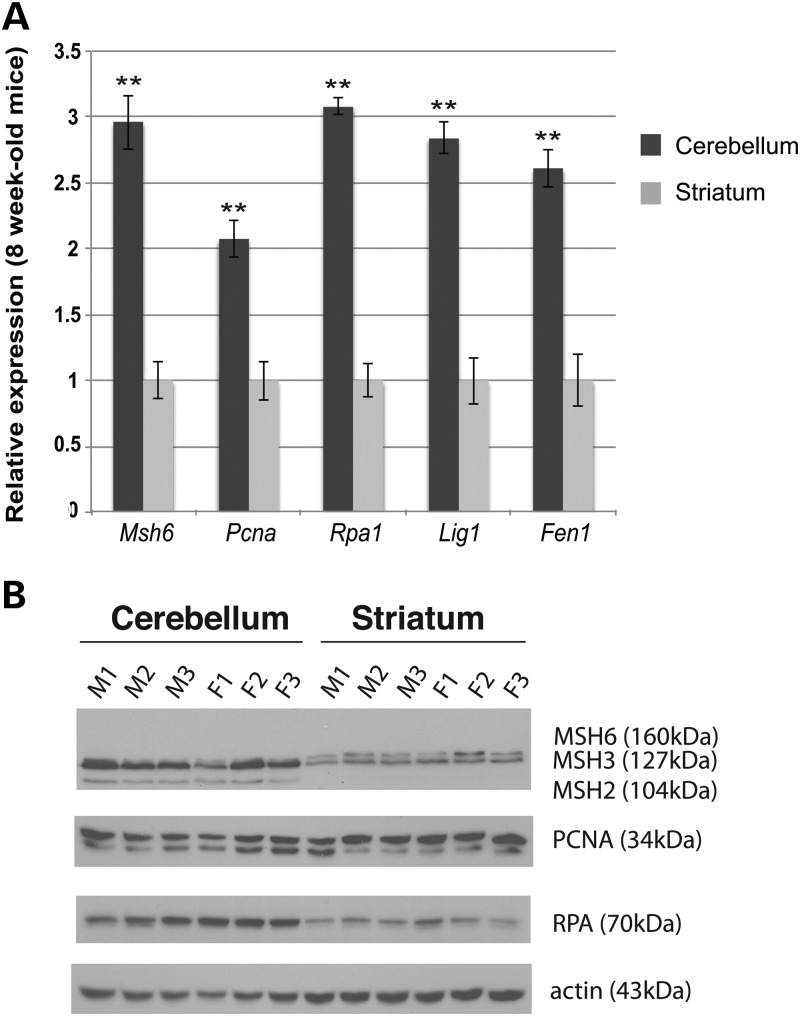
The cerebellum and striatum have very different cellular compositions; hence, expression differences in the two tissues could be due to age or to the intrinsic nature of the tissues. We therefore dissected the striatum and cerebellum from six 8-week-old C57BL/6J mice, three males and three females, using half of the tissue for RNA isolation and the other half for protein extraction. As in the aged tissues, we observed significantly increased RNA expression levels (P < 0.005) for Pcna, Rpa1, Msh6, Lig1 and Fen1 in the cerebellum in comparison to the striatum in 8-week-old mice (Fig. 3A). Western blot analysis of RPA and PCNA similarly revealed considerably higher protein expression levels in the cerebellum (Fig. 3B). We also found that MSH2 and MSH3 proteins are markedly enriched in the cerebellum compared with the striatum, while MSH6 expression levels were too low to permit quantitative comparison (Fig. 3B).
Figure 3.
DNA replication and repair genes display elevated RNA and protein expression in the cerebellum in young mice. (A) The RNA expression levels for Rpa1, Pcna, Msh6, Lig1 and Fen1 were quantified by real-time RT–PCR analysis for sets of cerebellum and striatum samples (n = 6/group) for 8-week-old mice. Significant mean elevations for all five genes were detected in the cerebellum when compared with the striatum (mean ± s.e.m., three independent experiments; **P < 0.01; t-test). β-actin served as an internal normalization control. (B) Western blot analysis of MSH2, MSH3, MSH6, PCNA and RPA1 proteins in the cerebellum of 8-week-old C57BL/6J mice confirmed elevated RNA expression levels.
MMR expression levels are not affected by polyglutamine disease protein expression or age
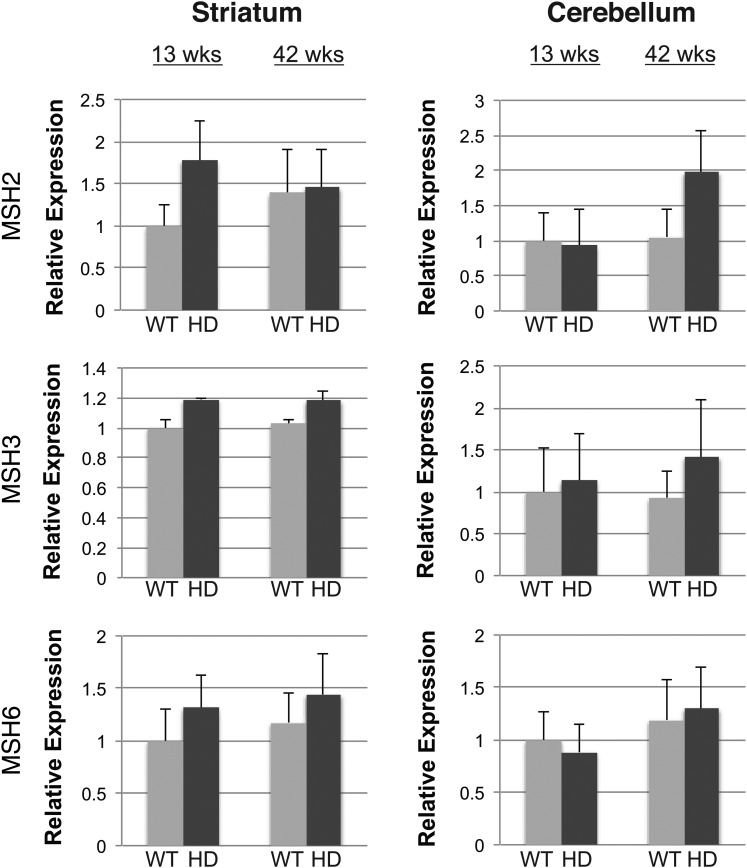
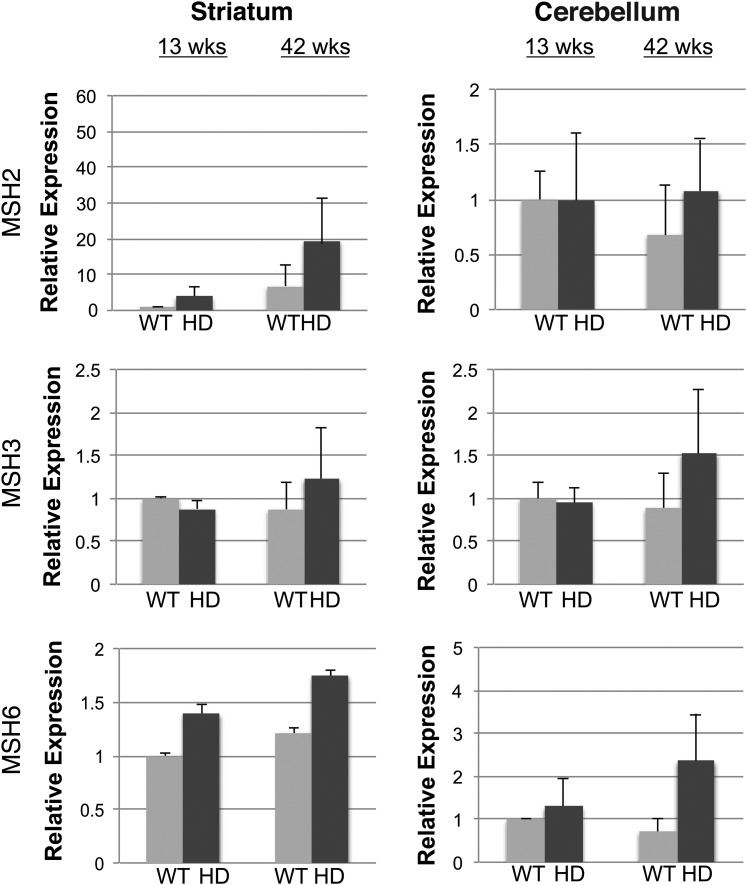
Although the expression levels of MMR genes did not change within a tissue with age, we wondered if the presence of an expressed CAG repeat expansion might alter the levels of these protein factors, especially since MMR proteins have been directly implicated in somatic repeat instability (17). To address this, we measured the expression levels of Msh2, Msh3 and Msh6 in the striatum and cerebellum of wild-type (WT) control mice and in HD R6/2 transgenic mice, a line that carries a 250 CAG repeat and shows marked age-dependent instability in CAG repeat length (16). We found that the RNA expression levels of Msh2, Msh3 and Msh6 do not significantly change in R6/2 mice at two different ages, 13 weeks (pre-symptomatic) and 42 weeks (symptomatic with documented somatic mosaicism), and noted that MMR RNA expression levels in HD R6/2 mice were similar to the levels measured in WT control mice (Fig. 4). These findings indicate that MMR gene expression is not altered by polyglutamine-expanded protein expression or on-going disease. We also performed quantitative western blot analysis, and did not observe any effect of the expressed polyglutamine-expanded protein on MSH2, MSH3 and MSH6 protein levels between 13-week-old or 42-week-old R6/2 mice and WT controls (Fig. 5).
Figure 4.
Polyglutamine-expanded huntingtin does not alter MMR RNA expression levels in cerebellum and striatum, even in aged mice. Real-time RT–PCR analysis of Msh2, Msh3 and Msh6 in the striatum and cerebellum of HD R6/2 mice (HD), carrying a hyper-expanded CAG repeat expansion (255–258 units at 13 weeks; 347–350 units at 43 weeks), and for littermate non-transgenic wild-type (WT) controls (n = 3/group). RNA expression levels for each tissue sample set for a given target (i.e. each ‘box’) are given relative to the 13-week-old WT result, which was arbitrarily set to 1 (mean ± s.e.m., three independent experiments; P = n.s.; t-test). 18S RNA served as the internal control.
Figure 5.
Polyglutamine-expanded huntingtin does not alter MMR protein expression levels in cerebellum and striatum, even in aged mice. Western blot analysis of MSH2, MSH3 and MSH6 proteins in the striatum and cerebellum for HD R6/2 mice (HD), carrying a hyper-expanded CAG repeat expansion (255–258 units at 13 weeks; 347–350 units at 43 weeks) and for littermate non-transgenic wild-type (WT) controls (n = 3/group). β-actin was used as a loading control for expression level normalization, and protein expression levels for each tissue sample set for a given target (i.e. each ‘box’) are given relative to the 13-week-old WT result, which was arbitrarily set to 1 (mean ± s.e.m., three independent experiments; P = n.s.; t-test).
Tissue-specific patterns of CAG instability and MMR expression levels show no correlation
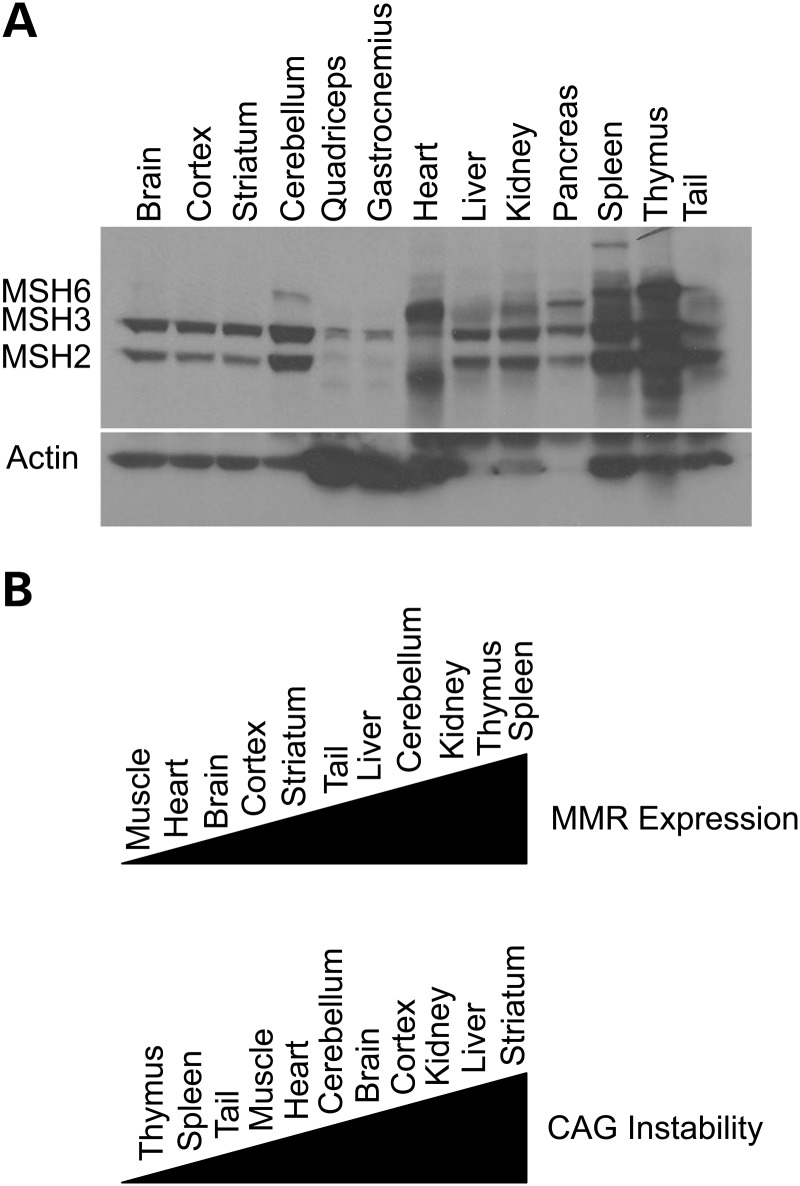
We next examined whether the expression of MMR protein levels might correlate with the levels or pattern of trinucleotide repeat instability in tissues of HD mice. Using simultaneous western blot analysis to obtain the relative expression levels of MSH2, MSH3 and MSH6 proteins in different tissue samples, we found that MSH2, MSH3 and MSH6 protein levels vary widely between tissues from WT control mice, with MMR protein expression highest in thymus and spleen, intermediate in striatum and liver, and lowest in skeletal muscle and heart (Fig. 6A). When we compared the level of MMR protein expression in different mouse tissues to the extent of somatic repeat instability documented for these same tissues in HD R6/2 mice (16), we found that for certain tissues (cerebellum, spleen, thymus), high MMR expression correlates with reduced somatic mosaicism, but for other tissues (kidney, muscle), MMR expression level does not correlate with somatic repeat instability (Fig. 6B). These results underscore the complexity of the repeat instability process, and indicate that MMR protein expression is but one factor in determining the extent of repeat instability in a given tissue.
Figure 6.
MMR protein levels in various tissues and their relationship to CAG instability. (A) Simultaneous western blot analysis of MMR protein levels in tissues from a 14-week-old WT CBA × C57BL/6J mouse. β-actin was used as the loading control. (B) Illustration of the rank order of MMR expression levels in 13 tissues from 14-week-old WT CBA × C57BL/6J mouse and the extent of CAG repeat instability in the same tissues of 13-week-old R6/2 transgenic mice (16). Muscle corresponds to quadriceps and gastrocnemius, which were comparable.
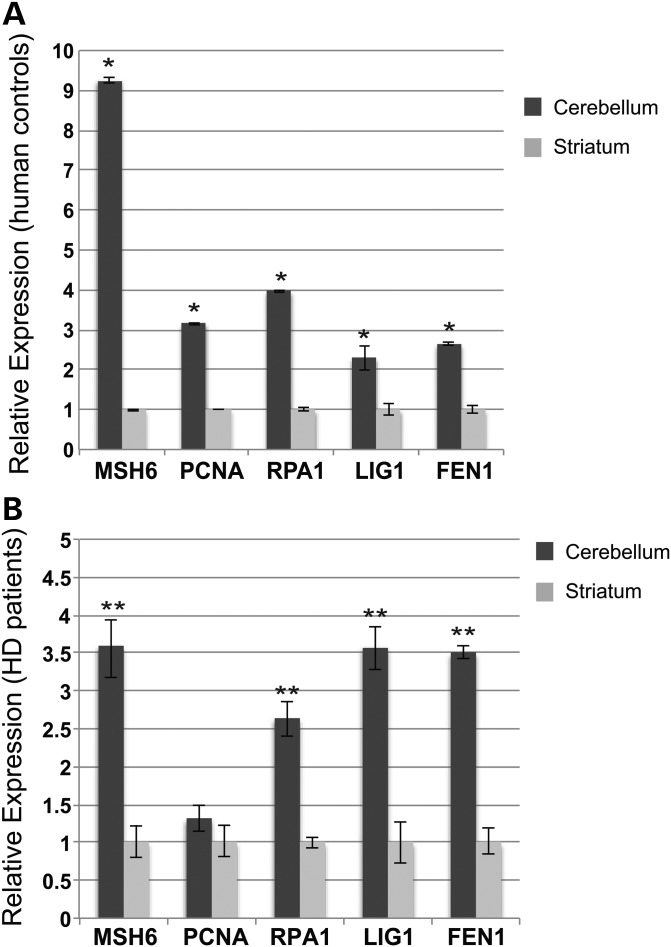
Expression levels of PCNA, RPA1, MSH6, LIG1 and FEN1 in human striatum and cerebellum parallel expression levels observed in mice
To determine if these findings are potentially relevant to human patients afflicted with CAG/CTG repeat expansion disorders, we evaluated the levels of candidate DNA metabolism genes in adult cerebellum and striatum RNA samples isolated from unaffected human individuals as well as from HD patients. Real-time RT–PCR analysis of human cerebellum and striatum samples revealed increased expression of RPA1, MSH6, LIG1 and FEN1 in the cerebellum in comparison to the striatum for these DNA metabolism and repair genes in both unaffected controls and in affected HD patients (Fig. 7A and B). Interestingly, although PCNA was more highly expressed in the cerebellum of unaffected controls in comparison to striatum, PCNA expression levels in HD cerebellum and striatum were comparable.
Figure 7.
DNA replication and repair gene expression is elevated in the cerebellum of human HD patients and unaffected controls. (A) Real-time RT–PCR analysis of MSH6, PCNA, RPA1, LIG1 and FEN1 expression levels in the striatum and cerebellum of unaffected human adults (n = 2; mean ± s.e.m., three independent experiments; *P < 0.05; t-test). β-actin served as the normalization control. (B) Real-time RT–PCR analysis of MSH6, PCNA, RPA1, LIG1 and FEN1 expression levels in the striatum and cerebellum of human HD patients (n = 2; mean ± s.e.m., three independent experiments; **P < 0.01; t-test). β-actin served as the normalization control.
DISCUSSION
Despite years of intensive study, the molecular basis of repeat instability in TNR disease remains enigmatic, particularly, with regard to tissue-selectivity. Investigations into the mechanistic basis of the repeat instability process have spanned a wide range of model organisms, in vitro systems, and cell-culture approaches. These studies have shown that expanded repeat tracts have the propensity to adopt aberrant structures at the DNA/RNA level, including hairpins, slipped-DNAs, triplexes and R-loops, and that the predisposition to adopt such altered DNA/RNA conformations is what renders expanded repeats susceptible to high rates of germ line and somatic instability (43–50). In light of the importance of DNA structures and biology for repeat instability, pathways of DNA metabolism and repair emerged as likely candidates for involvement in the molecular basis of repeat instability. DNA replication and MMR are thus viewed as central to the instability process, although the exact nature of how these processes promote repeat instability and whether they act independently or in concert to yield repeat length change remains uncertain (2,34,51–54). Experiments in both mouse models and human patients have shown that patterns of repeat instability in the different CAG/CTG repeat diseases are constant across specific brain regions, despite the fact that the different diseases exhibit distinct regional vulnerabilities and involve unique gene loci distributed throughout the genome. In this study, we applied microarray analysis to identify gene expression differences between the striatum and the cerebellum to determine potential molecular explanations for these observed repeat instability differences. We identified eight candidate factors, and then verified the differential expression of the leading candidates. We found that their expression did not change with advanced age or upon polyglutamine-expanded disease protein expression. Our findings provide insight into why repeat instability differences may exist between the striatum, a site of marked somatic mosaicism, and the cerebellum, a brain region with limited repeat length variation.
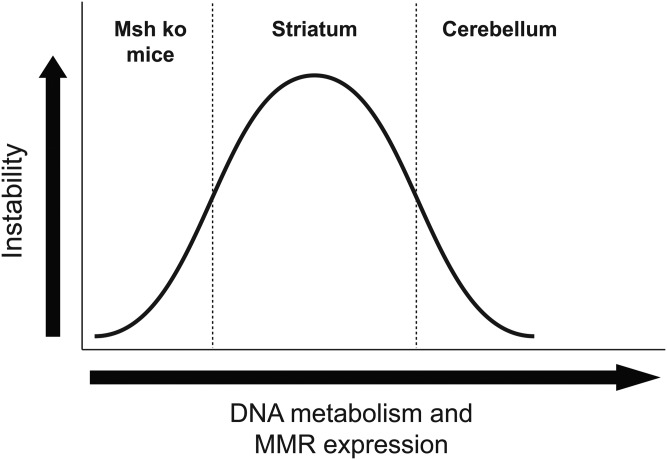
To delineate the molecular processes underlying repeat instability, we performed a microarray expression analysis comparison of striatum and cerebellum RNAs, and chose to compare aged tissues, since repeat instability increases with time in somatic tissues. Surprisingly, we found that certain DNA replication and DNA repair genes and proteins are more highly expressed in the cerebellum, where somatic mosaicism is limited. This was unexpected, as numerous repeat instability studies in mice have shown that MMR gene expression is required for enhanced somatic repeat expansion (17,35,55–64). These prior mouse studies demonstrated that contractions arise and expansions are suppressed in the absence of MMR, suggesting that MMR is required to promote CAG/CTG repeat expansion. In light of previous work, we conclude that a baseline level of MMR protein expression is required for dramatic expansion-biased repeat instability to occur, as is the case in the striatum, but when DNA metabolism enzymes and MMR proteins are expressed at much higher levels, as in the cerebellum, pronounced repeat instability does not occur. A biphasic model for MMR and DNA metabolism pathway regulation of TNR instability is presented in Figure 8. According to this model, high levels of MMR proteins and DNA metabolism factors in the cerebellum favor conditions that stabilize expanded CAG repeats and prevent their expansion, although the mechanisms for reduced repeat instability remain unclear. One possible explanation is that excessively high expression levels yield impaired enzymatic function, as direct modulation of MSH3 levels in a prior study demonstrated that MMR function is impaired when MSH3 is very highly expressed (39). Consequently, a threshold level of DNA metabolism and MMR gene expression might promote repeat instability, as it would yield initiation of a robust repair process destined to be unsuccessful, and instead produce CAG repeat expansions.
Figure 8.
Model for biphasic MMR and DNA metabolism pathway regulation of repeat instability. According to this model, a threshold level of DNA metabolism gene expression and MMR protein expression is required for pronounced repeat instability to occur, which is the situation in the striatum. However, when DNA replication and MMR expression levels are below this threshold, repeat instability does not occur, as is the case for repeat instability mice crossed onto a Msh knock-out background. Similarly, when elevated levels of expression for both DNA replication and MMR proteins are achieved, as observed for the cerebellum, repeat instability is also suppressed.
Another recent investigation into the role of trans-acting factors in somatic instability reported that differences in MMR protein expression or DNA metabolism pathways do not account for brain region variations in somatic mosaicism in HD model mice (65). However, in this work, western blot analysis of MSH2 revealed a dramatic increase in MSH2 protein levels in the cerebellum in comparison to striatum, though dual comparison of sets of stable and unstable tissues did not support a correlation between MSH2 levels and instability, when non-CNS tissues were included (65). Interestingly, of 74 down-regulated genes with Pearson coefficient correlations that were weak to moderate, 63 fell within the DNA metabolism gene class, suggesting that DNA metabolism gene expression patterns may correspond with regional instability differences (65). It is also noteworthy that MMR gene expression is markedly down-regulated in human embryonic stem cells derived from DM1 parents with expanded CTG repeats, a phenomenon coincident with the loss of spontaneous CTG instability (42). Similar to MSH2 and MSH3, perturbation of DNA Ligase 1, FEN1, XPA and other DNA repair proteins has been reported to alter CAG instability (66–70), and a recent screen for genes affecting GAA instability in dividing and non-dividing yeast cells also yielded DNA metabolism factors, including POL30 (Pcna), MCM7 and RAD27 (Fen1) (71), all of which were identified in our microarray expression comparison. These reported observations, along with data presented here, indicate that trinucleotide repeat instabilities of various natures and in various eukaryotic systems are all associated with perturbation of DNA repair mechanisms. This is intriguing and suggests a major and conserved role of DNA repair factors in monitoring repeat sequences. Tightly controlled expression of MMR genes could be essential for fine-tuning the DNA replication machinery and may thus underlie a universal mechanism to keep repeat expansions in check. Another approach for modeling repeat instability has been to develop induced pluripotent stem cell (iPSC) models, and this strategy has yielded pronounced expansion-biased instability in iPSCs and iPSC derivatives from Friedreich's ataxia (FRDA) patients (72). Directed studies of MMR using the FRDA iPSC model system found that reduced expression of MSH2 or MSH6 could diminish GAA repeat instability (72), which is consistent with the threshold model proposed here (Fig. 8).
To clarify the nature of MMR protein expression change as a function of age and in the face of polyglutamine neurodegeneration, we measured MMR expression levels in young mice and in the R6/2 HD mouse model. In both cases, we observed increased MMR expression in the cerebellum compared with the striatum, and did not detect altered MMR expression in young mice or in the brains of the HD R6/2 mice. This is consistent with a recent study that examined the role of trans-acting factors in regional instability differences in HD knock-in mice (65). This previous work also evaluated the effect of neurodegeneration in the cerebellum by crossing a HD knock-in mouse model with the Harlequin (Hq) model of cerebellum degeneration, and reported no increase in somatic repeat instability in the cerebella of HD knock-in mice carrying the Hq mutation (65). Hence, it appears that the cellular changes that accompany neurodegeneration do not further enhance the pathways that promote repeat instability in somatic tissues in the brain.
Although our study reinforces a role for DNA replication and MMR in regulating repeat instability, we found that differences in MMR protein expression cannot fully account for repeat instability differences resulting from advancing age or for somatic mosaicism differences between certain tissues. Our findings indicate that factors other than just MMR protein expression level must contribute to the complex process of repeat expansion. One possible explanation is that with age or in certain tissues, the ability for alternative structures to form increases. There are many reasons why DNA conformation change could be favored with age or in certain tissue milieus, including increasing oxidative damage to DNA or accumulation of epigenetic alterations at the level of histone modification status in chromatin (73). Hence, future studies of repeat instability will need to consider a variety of processes influencing DNA metabolism and repair, especially epigenetic regulatory pathways, as epigenetic processes are emerging as powerful regulators of MMR (74), and repeat instability differences due to parent-of-origin effects and aging likely reflect fundamentally distinct epigenetic processes at work in these varying milieus.
We have provided a possible molecular explanation for repeat stability in the cerebellum relative to repeat instability in the striatum. While HD suffers from striatal degeneration and the cerebellum is spared, many SCAs exhibit cerebellar degeneration. These paradoxical findings suggest that regional patterns of somatic CAG repeat instability do not account for selective neuronal vulnerability to degeneration. However, although there is commonality in patterns of CAG repeat instability among many of the CAG polyglutamine diseases, patterns of CAG repeat instability between different brain regions are not fully shared for all of the various CAG repeat disorders (11,75). Such variation suggests that locus-specific factors also contribute to the process of somatic instability (6,76). Hence, variations in somatic instability between different CAG repeat disease loci may reflect the influence of polymorphisms or flanking sequences surrounding each disease gene. Consequently, to understand the molecular basis of the CAG repeat instability process, we ultimately will need to understand how cis-acting elements and trans-acting factors interact at each locus and in each different tissue milieu.
Instead of only relying upon mouse models to examine the molecular basis of TNR instability, we obtained human cerebellum and striatum RNA samples to determine whether the changes identified in mouse brain also occur in the human brain. We confirmed that in HD patients, as well as in normal human controls, both DNA metabolism genes and MMR genes are more highly expressed in the cerebellum than in the striatum, with one exception—PCNA. These findings thus support a role for DNA metabolism and MMR gene expression in the somatic instability differences documented between the striatum and the cerebellum in post-mortem materials obtained from human patients afflicted with CAG/CTG repeat disease (8). Certainly, one fascinating aspect of the TNR disease field is the uniqueness of these diseases in the human, especially when one considers that the dramatic expansion-biased repeat instability seen in human disease does not occur naturally in related mammalian species or model organisms. While a set of examples of repeat-associated pathology has been reported in plants and in a canine species, neither of these repeats demonstrates the dramatic repeat instability seen in human patients (2,34). Hence, understanding the biology of repeat instability holds the potential for unlocking one of the most perplexing mysteries of this uniquely human disease category, and perhaps may shed light on the evolutionary processes that promoted the relatively rapid rise of the human species from a small founder population more than 150 000 years ago.
MATERIALS AND METHODS
Mice
WT C57BL/6J and CBA × C57BL/6J mice were obtained from the Jackson Laboratories. All animal experiments adhered to National Institute of Health (NIH) guidelines and were approved by the University of California, San Diego Institutional Animal Care and Use Committee (IACUC). HD and WT mice bred on a CBA × C57BL/6 F1 background were taken from an allelic series colony established at the University of Cambridge as previously described (77). Genotyping and repeat length measurement were performed by Laragen, Los Angeles, USA. All studies were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986. In the present study, we used R6/2 mice carrying a repeat length of either ∼256 or ∼346 CAG repeats.
Nucleic acid studies
Total RNA was extracted from striatum and cerebellum frozen tissues of 13- and 42-week-old WT and HD (R6/2) mice, using RNeasy mini kit from Qiagen. Reverse transcription was performed with 500 ng of total RNA and SuperScript™ II Reverse Transcriptase (Invitrogen), using hexamer primers. Total RNA was extracted from fresh striatum and cerebellum tissues of 8- and 40-week-old WT C57BL/6 mice, using TRIzol reagent from Invitrogen using their standard protocol. Reverse transcription was done using 1 μg of total RNA and MultiScribe™ Reverse Transcriptase (Life Technologies), using random primers. PCNA (Mm_00448100_g1), RPA1 (Mm_01253368_m1), Msh6 (Mm_00487761_m1), Lig1 (Mm_00495331_m1), and Fen1 (Mm_ 01700195_m1) real-time PCR experiments on C57BL/6J WT mice [Taqman (life sciences/ABI)] were performed using a 7500 real-time PCR system (Applied Biosystems). Experimental samples were diluted 50 times to analyze the expression of all genes of interest and β-actin (Mm_01205647_g1) as control. The PCR cycling parameters were 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s, 60°C for 1 min.
For human striatal and cerebellar studies experiments were carried out as described earlier using PCNA (Hs_00696862_m1), RPA1 (Hs_00161419_m1), Msh6 (Hs_00264721_m1), Lig1 (Hs_01553527_m1) and Fen1 (Hs_ 00748727_s1) and β-actin (Hs_01060665_g1) as control [Taqman (life sciences/ABI)] for real-time PCR analysis. Unaffected striatal and cerebellar RNA samples were purchased from Agilent Technologies, and HD affected tissue samples were obtained from the UCSD CNS Neurodegeneration Repository and were processed as described earlier for 8- and 40-week WT C57BL/6 mice.
MMR Real-time PCR experiments on HD and CBA WT [SybR green (Roche) with dissociation curve] were performed using an Mx3005P cycler (Strategen). Specific primers for each tested genes and for 18S mouse endogenous control were designed using NCBI primer-Blast (discussed subsequently). Standard curves were generated for each gene of interest using serial dilutions of testis mouse cDNAs. Experimental samples diluted 20 times to analyze MMR expression and 2000 times to analyze 18S rRNA expression were all run in triplicate. The PCR cycling parameters were 95°C for 10 min, and 40 cycles of 95°C for 30 s, 60°C for 1 min and 72°C for 1 min. Student's t-test to determine exact P-values, was used to determine differences in expression between wild-type (WT) and HD transgenic mice (biological triplicate and experimental duplicate for each tested gene). The pattern of MMR expression is reproducible between experiments for each tested gene.
Gene-specific primers are as follows:
.
| PCR | Name | Sequence | bp |
|---|---|---|---|
| Msh2 | Msh2ex14STA | AGCGCTCACTACTGAGGAGACCC | 23 |
| Msh2ex15STB | GCGCACGCTATCACGTGCCTC | 21 | |
| Msh3 | Msh3ex20STA | ATGGCTCAGATTGGCTCCTACG | 22 |
| Msh3ex21STB | TTCCGCTGTGTCCGTCAGTTCTTC | 24 | |
| Msh6 | Msh6ex3STA | AGGCTGCAGCTGGCAGTGTG | 20 |
| Msh6ex4STB | AGGCCCCTGAACACTGGGCT | 20 | |
| 18S | 18S3F | CAGTGAAACTGCGAATGG | 18 |
| 18S3R | CGGGTTGGTTTTGATCTG | 18 |
Heat map generation
The heat map of transformed intensities was generated using matlab with the following script:
>> [x,y,z]=xlsread(‘C:file’);
>> c=corrcoef(x);
>> imagesc(c);
Western blot analysis
We determined the MSH2, MSH3 and MSH6 protein expression in WT and HD mice (CBAxC57Bl/6) by simultaneously western blotting of MSH2, MSH3, MSH6 and Actin (as a loading control) (40). The different tissues were collected from WT C57BL/6 mice killed at 8 or 40 weeks or R6/2 and wild-type littermate control mice killed at either 13.7 or 42 weeks of age. Proteins were extracted by mechanical homogenisation in lysis buffer (0.125 M Tris–HCl pH 6.8, 4% SDS, 10% glycerol) containing complete Mini 7x protease inhibitor cocktail (Roche). Protein concentration was determined using the Pierce BCA protein assay kit. Forty micrograms of protein was denatured for 5 min at 95C° resolved by electrophoresis on a 8% polyacrylamide SDS–PAGE gel and electroblotted in transfer buffer (25 mm Tris–HCl pH 8.0, 192 mm glycine, 20% methanol and 0.1% SDS) at 300 mA at 4°C. Membranes were blocked for 1 h at room temperature in 5% milk in PBST ×1 then incubated overnight at 4°C in simultaneous primary antibodies. The membranes were washed three times for 20 min each in PBST, incubated for 1 h in secondary antibody (GE healthcare, α-mouse-HRP, 1:5000) at room temperature for MSH2, MSH3, MSH6, PCNA, RPA and for actin, and washed three times for 20 min each. Antibody binding was visualized using ECL plus western blotting detection system (Amersham). MSH2, MSH6, MSH3 and actin were detected using antibodies mouse anti-MSH2 (Calbiochem, 1:200), mouse anti-MSH6 (BD Laboratories, 1:200), MSH3 antibody, clone 2F11 (from Glenn Morris's lab, 1:750) (78), monoclonal mouse anti-PCNA (PC10) (Santa Cruz, cat. # Sc-56, dilution: 1/1000), monoclonal mouse anti-RPA 70 kDa subunit (B-6) (Santa Cruz, cat. no Sc-28304, dilution 1/1000) and Actin (BD Laboratories, 1:5000). Each experiment was reproduced at least three times for each protein tested.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the NIH (R01 GM059356 and R01 NS065874 to A.R.L., T32 award GM008666-14 to A.G.M. and P30-HD02274 to the UW Center on Human Development & Disability) and by funding from the Canadian Institutes of Health Research to C.E.P.
Supplementary Material
REFERENCES
- 1.Lopez Castel A., Cleary J.D., Pearson C.E. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 2.Pearson C.E., Nichol Edamura K., Cleary J.D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 3.La Spada A.R., Richards R.I., Wieringa B. Dynamic mutations on the move in Banff. Nat. Genet. 2004;36:667–670. doi: 10.1038/ng0704-667. [DOI] [PubMed] [Google Scholar]
- 4.Cleary J.D., Pearson C.E. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 5.La Spada A.R. Trinucleotide repeat instability: genetic features and molecular mechanisms. Brain Pathol. 1997;7:943–963. doi: 10.1111/j.1750-3639.1997.tb00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary J.D., Tome S., Lopez Castel A., Panigrahi G.B., Foiry L., Hagerman K.A., Sroka H., Chitayat D., Gourdon G., Pearson C.E. Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat. Struct. Mol. Biol. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- 7.Telenius H., Kremer B., Goldberg Y.P., Theilmann J., Andrew S.E., Zeisler J., Adam S., Greenberg C., Ives E.J., Clarke L.A., et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 8.Ishiguro H., Yamada K., Sawada H., Nishii K., Ichino N., Sawada M., Kurosawa Y., Matsushita N., Kobayashi K., Goto J., et al. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington's disease gene. J. Neurosci. Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- 9.Aoki M., Abe K., Tobita M., Kameya T., Watanabe M., Itoyama Y. Reduction of CAG expansions in cerebellar cortex and spinal cord of DRPLA. Clin. Genet. 1996;50:199–201. doi: 10.1111/j.1399-0004.1996.tb02625.x. [DOI] [PubMed] [Google Scholar]
- 10.Chong S.S., McCall A.E., Cota J., Subramony S.H., Orr H.T., Hughes M.R., Zoghbi H.Y. Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1995;10:344–350. doi: 10.1038/ng0795-344. [DOI] [PubMed] [Google Scholar]
- 11.Hashida H., Goto J., Kurisaki H., Mizusawa H., Kanazawa I. Brain regional differences in the expansion of a CAG repeat in the spinocerebellar ataxias: dentatorubral-pallidoluysian atrophy, Machado-Joseph disease, and spinocerebellar ataxia type 1. Ann. Neurol. 1997;41:505–511. doi: 10.1002/ana.410410414. [DOI] [PubMed] [Google Scholar]
- 12.Lopes-Cendes I., Maciel P., Kish S., Gaspar C., Robitaille Y., Clark H.B., Koeppen A.H., Nance M., Schut L., Silveira I., et al. Somatic mosaicism in the central nervous system in spinocerebellar ataxia type 1 and Machado-Joseph disease. Ann. Neurol. 1996;40:199–206. doi: 10.1002/ana.410400211. [DOI] [PubMed] [Google Scholar]
- 13.Takano H., Onodera O., Takahashi H., Igarashi S., Yamada M., Oyake M., Ikeuchi T., Koide R., Tanaka H., Iwabuchi K., et al. Somatic mosaicism of expanded CAG repeats in brains of patients with dentatorubral-pallidoluysian atrophy: cellular population-dependent dynamics of mitotic instability. Am. J. Hum. Genet. 1996;58:1212–1222. [PMC free article] [PubMed] [Google Scholar]
- 14.Ueno S., Kondoh K., Kotani Y., Komure O., Kuno S., Kawai J., Hazama F., Sano A. Somatic mosaicism of CAG repeat in dentatorubral-pallidoluysian atrophy (DRPLA) Hum. Mol. Genet. 1995;4:663–666. doi: 10.1093/hmg/4.4.663. [DOI] [PubMed] [Google Scholar]
- 15.Libby R.T., Monckton D.G., Fu Y.H., Martinez R.A., McAbney J.P., Lau R., Einum D.D., Nichol K., Ware C.B., Ptacek L.J., et al. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum. Mol. Genet. 2003;12:41–50. doi: 10.1093/hmg/ddg006. [DOI] [PubMed] [Google Scholar]
- 16.Mangiarini L., Sathasivam K., Mahal A., Mott R., Seller M., Bates G.P. Instability of highly expanded CAG repeats in mice transgenic for the Huntington's disease mutation. Nat. Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 17.Manley K., Shirley T.L., Flaherty L., Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 18.Richards R.I. Dynamic mutations: a decade of unstable expanded repeats in human genetic disease. Hum. Mol. Genet. 2001;10:2187–2194. doi: 10.1093/hmg/10.20.2187. [DOI] [PubMed] [Google Scholar]
- 19.Watase K., Venken K.J., Sun Y., Orr H.T., Zoghbi H.Y. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum. Mol. Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy L., Shelbourne P.F. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington's disease. Hum. Mol. Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- 21.Cha J.H. Transcriptional signatures in Huntington's disease. Prog. Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D., et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 23.Luthi-Carter R., Strand A., Peters N.L., Solano S.M., Hollingsworth Z.R., Menon A.S., Frey A.S., Spektor B.S., Penney E.B., Schilling G., et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 24.Luthi-Carter R., Strand A.D., Hanson S.A., Kooperberg C., Schilling G., La Spada A.R., Merry D.E., Young A.B., Ross C.A., Borchelt D.R., et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum. Mol. Genet. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- 25.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 26.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G., Leffak M. Instability of (CTG)n*(CAG)n trinucleotide repeats and DNA synthesis. Cell Biosci. 2012;2:7. doi: 10.1186/2045-3701-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 35.Slean M.M., Panigrahi G.B., Ranum L.P., Pearson C.E. Mutagenic roles of DNA "repair" proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst) 2008;7:1135–1154. doi: 10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Lenzmeier B.A., Freudenreich C.H. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 37.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak K.J., Flood S.J., Marmaro J., Giusti W., Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 39.Panigrahi G.B., Slean M.M., Simard J.P., Gileadi O., Pearson C.E. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc. Natl Acad. Sci. USA. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seriola A., Spits C., Simard J.P., Hilven P., Haentjens P., Pearson C.E., Sermon K. Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum. Mol. Genet. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 41.Tome S., Simard J.P., Slean M.M., Holt I., Morris G.E., Wojciechowicz K., te Riele H., Pearson C.E. Tissue-specific mismatch repair protein expression: MSH3 is higher than MSH6 in multiple mouse tissues. DNA Repair (Amst) 2013;12:46–52. doi: 10.1016/j.dnarep.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Seriola A., Spits C., Simard J.P., Hilven P., Haentjens P., Pearson C.E., Sermon K. Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum. Mol. Genet. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 43.Ireland M.J., Reinke S.S., Livingston D.M. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics. 2000;155:1657–1665. doi: 10.1093/genetics/155.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartenstine M.J., Goodman M.F., Petruska J. Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem. 2000;275:18382–18390. doi: 10.1074/jbc.275.24.18382. [DOI] [PubMed] [Google Scholar]
- 45.Kunkel T.A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 46.Kunkel T.A. Nucleotide repeats. Slippery DNA and diseases. Nature. 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- 47.Schweitzer J.K., Livingston D.M. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics. 1999;152:953–963. doi: 10.1093/genetics/152.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamori M., Pearson C.E., Thornton C.A. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum. Mol. Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy K., Tam M., Bowater R.P., Barber M., Tomlinson M., Nichol Edamura K., Wang Y.H., Pearson C.E. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonitel R., Moffitt H., Sathasivam K., Woodman B., Detloff P.J., Faull R.L., Bates G.P. DNA instability in postmitotic neurons. Proc. Natl Acad. Sci. USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parniewski P., Jaworski A., Wells R.D., Bowater R.P. Length of CTG.CAG repeats determines the influence of mismatch repair on genetic instability. J. Mol. Biol. 2000;299:865–874. doi: 10.1006/jmbi.2000.3796. [DOI] [PubMed] [Google Scholar]
- 53.Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 54.Wang G., Vasquez K.M. Models for chromosomal replication-independent non-B DNA structure-induced genetic instability. Mol. Carcinog. 2009;48:286–298. doi: 10.1002/mc.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dragileva E., Hendricks A., Teed A., Gillis T., Lopez E.T., Friedberg E.C., Kucherlapati R., Edelmann W., Lunetta K.L., MacDonald M.E., et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foiry L., Dong L., Savouret C., Hubert L., te Riele H., Junien C., Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 57.Gomes-Pereira M., Fortune M.T., Ingram L., McAbney J.P., Monckton D.G. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 58.Kovtun I.V., Thornhill A.R., McMurray C.T. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum. Mol. Genet. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 59.Savouret C., Brisson E., Essers J., Kanaar R., Pastink A., te Riele H., Junien C., Gourdon G. CTG Repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savouret C., Garcia-Cordier C., Megret J., te Riele H., Junien C., Gourdon G. MSH2-dependent Germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tome S., Holt I., Edelmann W., Morris G.E., Munnich A., Pearson C.E., Gourdon G. MSH2 ATPase domain mutation affects CTG*CAG repeat instability in transgenic mice. PLoS Genet. 2009;5:e1000482. doi: 10.1371/journal.pgen.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Broek W.J., Nelen M.R., Wansink D.G., Coerwinkel M.M., te Riele H., Groenen P.J., Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 63.Wheeler V.C., Lebel L.A., Vrbanac V., Teed A., te Riele H., MacDonald M.E. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 64.Kovalenko M., Dragileva E., St Claire J., Gillis T., Guide J.R., New J., Dong H., Kucherlapati R., Kucherlapati M.H., Ehrlich M.E., et al. Msh2 acts in medium-spiny striatal neurons as an enhancer of CAG instability and mutant huntingtin phenotypes in Huntington's disease knock-In mice. PLoS One. 2012;7:e44273. doi: 10.1371/journal.pone.0044273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J.M., Zhang J., Su A.I., Walker J.R., Wiltshire T., Kang K., Dragileva E., Gillis T., Lopez E.T., Boily M.J., et al. A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst. Biol. 2010;4:29. doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goula A.V., Berquist B.R., Wilson D.M., III, Wheeler V.C., Trottier Y., Merienne K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goula A.V., Pearson C.E., Della Maria J., Trottier Y., Tomkinson A.E., Wilson D.M., III, Merienne K. The nucleotide sequence, DNA damage location, and protein stoichiometry influence the base excision repair outcome at CAG/CTG repeats. Biochemistry. 2012;51:3919–3932. doi: 10.1021/bi300410d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hubert L., Jr, Lin Y., Dion V., Wilson J.H. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum. Mol. Genet. 2011;20:4822–4830. doi: 10.1093/hmg/ddr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez Castel A., Tomkinson A.E., Pearson C.E. CTG/CAG repeat instability is modulated by the levels of human DNA ligase I and its interaction with proliferating cell nuclear antigen: a distinction between replication and slipped-DNA repair. J. Biol. Chem. 2009;284:26631–26645. doi: 10.1074/jbc.M109.034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J., Freudenreich C.H. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene. 2007;393:110–115. doi: 10.1016/j.gene.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y., Shishkin A.A., Nishida Y., Marcinkowski-Desmond D., Saini N., Volkov K.V., Mirkin S.M., Lobachev K.S. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell. 2012;48:254–265. doi: 10.1016/j.molcel.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du J., Campau E., Soragni E., Ku S., Puckett J.W., Dervan P.B., Gottesfeld J.M. Role of mismatch repair enzymes in GAA-TTC triplet-repeat expansion in Friedreich's ataxia induced pluripotent stem cells (iPSCs) J. Biol. Chem. 2012;287:29861–29872. doi: 10.1074/jbc.M112.391961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Debacker K., Frizzell A., Gleeson O., Kirkham-McCarthy L., Mertz T., Lahue R.S. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaidi A., Jackson S.P. KAT5 Tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–74. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Tanaka F., Sobue G., Doyu M., Ito Y., Yamamoto M., Shimada N., Yamamoto K., Riku S., Hshizume Y., Mitsuma T. Differential pattern in tissue-specific somatic mosaicism of expanded CAG trinucleotide repeat in dentatorubral-pallidoluysian atrophy, Machado-Joseph disease, and X-linked recessive spinal and bulbar muscular atrophy. J. Neurol. Sci. 1996;135:43–50. doi: 10.1016/0022-510x(95)00249-2. [DOI] [PubMed] [Google Scholar]
- 76.Libby R.T., Hagerman K.A., Pineda V.V., Lau R., Cho D.H., Baccam S.L., Axford M.M., Cleary J.D., Moore J.M., Sopher B.L., et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duzdevich D., Li J., Whang J., Takahashi H., Takeyasu K., Dryden D.T., Morton A.J., Edwardson J.M. Unusual structures are present in DNA fragments containing super-long Huntingtin CAG repeats. PLoS One. 6:e17119. doi: 10.1371/journal.pone.0017119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holt I., Thanh Lam L., Tome S., Wansink D.G., Te Riele H., Gourdon G., Morris G.E. The mouse mismatch repair protein, MSH3, is a nucleoplasmic protein that aggregates into denser nuclear bodies under conditions of stress. J. Cell Biochem. 2011;112:1612–1621. doi: 10.1002/jcb.23075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.